Comparative Transcriptomics Analysis Reveals Genes Associated with a Dehiscent-Corolla Mutant in Sesame (Sesamum indicum L.)
Abstract
1. Introduction
2. Results
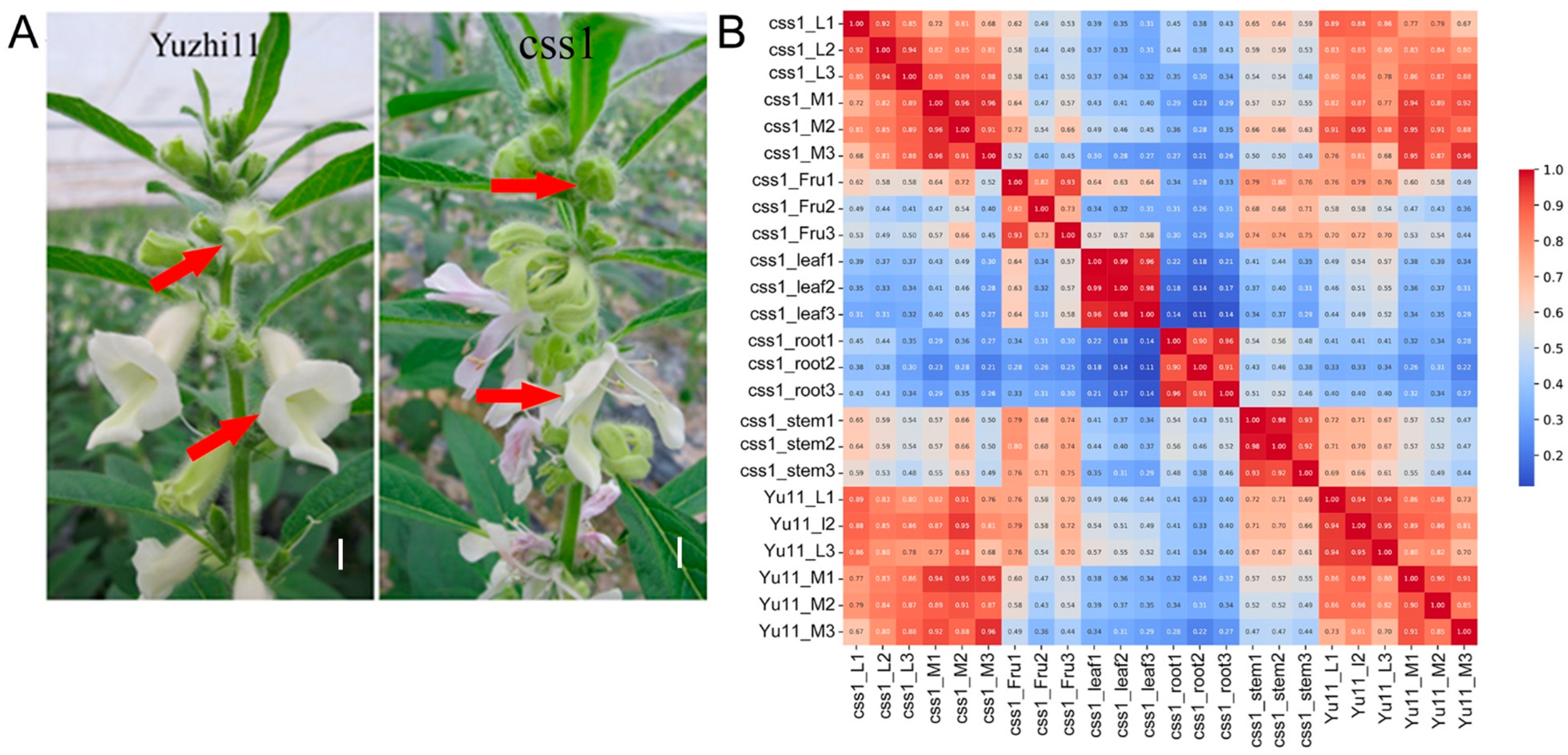
2.1. Transcriptomic Data Assessment
2.2. Corolla-Specific Expression Gene Analysis
2.3. DEG Analysis Between Yuzhi11 and the Dehiscent-Corolla Mutant css1
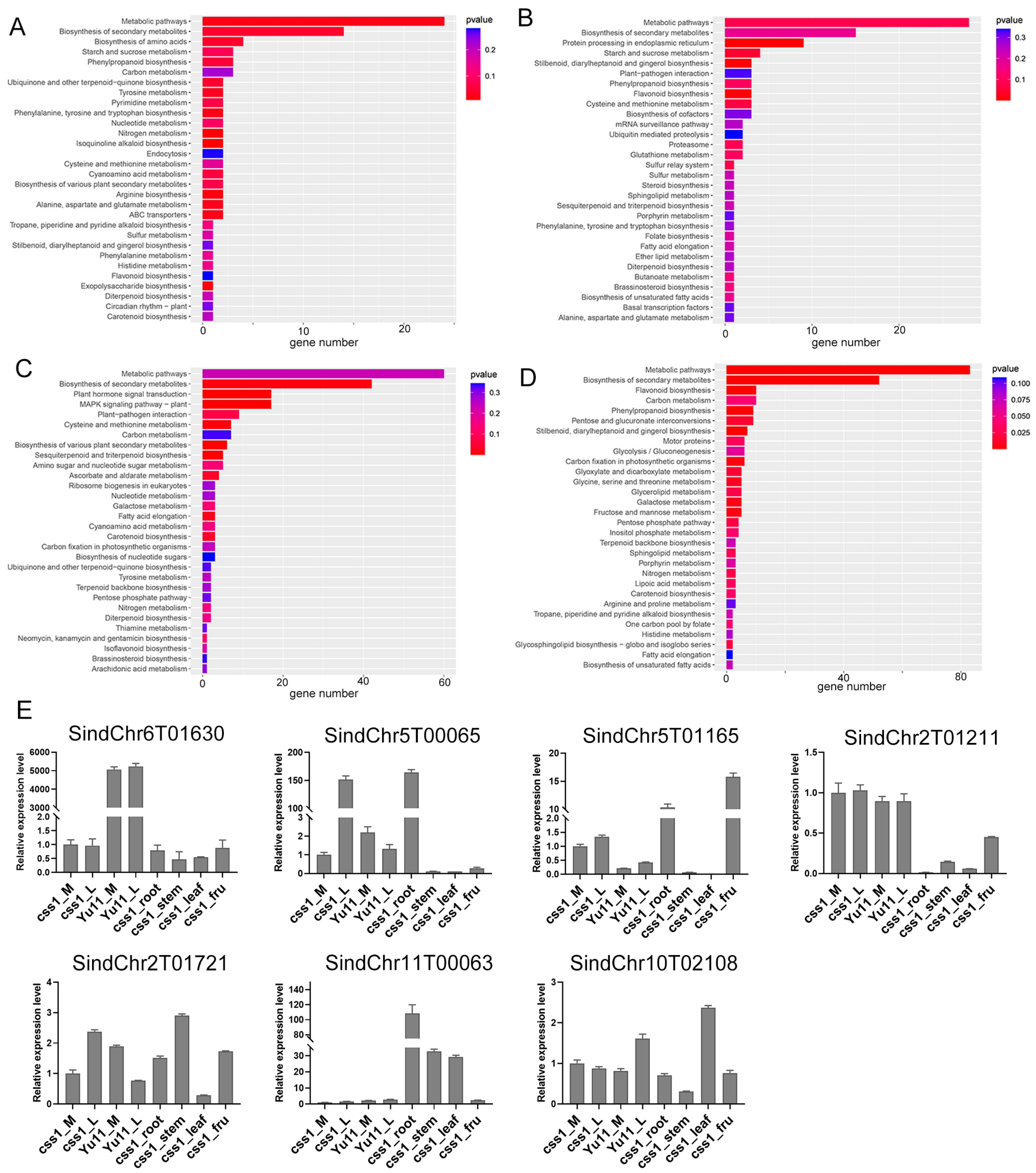
2.4. Analysis of DEGs Associated with the Dehiscent-Corolla Phenotype
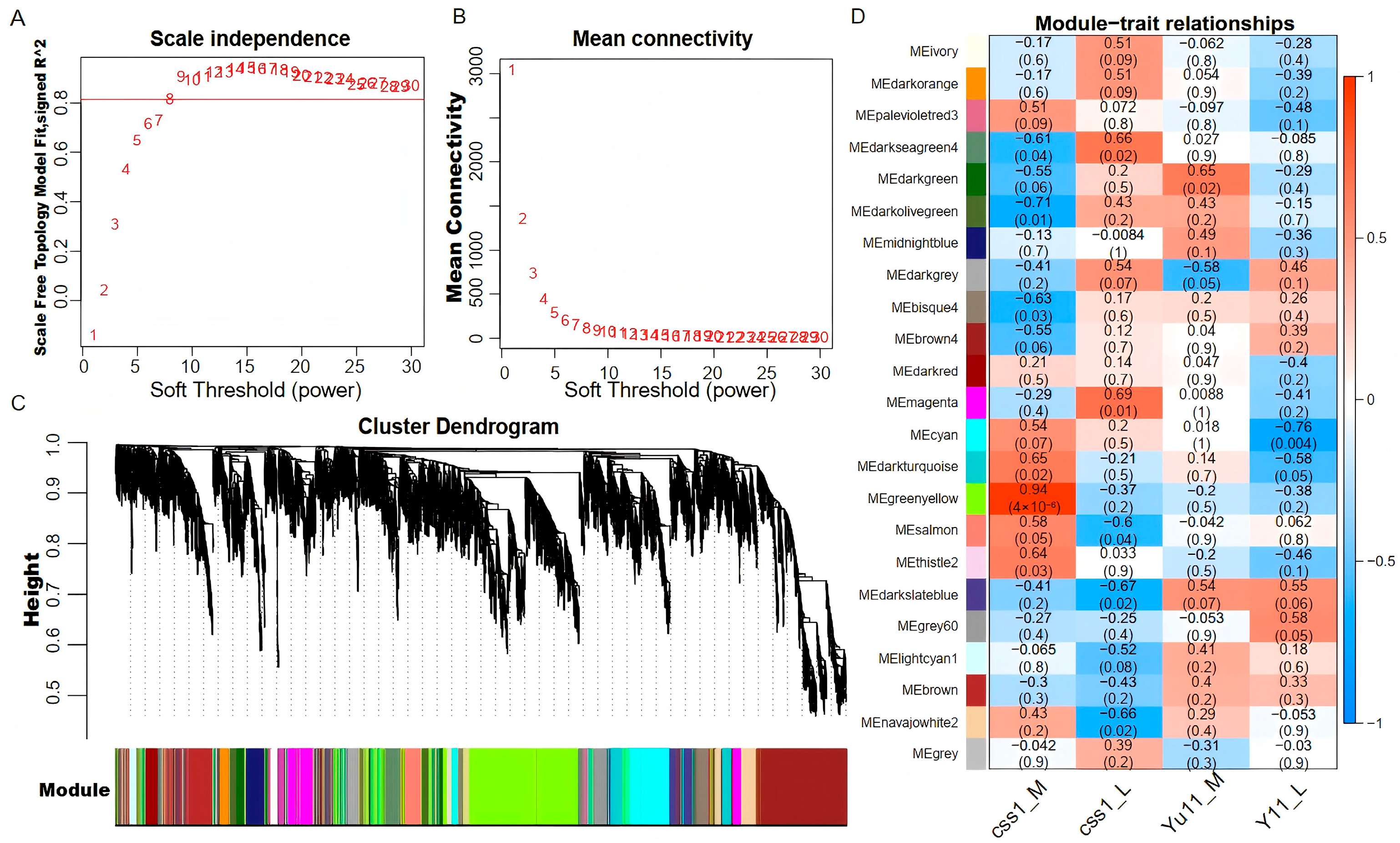
2.5. Gene Co-Expression Network Analysis
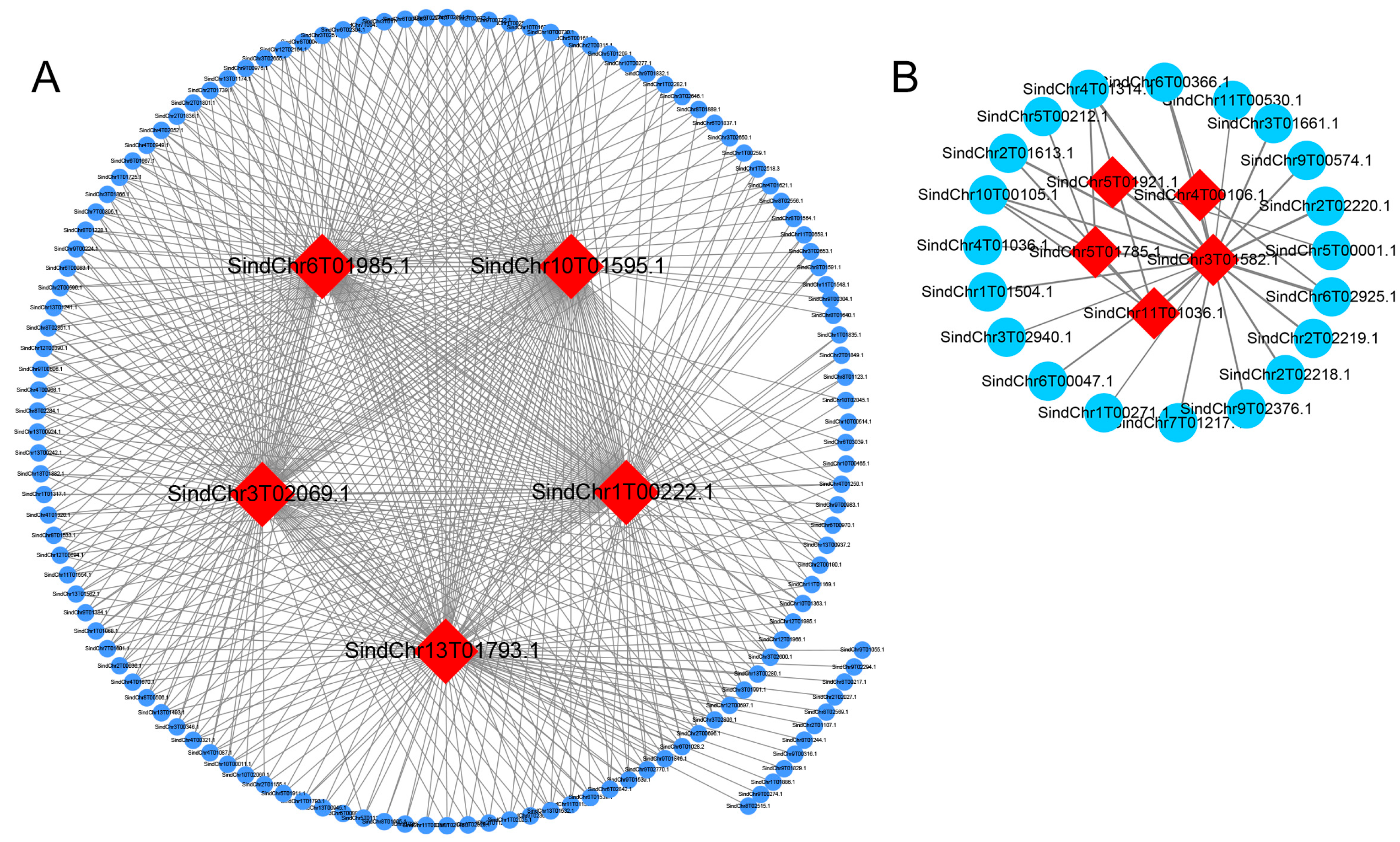
2.6. Expression Analysis and Interaction Network Prediction of Hub Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Isolation and Transcriptome Sequencing
4.3. RNA-Seq Analysis
4.4. Quantitative Real-Time PCR (qPCR)
4.5. Weighted Gene Co-Expression Network Analysis
4.6. Protein–Protein Interaction Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ashri, A. Sesame breeding. In Plant Breeding Reviews; Jules, J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1998; pp. 179–228. [Google Scholar]
- Namiki, M. Nutraceutical functions of sesame: A review. Crit. Rev. Food Sci. Nutr. 2007, 47, 651–673. [Google Scholar] [CrossRef]
- Fonseca, R.; Capel, C.; Lebrón, R.; Ortiz-Atienza, A.; Yuste-Lisbona, F.J.; Angosto, T.; Capel, J.; Lozano, R. Insights into the functional role of tomato TM6 as a transcriptional regulator of flower development. Hortic. Res. 2024, 11, uhae019. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Zhang, D.; Luo, D.; Sun, W.; Zhou, R.; Hong, Z.; Munir, S.; Ye, Z.; Yang, C.; Zhang, J.; et al. SlTCP24 and SlTCP29 synergistically regulate compound leaf development through interacting with SlAS2 and activating transcription of SlCKX2 in tomato. New Phytol. 2023, 240, 1275–1291. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, Q.; Lin, S.; Li, D.; Bao, M.; Fu, X. Identification and characterization of class E genes involved in floral organ development in Dianthus chinensis. Ornam. Plant Res. 2023, 3, 1–10. [Google Scholar] [CrossRef]
- Theißen, G.; Melzer, R.; Rümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef]
- Chopy, M.; Cavallini-Speisser, Q.; Chambrier, P.; Morel, P.; Just, J.; Hugouvieux, V.; Rodrigues Bento, S.; Zubieta, C.; Vandenbussche, M.; Monniaux, M. Cell layer–specific expression of the homeotic MADS-box transcription factor PhDEF contributes to modular petal morphogenesis in petunia. Plant Cell 2024, 36, 324–345. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; He, X.; Tang, Y.; Chen, Z.; Zhou, L.; Li, X.; Zhang, C.; Huang, X.; Yang, Y.; Zhang, W.; et al. Photoperiod controls plant seed size in a CONSTANS-dependent manner. Nat. Plants 2023, 9, 343–354. [Google Scholar] [CrossRef]
- Zhang, H.; Xue, F.; Guo, L.; Cheng, J.; Jabbour, F.; DuPasquier, P.-E.; Xie, Y.; Zhang, P.; Wu, Y.; Duan, X.; et al. The mechanism underlying asymmetric bending of lateral petals in Delphinium (Ranunculaceae). Curr. Biol. 2024, 34, 755–768.e4. [Google Scholar] [CrossRef]
- Tröbner, W.; Ramirez, L.; Motte, P.; Hue, I.; Huijser, P.; Lönnig, W.E.; Saedler, H.; Sommer, H.; Schwarz-Sommer, Z. GLOBOSA: A homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 1992, 11, 4693–4704. [Google Scholar] [CrossRef]
- Gong, P.; Song, C.; Liu, H.; Li, P.; Zhang, M.; Zhang, J.; Zhang, S.; He, C.; Wilson, Z. Physalis floridana CRABS CLAW mediates neofunctionalization of GLOBOSA genes in carpel development. J. Exp. Bot. 2021, 72, 6882–6903. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhu, W.; Yang, L.; Liang, W.; Li, H.; Yang, L.; Chen, M.; Luo, Z.; Huang, G.; Duan, L.; et al. SMALL REPRODUCTIVE ORGANS, a SUPERMAN-like transcription factor, regulates stamen and pistil growth in rice. New Phytol. 2021, 233, 1701–1718. [Google Scholar] [CrossRef]
- Li, X.; Sun, M.; Jia, Y.; Qiu, D.; Peng, Q.; Zhuang, L. Genetic control of the lateral petal shape and identity of asymmetric flowers in mungbean (Vigna radiata L.). Front. Plant Sci. 2022, 13, 996239. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Zhao, W.; Kong, S.; Li, L.; Lin, S. Overview of molecular mechanisms of plant leaf development: A systematic review. Front. Plant Sci. 2023, 14, 1293424. [Google Scholar] [CrossRef]
- Yu, Q.; Ge, L.; Ahmad, S.; Luo, D.; Li, X. A perspective on the molecular mechanism in the control of organ internal (IN) asymmetry during petal development. Hortic. Res. 2022, 9, uhac202. [Google Scholar] [CrossRef]
- Zhao, H.; Liao, H.; Li, S.; Zhang, R.; Dai, J.; Ma, P.; Wang, T.; Wang, M.; Yuan, Y.; Fu, X.; et al. Delphinieae flowers originated from the rewiring of interactions between duplicated and diversified floral organ identity and symmetry genes. Plant Cell 2023, 35, 994–1012. [Google Scholar] [CrossRef]
- Miao, H.; Wang, L.; Qu, L.; Liu, H.; Sun, Y.; Le, M.; Wang, Q.; Wei, S.; Zheng, Y.; Lin, W.; et al. Genomic evolution and insights into agronomic trait innovations of Sesamum species. Plant Commun. 2023, 5, 100729. [Google Scholar] [CrossRef]
- Cui, C.; Mei, H.; Liu, Y.; Zhang, H.; Zheng, Y. Genetic diversity, population structure, and linkage disequilibrium of an association-mapping panel revealed by genome-wide SNP markers in sesame. Front. Plant Sci. 2017, 8, 1189, Correction in Front. Plant Sci. 2017, 8, 2083. [Google Scholar]
- Zhang, H.; Miao, H.; Li, C.; Wei, L.; Duan, Y.; Ma, Q.; Kong, J.; Xu, F.; Chang, S. Ultra-dense SNP genetic map construction and identification of SiDt gene controlling the determinate growth habit in Sesamum indicum L. Sci. Rep. 2016, 6, 31556. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L.; Gao, Y.; Li, D.; Yu, J.; Zhou, R.; Zhang, X. Genetic dissection and fine mapping of a novel dt gene associated with determinate growth habit in sesame. BMC Genet. 2018, 19, 38. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Dossou, S.S.K.; Meng, M.; Sheng, C.; Li, H.; Zhou, R.; Li, D.; Xu, P.; You, J.; Wang, L. Five improved sesame reference genomes and genome resequencing unveil the contribution of structural variants to genetic diversity and yield-related traits variation. Plant Biotechnol. J. 2023, 21, 1722–1724. [Google Scholar] [CrossRef]
- Zhou, R.; Liu, P.; Li, D.; Zhang, X.; Wei, X. Photoperiod response-related gene SiCOL1 contributes to flowering in sesame. BMC Plant Biol. 2018, 18, 343. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Cui, C.; Wei, W.; Du, Z.; Wu, K.; Jiang, X.; Zheng, Y.; Liu, Y.; Mei, H.; Zhang, H. The candidate gene SibHLHA regulates anthocyanin-driven purple pigmentation in Sesamum indicum flowers. Theor. Appl. Genet. 2025, 138, 40. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Cao, H.; Ma, Q.; Ju, M.; Wang, H.; Tian, Q.; Feng, X.; Zhang, X.; Kong, J.; Zhang, H.; et al. An ethyl methanesulfonate-induced GIF1 splicing site mutation in sesame is associated with floral malformation and small seed size. Plants 2024, 13, 3294. [Google Scholar] [CrossRef]
- Wang, H.; Sha, G.; Gao, R.; Pang, J.; Zhai, R.; Yang, C.; Wang, Z.; Xu, L. PbGIF1 promoting cell-proliferation in pear fruit is transcriptionally activated by PbRR1. Hortic. Plant J. 2024, 10, 689–697. [Google Scholar] [CrossRef]
- Zheng, L.; Wu, H.; Wang, A.; Zhang, Y.; Liu, Z.; Ling, H.; Song, X.; Li, Y. The SOD7/DPA4–GIF1 module coordinates organ growth and iron uptake in Arabidopsis. Nat. Plants 2023, 9, 1318–1332. [Google Scholar] [CrossRef]
- Li, S.; Gao, F.; Xie, K.; Zeng, X.; Cao, Y.; Zeng, J.; He, Z.; Ren, Y.; Li, W.; Deng, Q.; et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 2016, 14, 2134–2146. [Google Scholar] [CrossRef]
- Kang, I.H.; Steffen, J.G.; Portereiko, M.F.; Lloyd, A.; Drews, G.N. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 2008, 20, 635–647. [Google Scholar] [CrossRef]
- Li, X.; Xie, Z.; Qin, T.; Zhan, C.; Jin, L.; Huang, J. The SLR1-OsMADS23-D14 module mediates the crosstalk between strigolactone and gibberellin signaling to control rice tillering. New Phytol. 2025, 246, 2137–2154. [Google Scholar] [CrossRef]
- Li, H.; Liang, W.; Jia, R.; Yin, C.; Zong, J.; Kong, H.; Zhang, D. The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res. 2010, 20, 299–313. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.; Shen, L.; Wu, Y.; Chen, H.; Robertson, M.; Helliwell, C.A.; Ito, T.; Meyerowitz, E.; Yu, H. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 2008, 15, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Xia, Q.; Xie, W.; Datla, R.; Selvaraj, G. The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proc. Natl. Acad. Sci. USA 2003, 100, 14487–14492. [Google Scholar] [CrossRef]
- Park, J.; Cui, Y.; Kang, B.H. AtPGL3 is an Arabidopsis BURP domain protein that is localized to the cell wall and promotes cell enlargement. Front. Plant Sci. 2015, 6, 412. [Google Scholar] [CrossRef] [PubMed]
- Vercesi, A.; Borecký, J.; Maia Ide, G.; Arruda, P.; Cuccovia, I.; Chaimovich, H. Plant uncoupling mitochondrial proteins. Annu. Rev. Plant Biol. 2006, 57, 383–404. [Google Scholar] [CrossRef]
- de Jesús-Pires, C.; Ferreira-Neto, J.; Pacifico Bezerra-Neto, J.; Kido, E.; de Oliveira Silva, R.; Pandolfi, V.; Wanderley-Nogueira, A.; Binneck, E.; da Costa, A.; Pio-Ribeiro, G.; et al. Plant thaumatin-like proteins: Function, evolution and biotechnological applications. Curr. Protein Pept. Sci. 2020, 21, 36–51. [Google Scholar] [CrossRef]
- Sweetman, C.; Waterman, C.; Rainbird, B.; Smith, P.; Jenkins, C.; Day, D.; Soole, K. AtNDB2 is the main external NADH dehydrogenase in mitochondria and is important for tolerance to environmental stress. Plant Physiol. 2019, 181, 774–788. [Google Scholar] [CrossRef]
- Zheng, J.; Xie, X.; Li, C.; Wang, H.; Yu, Y.; Huang, B. Regulation mechanism of plant response to heavy metal stress mediated by endophytic fungi. Int. J. Phytoremediation 2023, 25, 1596–1613. [Google Scholar] [CrossRef]
- Zou, T.; Pu, L.; Lin, R.; Mo, H.; Wang, Z.; Jian, S.; Zhang, M. Roles of Canavalia rosea metallothioneins in metal tolerance and extreme environmental adaptation to tropical coral reefs. J. Plant Physiol. 2022, 268, 153559. [Google Scholar] [CrossRef]
- Liebsch, D.; Palatnik, J. MicroRNA miR396, GRF transcription factors and GIF co-regulators: A conserved plant growth regulatory module with potential for breeding and biotechnology. Curr. Opin. Plant Biol. 2020, 53, 31–42. [Google Scholar] [CrossRef]
- Kim, J.H.; Kende, H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 13374–13379. [Google Scholar] [CrossRef] [PubMed]
- Debernardi, J.; Tricoli, D.; Ercoli, M.; Hayta, S.; Ronald, P.; Palatnik, J.; Dubcovsky, J. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 2020, 38, 1274–1279. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, N.; Zhang, Y.; Li, Y. Transcriptional repression of GIF1 by the KIX-PPD-MYC repressor complex controls seed size in Arabidopsis. Nat. Commun. 2020, 11, 1846. [Google Scholar] [CrossRef]
- Jin, W.; Gong, F.; Zhang, Y.; Wang, R.; Liu, H.; Wei, Y.; Tang, K.; Jiang, Y.; Gao, J.; Sun, X. Cytokinin-responsive RhRR1-RhSCL28 transcription factor module positively regulates petal size by promoting cell division in rose. J. Exp. Bot. 2025, 76, 381–392. [Google Scholar] [CrossRef]
- Jing, W.; Gong, F.; Liu, G.; Deng, Y.; Liu, J.; Yang, W.; Sun, X.; Li, Y.; Gao, J.; Zhou, X.; et al. Petal size is controlled by the MYB73/TPL/HDA19-miR159-CKX6 module regulating cytokinin catabolism in Rosa hybrida. Nat. Commun. 2023, 14, 7106. [Google Scholar] [CrossRef]
- Guan, Y.; Wong, C.; Zhang, Q.; Peng, D.; Lan, S.; Chen, F.; Liu, Z.; Yu, H. Sizing up beauty: Mechanisms of petal size regulation in ornamental plants. Plant Physiol. 2025, 198, 3. [Google Scholar] [CrossRef]
- Xu, Q.; Yang, Z.; Jia, Y.; Wang, R.; Zhang, Q.; Gai, R.; Wu, Y.; Yang, Q.; He, G.; Wu, J.; et al. PeNAC67-PeKAN2-PeSCL23 and B-class MADS-box transcription factors synergistically regulate the specialization process from petal to lip in Phalaenopsis equestris. Mol. Hortic. 2024, 4, 15. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Song, X.; Miao, H.; Feng, S.; Wu, G. Natural variation in CYCLIC NUCLEOTIDE-GATED ION CHANNEL 4 reveals a novel role of calcium signaling in vegetative phase change in Arabidopsis. New Phytol. 2024, 242, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Thomé, V.; Ferreira, P.; Lubini, G.; Nogueira, F.; Strini, E.; Pinoti, V.; Cruz, J.; San Martin, J.; Quiapim, A.; daSilva, L.; et al. Unveiling the movement of ranbp1 during the cell cycle and its interaction with a Cyclin-Dependent Kinase (CDK) in Plants. Int. J. Mol. Sci. 2024, 26, 46. [Google Scholar] [CrossRef] [PubMed]
- DePaoli, H.; Brito, M.; Quiapim, A.; Teixeira, S.; Goldman, G.; Dornelas, M.; Goldman, M. Stigma/style cell cycle inhibitor 1 (SCI1), a tissue-specific cell cycle regulator that controls upper pistil development. New Phytol. 2011, 190, 882–895. [Google Scholar] [CrossRef]
- Wang, G.; Wu, Z.; Sun, B. KNUCKLES regulates floral meristem termination by controlling auxin distribution and cytokinin activity. Plant Cell 2024, 37, koae312. [Google Scholar] [CrossRef]
- Xu, B.; Gou, J.; Li, F.; Shangguan, X.; Zhao, B.; Yang, C.; Wang, L.; Yuan, S.; Liu, C.; Chen, X. A cotton BURP domain protein interacts with α-Expansin and their co-expression promotes plant growth and fruit production. Mol Plant. 2013, 6, 945–958. [Google Scholar] [CrossRef]
- Yu, S.; Yang, F.; Zou, Y.; Yang, Y.; Li, T.; Chen, S.; Wang, Y.; Xu, K.; Xia, H.; Luo, L. Overexpressing PpBURP2 in rice increases plant defense to abiotic stress and bacterial leaf blight. Front. Plant Sci. 2022, 6, 812279. [Google Scholar] [CrossRef] [PubMed]
- Escobar, M.; Geisler, D.; Rasmusson, A. Reorganization of the alternative pathways of the Arabidopsis respiratory chain by nitrogen supply: Opposing effects of ammonium and nitrate. Plant J. 2006, 45, 775–788. [Google Scholar] [CrossRef]
- Wallström, S.; Florez-Sarasa, I.; Araújo, W.; Escobar, M.; Geisler, D.; Aidemark, M.; Lager, I.; Fernie, A.; Ribas-Carbó, M.; Rasmusson, A. Suppression of NDA-type alternative mitochondrial NAD(P)H dehydrogenases in Arabidopsis Thaliana modifies growth and metabolism, but not high light stimulation of mitochondrial electron transport. Plant Cell Physiol. 2014, 55, 881–896. [Google Scholar] [CrossRef]
- Ju, M.; Miao, H.; Wang, H.; Zhang, H. Mutagenesis for Creation of Genetic Variability in Sesame. In The Sesame Genome; Miao, H., Zhang, H., Kole, C., Eds.; Compendium of Plant Genomes; Springer: Cham, Switzerland, 2021; pp. 121–130. [Google Scholar]
- Kim, D.; Paggi, J.; Park, C.; Bennett, C.; Salzberg, S. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]

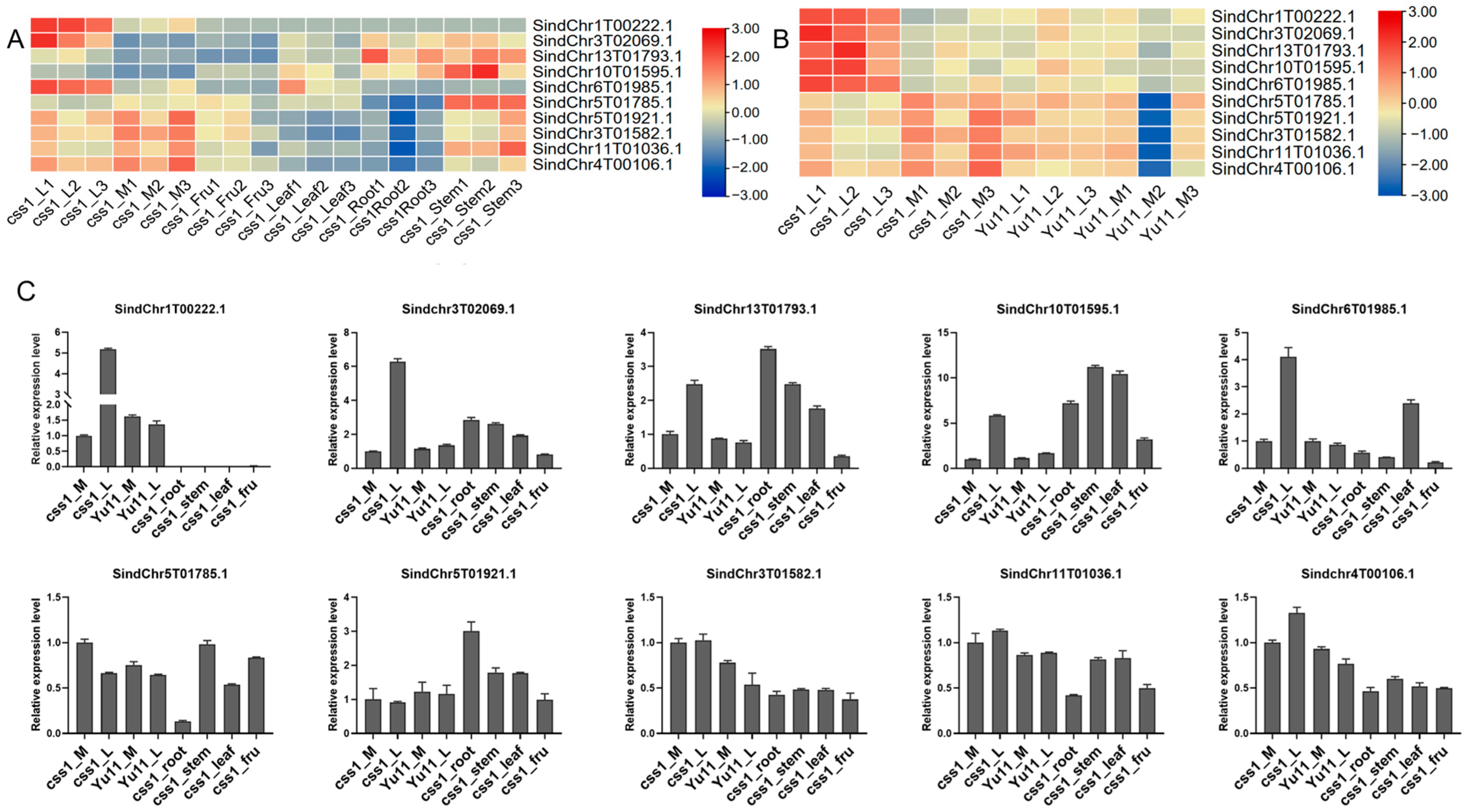
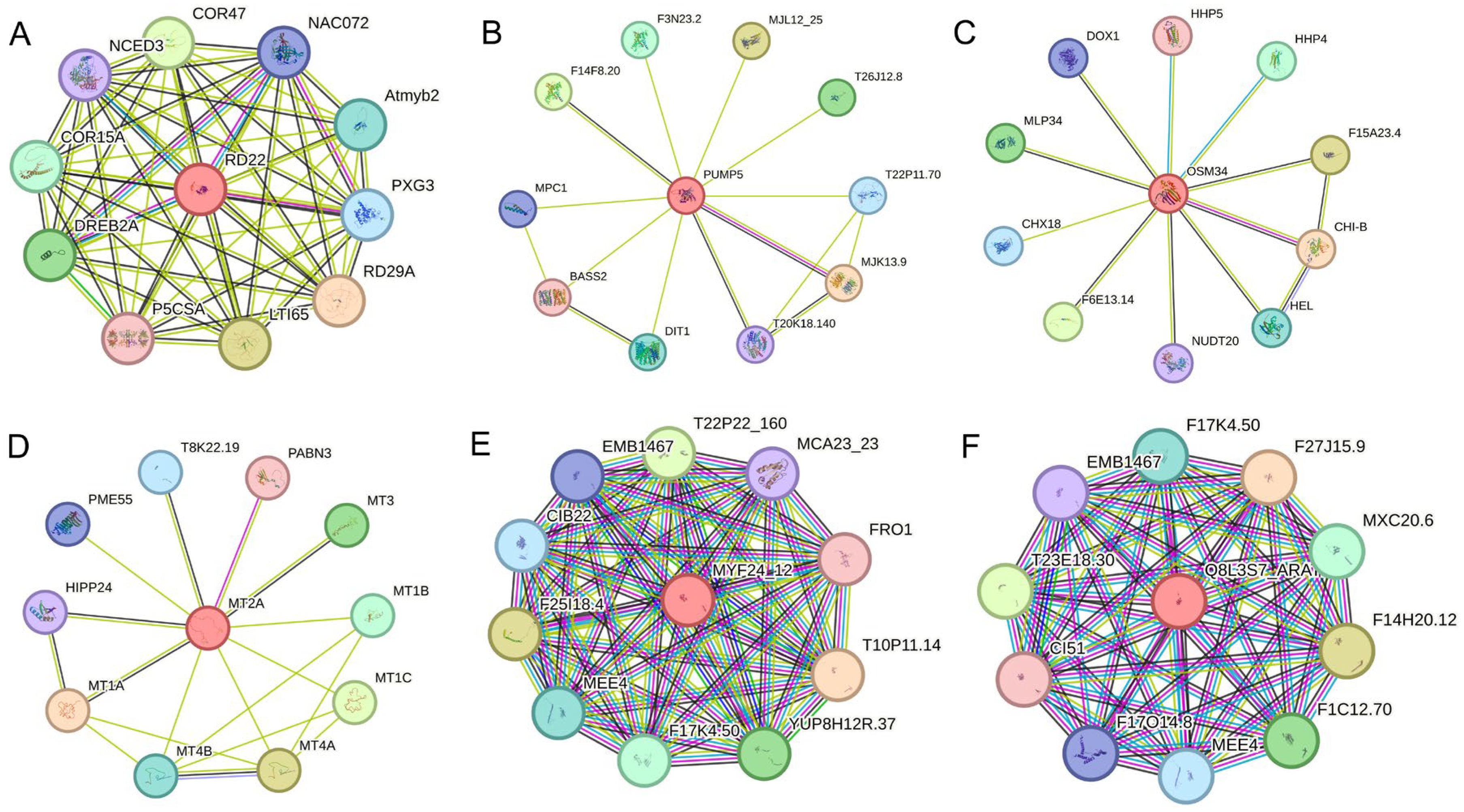
| Module | Hub Genes | Annotation |
|---|---|---|
| MEgreenyellow | SindChr1T00222 | BURP domain-containing protein 3-like |
| MEgreenyellow | SindChr3T02069 | Pathogenesis-related protein STH-2-like |
| MEgreenyellow | SindChr13T01793 | Mitochondrial uncoupling protein 5 |
| MEgreenyellow | SindChr10T01595 | Major allergen Pru ar 1-like |
| MEgreenyellow | SindChr6T01985 | thaumatin-like protein |
| MEmagenta | SindChr5T01785 | metallothionein-like protein |
| MEmagenta | SindChr5T01921 | - |
| MEmagenta | SindChr3T01582 | NADH dehydrogenase ubiquinone 1 beta subcomplex subunit |
| MEmagenta | SindChr11T01036 | - |
| MEmagenta | SindChr4T00106 | ESSS subunit of NADH: ubiquinone oxidoreductase (Complex I) protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, X.; Yang, W.; Cao, H.; Ma, Q.; Ju, M.; Hou, W.; Mu, C.; Chang, P.; Duan, Y.; Zhang, Z.; et al. Comparative Transcriptomics Analysis Reveals Genes Associated with a Dehiscent-Corolla Mutant in Sesame (Sesamum indicum L.). Int. J. Mol. Sci. 2025, 26, 11841. https://doi.org/10.3390/ijms262411841
Feng X, Yang W, Cao H, Ma Q, Ju M, Hou W, Mu C, Chang P, Duan Y, Zhang Z, et al. Comparative Transcriptomics Analysis Reveals Genes Associated with a Dehiscent-Corolla Mutant in Sesame (Sesamum indicum L.). International Journal of Molecular Sciences. 2025; 26(24):11841. https://doi.org/10.3390/ijms262411841
Chicago/Turabian StyleFeng, Xiaoxu, Weifei Yang, Hengchun Cao, Qin Ma, Ming Ju, Weixiu Hou, Cong Mu, Pengjie Chang, Yinghui Duan, Zhanyou Zhang, and et al. 2025. "Comparative Transcriptomics Analysis Reveals Genes Associated with a Dehiscent-Corolla Mutant in Sesame (Sesamum indicum L.)" International Journal of Molecular Sciences 26, no. 24: 11841. https://doi.org/10.3390/ijms262411841
APA StyleFeng, X., Yang, W., Cao, H., Ma, Q., Ju, M., Hou, W., Mu, C., Chang, P., Duan, Y., Zhang, Z., Li, G., Tian, Q., Zhang, H., & Miao, H. (2025). Comparative Transcriptomics Analysis Reveals Genes Associated with a Dehiscent-Corolla Mutant in Sesame (Sesamum indicum L.). International Journal of Molecular Sciences, 26(24), 11841. https://doi.org/10.3390/ijms262411841






