Mutation of OsKIF14.3, a Kinesin-14 Subfamily Protein, Altered Starch Metabolism and Caused Yellowish Leaf in Rice
Abstract
1. Introduction
2. Results
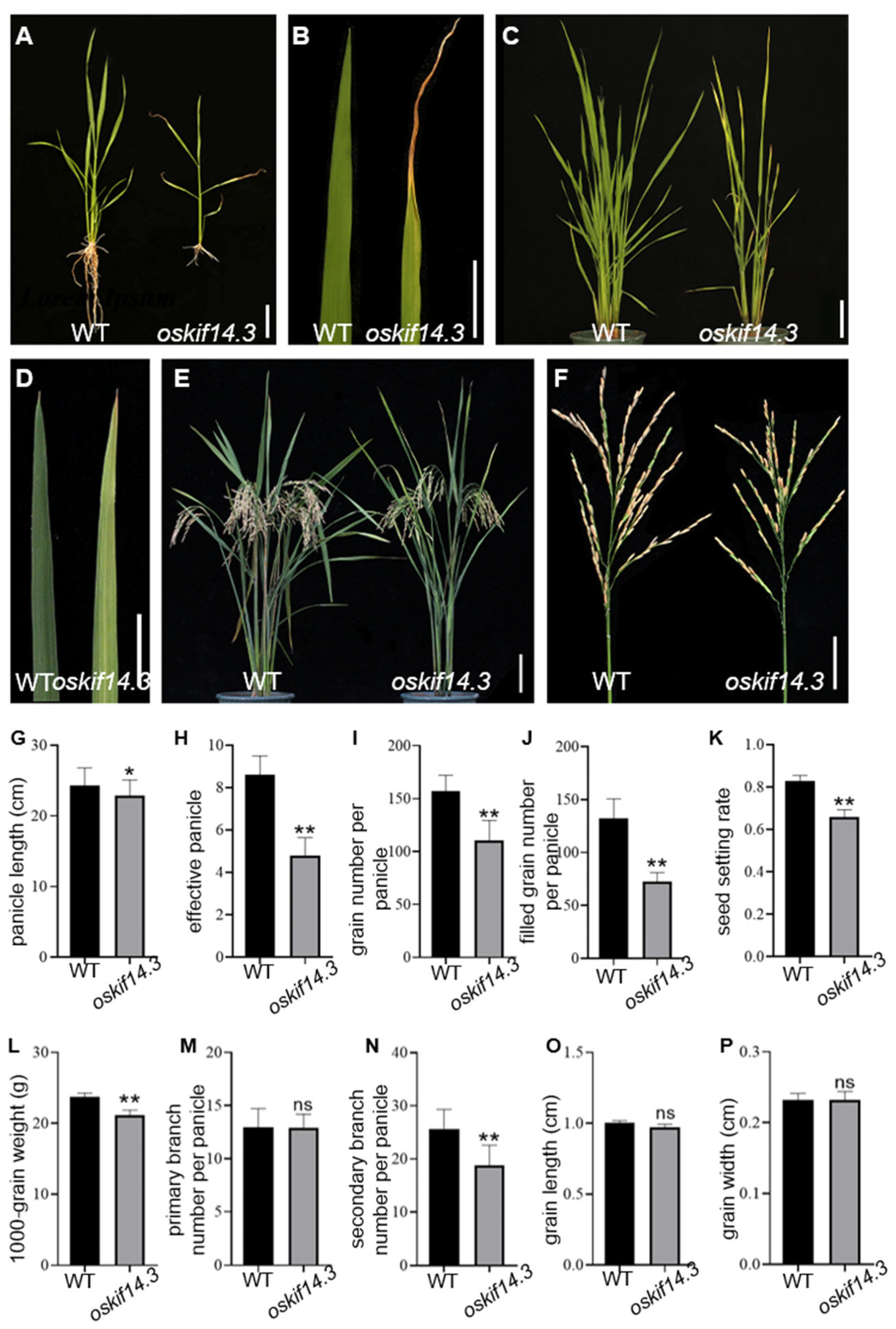
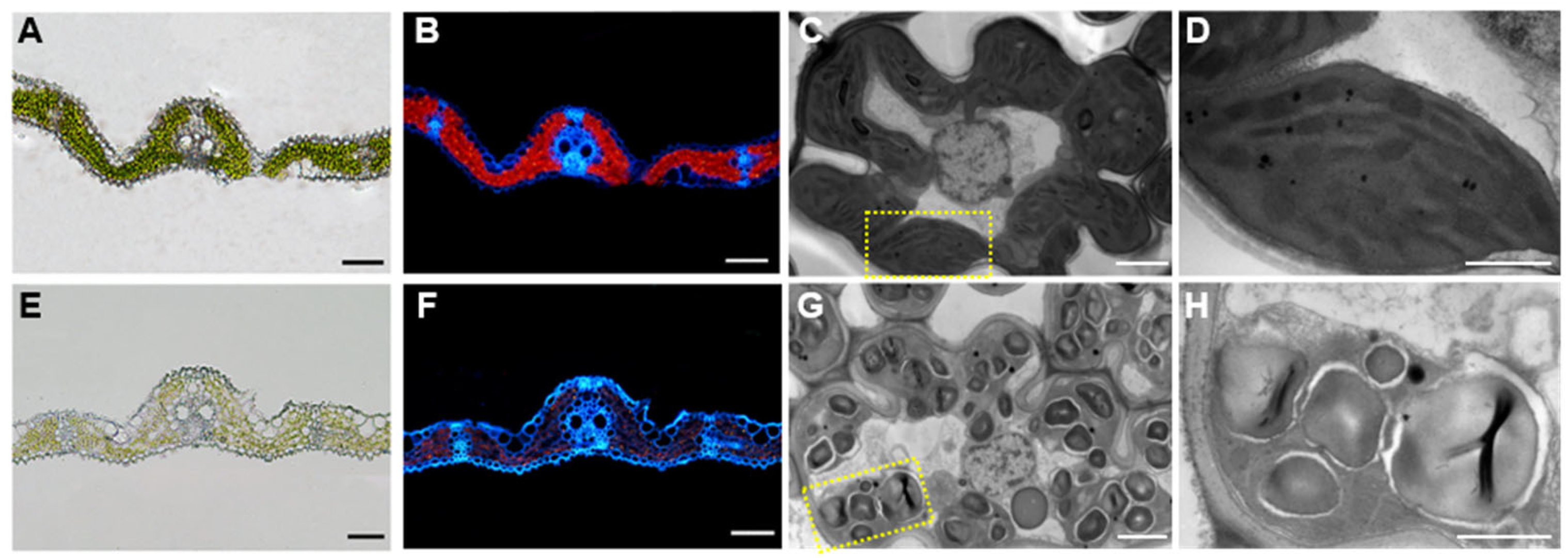
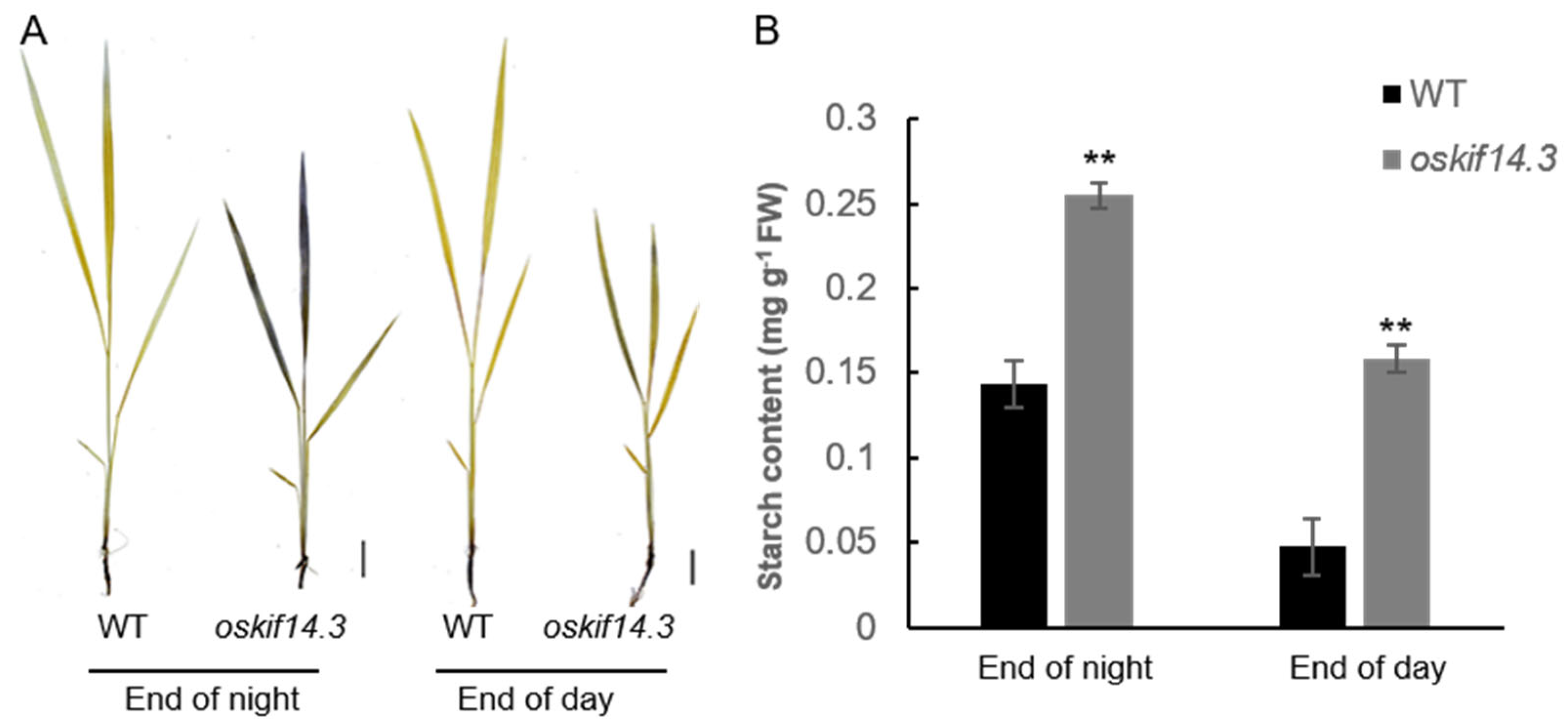
2.1. Phenotype Analysis of oskif14.3 Mutant
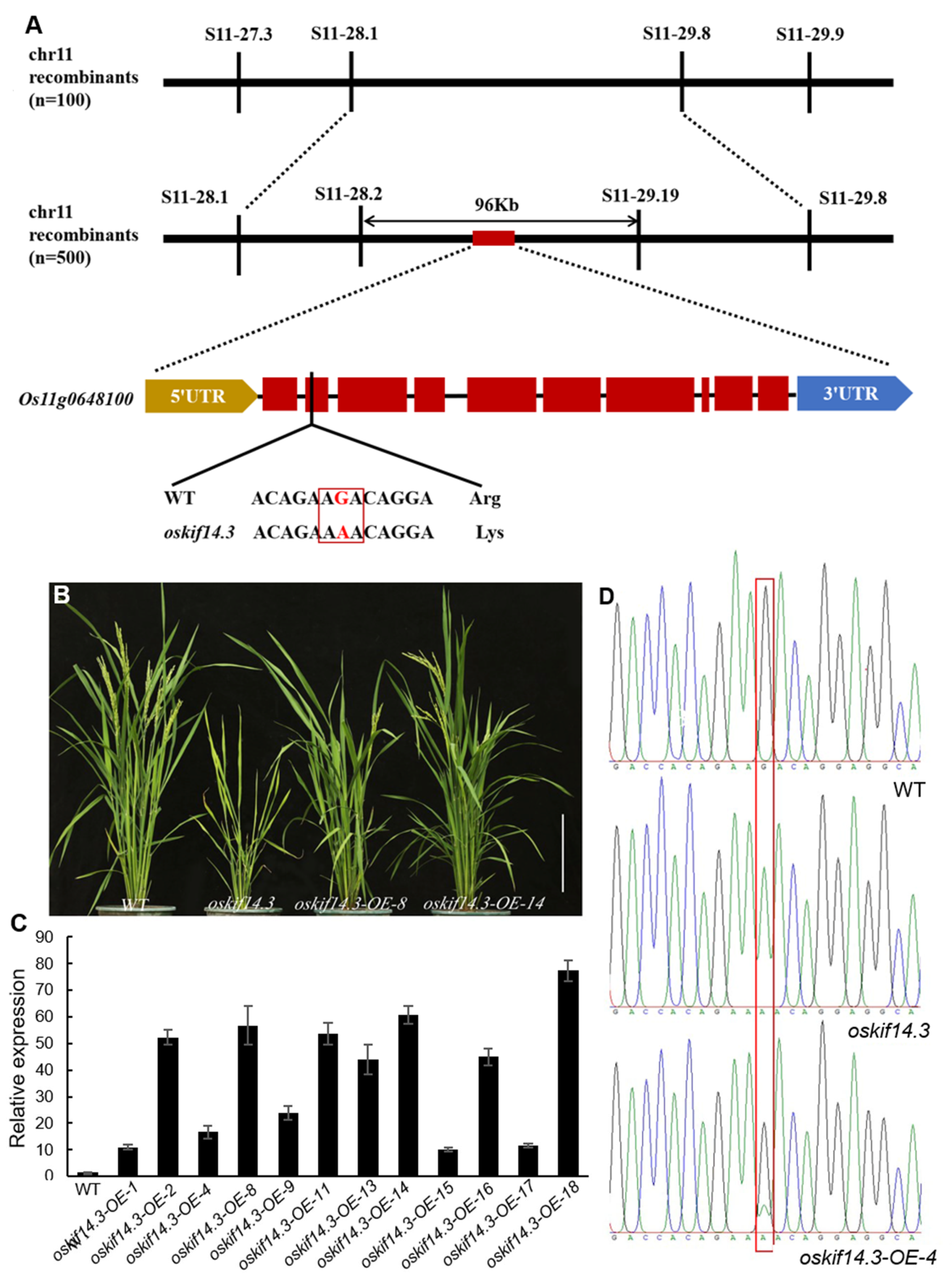
2.2. Map-Based Clone of OsKIF14.3
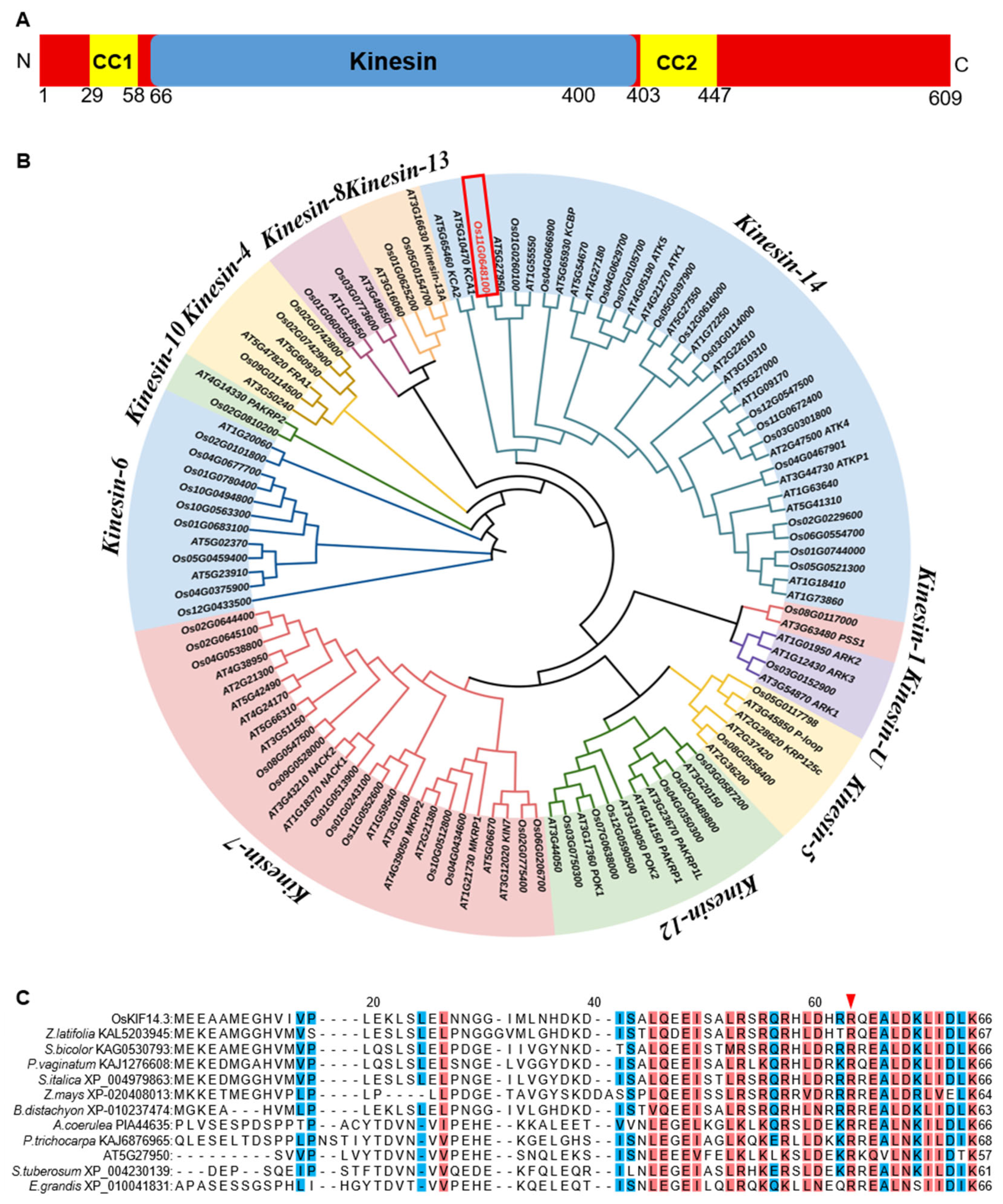
2.3. Characteristic Analysis of OsKIF14.3 Gene
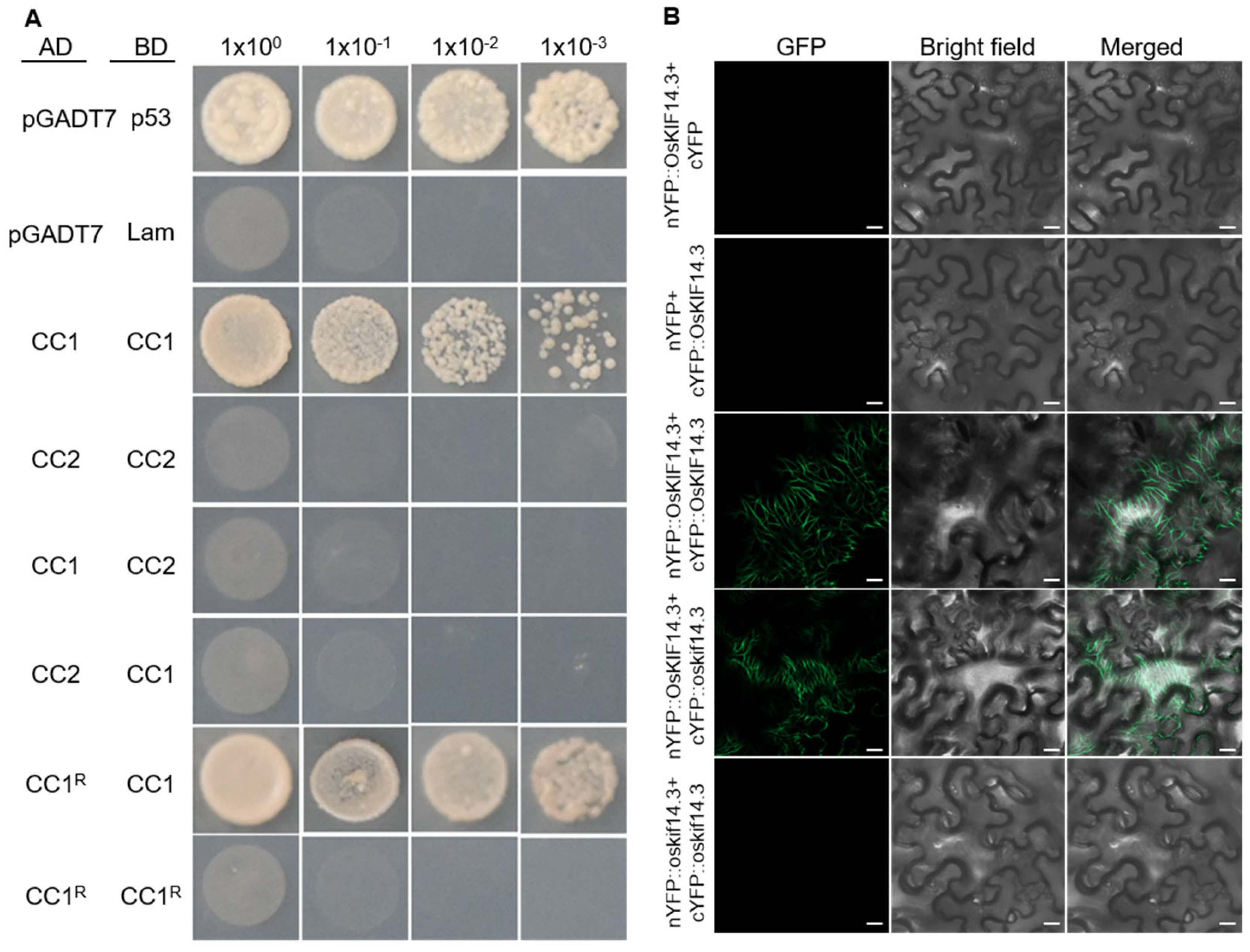
2.4. OsKIF14.3 Functions as Homodimer
2.5. OsKIF14.3 Participates in the Plasma Membrane Location of OsSWEET11
3. Discussion
3.1. The oskif14.3 Is a New Leaf Color Mutant
3.2. OsKIF14.3 Participates in Carbohydrate Distribution by Regulating OsSWEET11’s Location
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Physical and Chemical Analysis
4.3. Frozen Section
4.4. Transmission Electron Microscope (TEM)
4.5. Map-Based Cloning
4.6. Statistical Analysis
4.7. Multiple Sequence Alignment and Evolutionary Analysis
4.8. Function Analysis of Os11g0648100
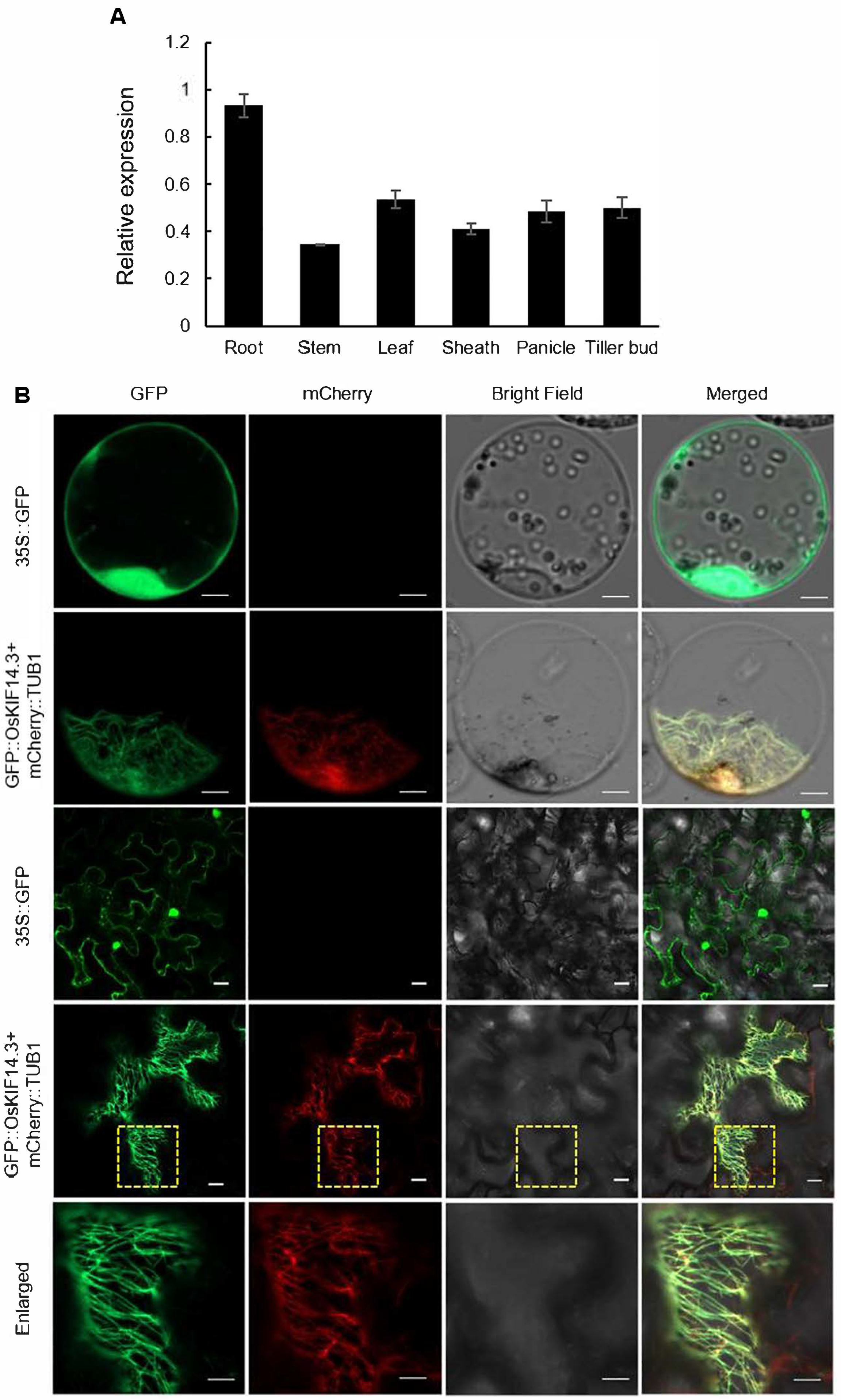
4.9. Gene Expression and RNA Isolation
4.10. Subcelluar Localization of GFP::OsKIF14.3 in Protoplast and N. benthamiana Cells
4.11. Bimolecular Fluorescence Complementation Assay
4.12. Protein Interaction Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, S.; Kim, J.; Yoo, E.; Lee, C.; Hirochika, H.; An, G. Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol. Biol. 2005, 57, 805–818. [Google Scholar] [CrossRef]
- Ruan, B.; Gao, Z.; Zhao, J.; Zhang, B.; Zhang, A.; Hong, K.; Yang, S.; Jiang, H.; Liu, C.; Chen, G.; et al. The Rice YGL Gene Encoding an Mg2+-chelatase ChlD Subunit is Affected by Temperature for Chlorophyll Biosynthesis. J. Plant Biol. 2017, 60, 314–321. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Rahman, M.; Cho, S.; Kim, Y.; Koh, H.; Yoo, S.; Paek, N. The rice faded green leaf locus encodes protochlorophyllide oxidoreductaseB and is essential for chlorophyll synthesis under high light conditions. Plant J. 2013, 74, 122–133. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, P.; Wang, H.; Chao, S.; Guo, W.; Zhang, Y.; Miao, C.; Yuan, H.; Peng, B. Physiological and transcriptomic analysis of OsLHCB3 knockdown lines in rice. Mol. Breed. 2023, 43, 15. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.; Cho, S.; Sugimoto, H.; Li, J.; Kusumi, K.; Koh, H.; Iba, K.; Paek, N. Rice Virescent3 and Stripe1 Encoding the Large and Small Subunits of Ribonucleotide Reductase Are Required for Chloroplast Biogenesis during Early Leaf Development. Plant Physiol. 2009, 150, 388–401. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Y.; Mazumder, M.A.R.; Pan, R.; Akhter, D. Research progresses on rice leaf color mutants. Crop Des. 2022, 1, 100015. [Google Scholar] [CrossRef]
- Xie, Z.; Zhao, B.; Zhang, M.; Sang, X.; Zhao, F.; Feng, P.; He, G.; Zhu, X. Mutation of OsSAC3, Encoding the Xanthine Dehydrogenase, Caused Early Senescence in Rice. Int. J. Mol. Sci. 2022, 23, 11053. [Google Scholar] [CrossRef] [PubMed]
- Endow, S.; Kull, F.; Liu, H. Kinesins at a glance. J. Cell Sci. 2010, 123, 3420–3424. [Google Scholar] [CrossRef]
- Vale, R.; Reese, T.; Sheetz, M. Identification of a Novel Force-Generating Protein, Kinesin, Involved in Microtubule-Based Motility. Cell 1985, 42, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, Y.; Chong, K. The novel functions of kinesin motor proteins in plants. Protoplasma 2012, 249, 95–100. [Google Scholar] [CrossRef]
- Vale, R.; Fletterick, R. The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 1997, 13, 745–777. [Google Scholar] [CrossRef]
- Lawrence, C.; Malmberg, R.; Muszynski, M.; Dawe, R. Maximum likelihood methods reveal conservation of function among closely related kinesin families. J. Mol. Evol. 2002, 54, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Marcus, A.; Li, W.; Hu, Y.; Calzada, J.; Grossniklaus, U.; Cyr, R.; Ma, H. The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 2002, 129, 2401–2409. [Google Scholar] [CrossRef] [PubMed]
- Marcus, A.; Li, W.; Ma, H.; Cyr, R. A kinesin mutant with an atypical bipolar spindle undergoes normal mitosis. Mol. Biol. Cell 2003, 14, 1717–1726. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Han, L.; Feng, Z.; Wang, G.; Liu, W.; Ma, Y.; Yu, Y.; Kong, Z. Orchestration of microtubules and the actin cytoskeleton in trichome cell shape determination by a plant-unique kinesin. eLife 2015, 4, 22. [Google Scholar] [CrossRef]
- Ganguly, A.; Zhu, C.; Chen, W.; Dixit, R. FRA1 Kinesin Modulates the Lateral Stability of Cortical Microtubules through Cellulose Synthase-Microtubule Uncoupling Proteins. Plant Cell 2020, 32, 2508–2524. [Google Scholar] [CrossRef]
- Suetsugu, N.; Sato, Y.; Tsuboi, H.; Kasahara, M.; Imaizumi, T.; Kagawa, T.; Hiwatashi, Y.; Hasebe, M.; Wada, M. The KAC Family of Kinesin-Like Proteins is Essential for the Association of Chloroplasts with the Plasma Membrane in Land Plants. Plant Cell Physiol. 2012, 53, 1854–1865. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, Y.; Li, W.; Zhao, Z.; Ren, Y.; Wang, Y.; Gu, S.; Lin, Q.; Wang, D.; Jiang, L.; et al. Pollen Semi-Sterility1 Encodes a Kinesin-1-Like Protein Important for Male Meiosis, Anther Dehiscence, and Fertility in Rice. Plant Cell 2011, 23, 111–129. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, Y.; You, H.; Chen, J.; Wang, B.; Wen, M.; Zhang, Y.; Tang, D.; Shen, Y.; Yu, H.; et al. Kinesin-1-like protein PSS1 is essential for full-length homologous pairing and synapsis in rice meiosis. Plant J. 2024, 120, 928–940. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, B.; Qian, Q.; Yu, Y.; Li, R.; Zhang, J.; Liu, X.; Zeng, D.; Li, J.; Zhou, Y. Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J. 2010, 63, 312–328. [Google Scholar] [CrossRef]
- Deng, Z.; Liu, L.; Li, T.; Yan, S.; Kuang, B.; Huang, S.; Yan, C.; Wang, T. OsKinesin-13A Is an Active Microtubule Depolymerase Involved in Glume Length Regulation via Affecting Cell Elongation. Sci. Rep. 2015, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Yuan, S.; Li, C.; Jiang, D.; Li, X. Reduction of ATPase activity in the rice kinesin protein Stemless Dwarf1 inhibits cell division and organ development. Plant J. 2018, 96, 620–634. [Google Scholar] [CrossRef]
- Tseng, K.; Wang, P.; Lee, Y.; Bowen, J.; Gicking, A.; Guo, L.; Liu, B.; Qiu, W. The preprophase band-associated kinesin-14 OsKCH2 is a processive minus-end-directed microtubule motor. Nat. Commun. 2018, 9, 11. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, J.; Wang, X.; Sun, J.; Sha, W. Molecular cloning and expression analysis of the rice triose phosphate/phosphate translocator gene. Plant Sci. 2002, 162, 785–790. [Google Scholar] [CrossRef]
- Akihiro, T.; Mizuno, K.; Fujimura, T. Gene expression of ADP-glucose pyrophosphorylase and starch contents in rice cultured cells are cooperatively regulated by sucrose and ABA. Plant Cell Physiol. 2005, 46, 937–946. [Google Scholar] [CrossRef]
- Lee, S.; Hwang, S.; Han, M.; Eom, J.; Kang, H.; Han, Y.; Choi, S.; Cho, M.; Bhoo, S.; An, G.; et al. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol. Biol. 2007, 65, 531–546. [Google Scholar] [CrossRef]
- Hirose, T.; Aoki, N.; Harada, Y.; Okamura, M.; Hashida, Y.; Ohsugi, R.; Miyao, A.; Hirochika, H.; Terao, T. Disruption of a rice gene for α-glucan water dikinase, OsGWD1, leads to hyperaccumulation of starch in leaves but exhibits limited effects on growth. Front. Plant Sci. 2013, 4, 9. [Google Scholar] [CrossRef]
- Yun, M.; Umemoto, T.; Kawagoe, Y. Rice Debranching Enzyme Isoamylase3 Facilitates Starch Metabolism and Affects Plastid Morphogenesis. Plant Cell Physiol. 2011, 52, 1068–1082. [Google Scholar] [CrossRef]
- Allingham, J.; Sproul, L.; Rayment, I.; Gilbert, S. Vik1 modulates microtubule-Kar3 interactions through a motor domain that lacks an active site. Cell 2007, 128, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.; Fu, B.; Yang, H.; Xu, C.; Li, Z.; Sanchez, A.; Park, Y.; Bennetzen, J.; Zhang, Q.; Wang, S. Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor. Appl. Genet. 2006, 112, 455–461. [Google Scholar] [CrossRef]
- Sugimoto, H.; Kusumi, K.; Tozawa, Y.; Yazaki, J.; Kishimoto, N.; Kikuchi, S.; Iba, K. The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant Cell Physiol. 2004, 45, 985–996. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Li, M.; Yang, W.; Xu, W.; Xue, Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 2006, 141, 1376–1388. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ruiz, J.; Spínola, M.; Kirchsteiger, K.; Moreno, J.; Sahrawy, M.; Cejudo, F. Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 2006, 18, 2356–2368. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, X.; He, B.; Diao, L.; Sheng, S.; Wang, J.; Guo, X.; Su, N.; Wang, L.; Jiang, L.; et al. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 2007, 145, 29–40. [Google Scholar] [CrossRef]
- Fang, J.; Chai, C.; Qian, Q.; Li, C.; Tang, J.; Sun, L.; Huang, Z.; Guo, X.; Sun, C.; Liu, M.; et al. Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 2008, 54, 177–189. [Google Scholar] [CrossRef]
- Kusumi, K.; Sakata, C.; Nakamura, T.; Kawasaki, S.; Yoshimura, A.; Iba, K. A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. Plant J. 2011, 68, 1039–1050. [Google Scholar] [CrossRef]
- Zhou, S.; Sawicki, A.; Willows, R.; Luo, M. C-terminal residues of Oryza sativa GUN4 are required for the activation of the ChlH subunit of magnesium chelatase in chlorophyll synthesis. FEBS Lett. 2012, 586, 205–210. [Google Scholar] [CrossRef]
- Kong, W.; Yu, X.; Chen, H.; Liu, L.; Xiao, Y.; Wang, Y.; Wang, C.; Lin, Y.; Yu, Y.; Wang, C.; et al. The catalytic subunit of magnesium-protoporphyrin IX monomethyl ester cyclase forms a chloroplast complex to regulate chlorophyll biosynthesis in rice. Plant Mol. Biol. 2016, 92, 177–191. [Google Scholar] [CrossRef]
- Wang, G.; Wu, Y.; Ma, L.; Lin, Y.; Hu, Y.; Li, M.; Li, W.; Ding, Y.; Chen, L. Phloem loading in rice leaves depends strongly on the apoplastic pathway. J. Exp. Bot. 2021, 72, 3723–3738. [Google Scholar] [CrossRef]
- Eom, J.; Cho, J.; Reinders, A.; Lee, S.; Yoo, Y.; Tuan, P.; Choi, S.; Bang, G.; Park, Y.; Cho, M.; et al. Impaired Function of the Tonoplast-Localized Sucrose Transporter in Rice, OsSUT2, Limits the Transport of Vacuolar Reserve Sucrose and Affects Plant Growth. Plant Physiol. 2011, 157, 109–119. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, L.; Huang, W.; Yuan, M.; Zhou, F.; Li, X.; Lin, Y. Overexpression of OsSWEET5 in Rice Causes Growth Retardation and Precocious Senescence. PLoS ONE 2014, 9, e94210. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Qu, X.; Hou, B.; Sosso, D.; Osorio, S.; Fernie, A.; Frommer, W. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.; Yuan, M.; Yao, L.; Ge, X.; Yuan, B.; Xu, C.; Li, X.; Fu, B.; Li, Z.; Bennetzen, J.; et al. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 2006, 20, 1250–1255. [Google Scholar] [CrossRef] [PubMed]
| Leaf Position | Materials | Chla | Chlb | Total chl | Car |
|---|---|---|---|---|---|
| The first leaf from the top | WT | 5.27 ± 0.16 | 2.36 ± 0.51 | 7.26 ± 0.60 | 2.18 ± 0.09 |
| oskif14.3 | 3.32 ± 0.19 ** | 0.73 ± 0.03 * | 4.05 ± 0.23 ** | 1.30 ± 0.07 ** | |
| The second leaf from the top | WT | 5.25 ± 0.05 | 2.41 ± 0.10 | 4.05 ± 0.14 | 2.18 ± 0.04 |
| oskif14.3 | 3.82 ± 0.35 * | 0.79 ± 0.02 * | 4.68 ± 0.48 ** | 1.46 ± 0.18 * | |
| The third leaf from the top | WT | 5.90 ± 0.44 | 1.56 ± 0.26 | 7.45 ± 0.57 | 2.12 ± 0.33 |
| oskif14.3 | 4.61 ± 0.20 * | 0.96 ± 0.04 * | 5.56 ± 0.22 * | 1.49 ± 0.06 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Jiang, W.; Xie, Z.; Liu, C.; Li, Q.; Shen, W.; He, G.; Zhu, X. Mutation of OsKIF14.3, a Kinesin-14 Subfamily Protein, Altered Starch Metabolism and Caused Yellowish Leaf in Rice. Int. J. Mol. Sci. 2025, 26, 11577. https://doi.org/10.3390/ijms262311577
Zhang M, Jiang W, Xie Z, Liu C, Li Q, Shen W, He G, Zhu X. Mutation of OsKIF14.3, a Kinesin-14 Subfamily Protein, Altered Starch Metabolism and Caused Yellowish Leaf in Rice. International Journal of Molecular Sciences. 2025; 26(23):11577. https://doi.org/10.3390/ijms262311577
Chicago/Turabian StyleZhang, Mengxue, Wenchang Jiang, Ziyu Xie, Chang Liu, Qiyu Li, Wenqiang Shen, Guanghua He, and Xiaoyan Zhu. 2025. "Mutation of OsKIF14.3, a Kinesin-14 Subfamily Protein, Altered Starch Metabolism and Caused Yellowish Leaf in Rice" International Journal of Molecular Sciences 26, no. 23: 11577. https://doi.org/10.3390/ijms262311577
APA StyleZhang, M., Jiang, W., Xie, Z., Liu, C., Li, Q., Shen, W., He, G., & Zhu, X. (2025). Mutation of OsKIF14.3, a Kinesin-14 Subfamily Protein, Altered Starch Metabolism and Caused Yellowish Leaf in Rice. International Journal of Molecular Sciences, 26(23), 11577. https://doi.org/10.3390/ijms262311577





