Molecular Insights into Central Core Disease: Proteomic Signatures and Potential Therapeutic Biomarkers in RYR1 I4895T Mice
Abstract
1. Introduction
2. Results and Discussion
2.1. WT and IT Mice Muscles Present a Differential Protein Pattern
2.2. Enrichment Analysis of the Proteomic Data
2.3. Structural, Contractile, and Cytoskeletal Stabilization Proteins Were Altered
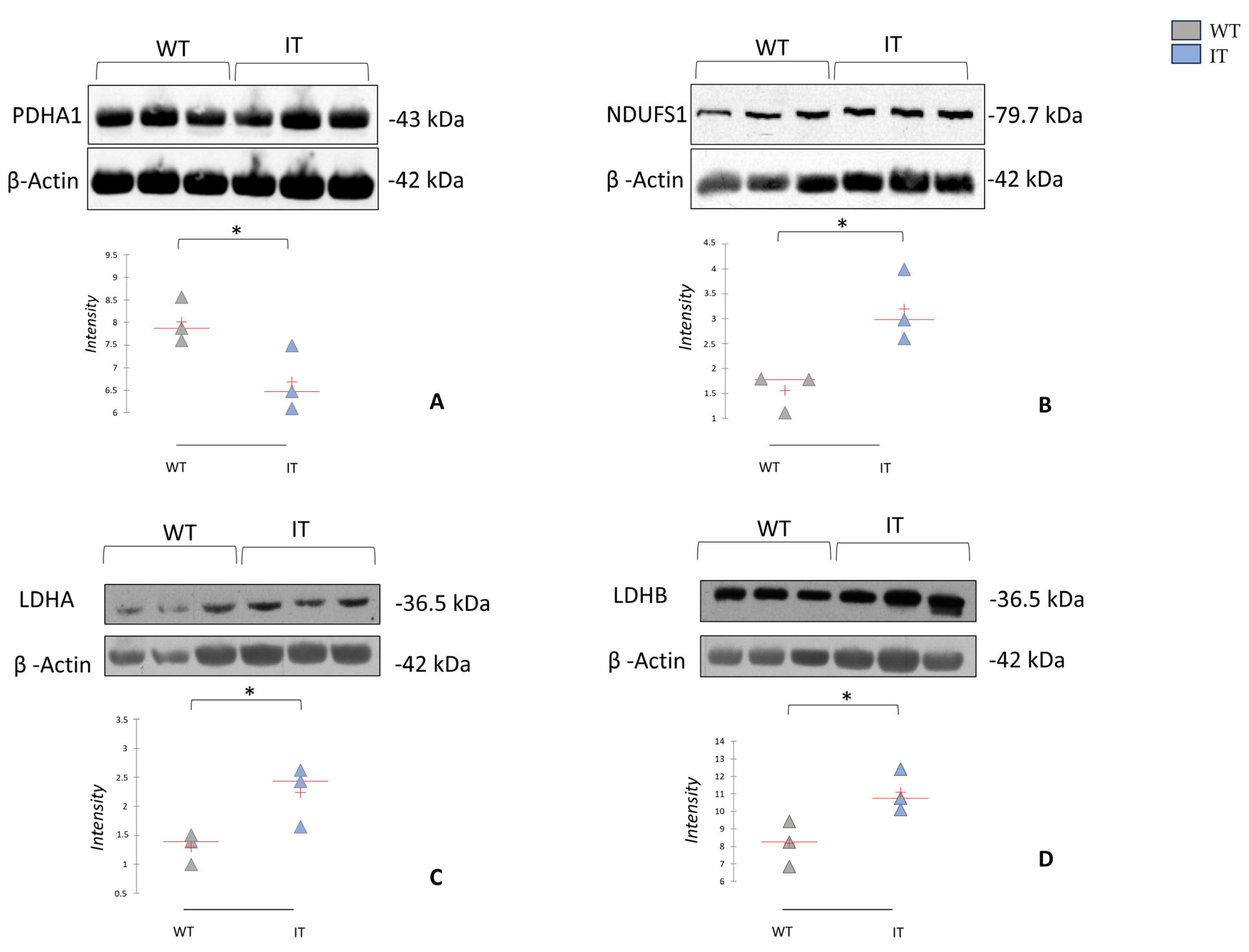
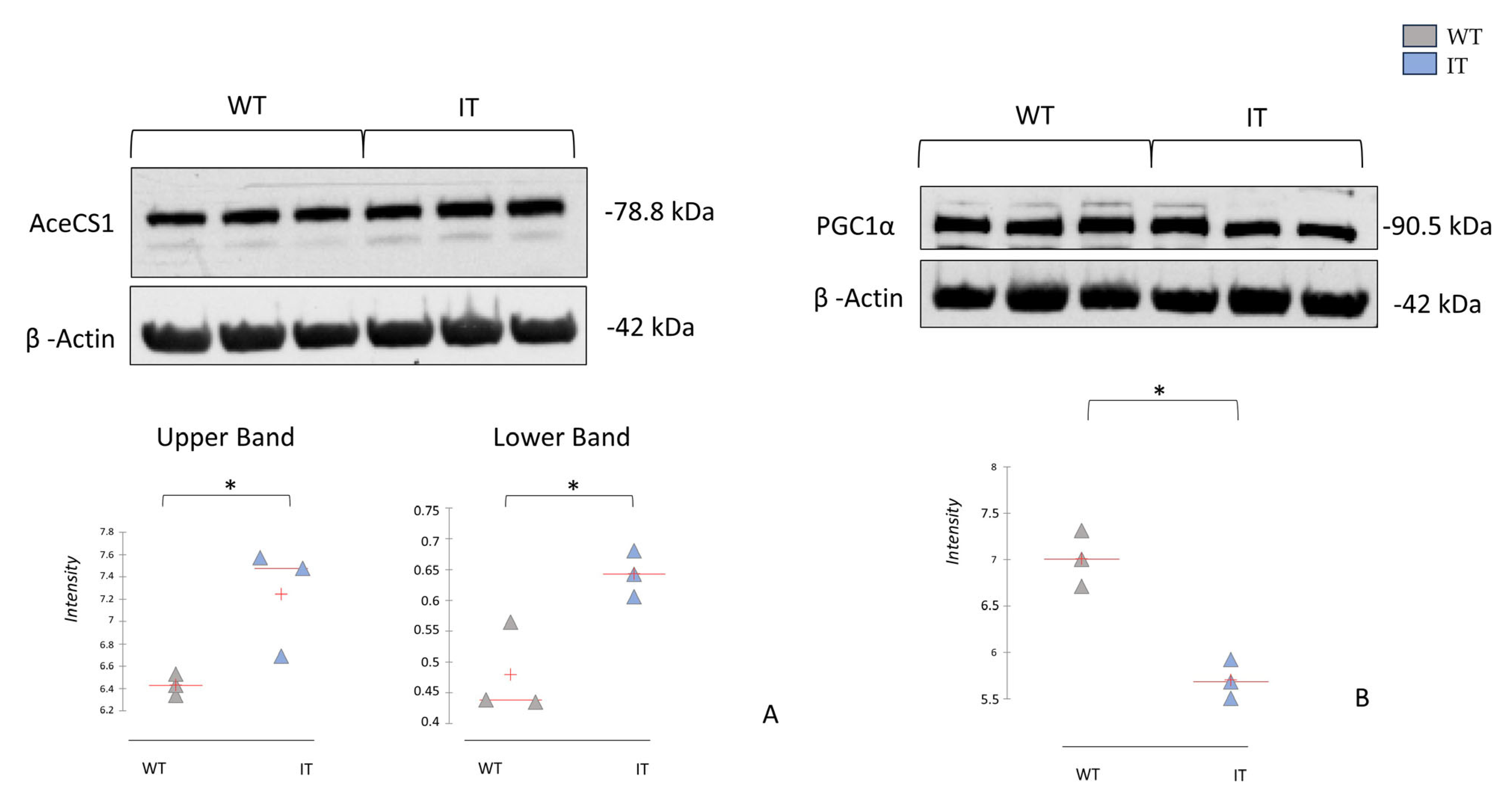
2.4. RYR1-I4898T Mutation May Induce Alteration in Metabolism
2.5. RYR1-I4898T Mutation May Induce an Alteration in Oxidative Stress Response
3. Materials and Methods
3.1. Animals
3.2. Sample Preparation for Two-Dimensional Electrophoresis
3.3. Two-Dimensional Electrophoresis
3.4. Image and Statistical Analysis
3.5. MALDI-TOF Identification by PMF
3.6. MetaCore Enrichment Analysis
3.7. Western Blot Analysis
3.8. Antibodies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Franzini-Armstrong, C.; Protasi, F. Ryanodine Receptors of Striated Muscles: A Complex Channel Capable of Multiple Interactions. Physiol. Rev. 1997, 77, 699–729. [Google Scholar] [CrossRef]
- Schneider, M.F. Control of Calcium Release in Functioning Skeletal Muscle Fibers. Annu. Rev. Physiol. 1994, 56, 463–484. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Catallo, M.R.; Pierantozzi, E.; Sorrentino, V. Mutations in Proteins Involved in E-C Coupling and SOCE and Congenital Myopathies. J. Gen. Physiol. 2022, 154, e202213115. [Google Scholar] [CrossRef]
- Lawal, T.A.; Todd, J.J.; Witherspoon, J.W.; Bönnemann, C.G.; Dowling, J.J.; Hamilton, S.L.; Meilleur, K.G.; Dirksen, R.T. Ryanodine Receptor 1-Related Disorders: An Historical Perspective and Proposal for a Unified Nomenclature. Skelet. Muscle 2020, 10, 32. [Google Scholar] [CrossRef]
- Hopkins, P.M.; Gupta, P.K.; Bilmen, J.G. Malignant Hyperthermia. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2018; Volume 157, pp. 645–661. [Google Scholar] [CrossRef]
- Galli, L.; Orrico, A.; Lorenzini, S.; Censini, S.; Falciani, M.; Covacci, A.; Tegazzin, V.; Sorrentino, V. Frequency and Localization of Mutations in the 106 Exons of the RYR1 Gene in 50 Individuals with Malignant Hyperthermia. Hum. Mutat. 2006, 27, 830. [Google Scholar] [CrossRef] [PubMed]
- Magee, K.R.; Shy, G.M. A New Congenital Non-Progressive Myopathy. Brain 1956, 79, 610–621. [Google Scholar] [CrossRef]
- Wu, S.; Ibarra, M.C.A.; Malicdan, M.C.V.; Murayama, K.; Ichihara, Y.; Kikuchi, H.; Nonaka, I.; Noguchi, S.; Hayashi, Y.K.; Nishino, I. Central Core Disease Is Due to RYR1 Mutations in More than 90% of Patients. Brain 2006, 129, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Lawal, T.A.; Todd, J.J.; Meilleur, K.G. Ryanodine Receptor 1-Related Myopathies: Diagnostic and Therapeutic Approaches. Neurotherapeutics 2018, 15, 885–899. [Google Scholar] [CrossRef]
- MacLennan, D.H.; Zvaritch, E. Mechanistic Models for Muscle Diseases and Disorders Originating in the Sarcoplasmic Reticulum. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2011, 1813, 948–964. [Google Scholar] [CrossRef]
- Jungbluth, H. Central Core Disease. Orphanet J. Rare Dis. 2007, 2, 25. [Google Scholar] [CrossRef]
- Topaloglu, H. Core Myopathies—A Short Review. Acta Myol. 2020, 39, 266–273. [Google Scholar] [CrossRef]
- Bharucha-Goebel, D.X.; Santi, M.; Medne, L.; Zukosky, K.; Dastgir, J.; Shieh, P.B.; Winder, T.; Tennekoon, G.; Finkel, R.S.; Dowling, J.J.; et al. Severe Congenital RYR1-Associated Myopathy: The Expanding Clinicopathologic and Genetic Spectrum. Neurology 2013, 80, 1584–1589. [Google Scholar] [CrossRef] [PubMed]
- Romero, N.B.; Monnier, N.; Viollet, L.; Cortey, A.; Chevallay, M.; Leroy, J.P.; Lunardi, J.; Fardeau, M. Dominant and Recessive Central Core Disease Associated with RYR1 Mutations and Fetal Akinesia. Brain 2003, 126, 2341–2349. [Google Scholar] [CrossRef]
- Iodice, P.; Boncompagni, S.; Pietrangelo, L.; Galli, L.; Pierantozzi, E.; Rossi, D.; Fusella, A.; Caulo, M.; Kern, H.; Sorrentino, V.; et al. Functional Electrical Stimulation: A Possible Strategy to Improve Muscle Function in Central Core Disease? Front. Neurol. 2019, 10, 479. [Google Scholar] [CrossRef]
- Vattemi, G.N.A.; Rossi, D.; Galli, L.; Catallo, M.R.; Pancheri, E.; Marchetto, G.; Cisterna, B.; Malatesta, M.; Pierantozzi, E.; Tonin, P.; et al. Ryanodine Receptor 1 (RYR1) Mutations in Two Patients with Tubular Aggregate Myopathy. Eur. J. Neurosci. 2022, 56, 4214–4223. [Google Scholar] [CrossRef]
- Loy, R.E.; Orynbayev, M.; Xu, L.; Andronache, Z.; Apostol, S.; Zvaritch, E.; MacLennan, D.H.; Meissner, G.; Melzer, W.; Dirksen, R.T. Muscle Weakness in Ryr1I4895T/WT Knock-in Mice as a Result of Reduced Ryanodine Receptor Ca2+ Ion Permeation and Release from the Sarcoplasmic Reticulum. J. Gen. Physiol. 2011, 137, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Zvaritch, E.; Depreux, F.; Kraeva, N.; Loy, R.E.; Goonasekera, S.A.; Boncompagni, S.; Kraev, A.; Gramolini, A.O.; Dirksen, R.T.; Franzini-Armstrong, C.; et al. An Ryr1I4895T Mutation Abolishes Ca2+ Release Channel Function and Delays Development in Homozygous Offspring of a Mutant Mouse Line. Proc. Natl. Acad. Sci. USA 2007, 104, 18537–18542. [Google Scholar] [CrossRef] [PubMed]
- Zvaritch, E.; Kraeva, N.; Bombardier, E.; McCloy, R.A.; Depreux, F.; Holmyard, D.; Kraev, A.; Seidman, C.E.; Seidman, J.G.; Tupling, A.R.; et al. Ca2+ Dysregulation in Ryr1I4895T/Wt Mice Causes Congenital Myopathy with Progressive Formation of Minicores, Cores, and Nemaline Rods. Proc. Natl. Acad. Sci. USA 2009, 106, 21813–21818. [Google Scholar] [CrossRef]
- Lee, C.S.; Hanna, A.D.; Wang, H.; Dagnino-Acosta, A.; Joshi, A.D.; Knoblauch, M.; Xia, Y.; Georgiou, D.K.; Xu, J.; Long, C.; et al. A Chemical Chaperone Improves Muscle Function in Mice with a RyR1 Mutation. Nat. Commun. 2017, 8, 14659. [Google Scholar] [CrossRef]
- Boncompagni, S.; Loy, R.E.; Dirksen, R.T.; Franzini-Armstrong, C. The I4895T Mutation in the Type 1 Ryanodine Receptor Induces Fiber-Type Specific Alterations in Skeletal Muscle That Mimic Premature Aging. Aging Cell 2010, 9, 958–970. [Google Scholar] [CrossRef]
- Melka, M.; Rotard, L.; Benstaali, C.; Brocard, J.; Giannesini, B.; Jouve, F.; Pelletier, L.; Fauré, J.; Rendu, J.; Jacquemond, V.; et al. Limited Pre-Clinical Relevance of the Heterozygous RYR1-I4895T/+ Mouse Model Due to Its Mild Phenotype. J. Neuromuscul. Dis. 2025. [Google Scholar] [CrossRef]
- Wang, C.H.; Dowling, J.J.; North, K.; Schroth, M.K.; Sejersen, T.; Shapiro, F.; Bellini, J.; Weiss, H.; Guillet, M.; Amburgey, K.; et al. Consensus Statement on Standard of Care for Congenital Myopathies. J. Child Neurol. 2012, 27, 363–382. [Google Scholar] [CrossRef]
- Jungblut, P.R. The Proteomics Quantification Dilemma. J. Proteom. 2014, 107, 98–102. [Google Scholar] [CrossRef]
- Vantaggiato, L.; Landi, C.; Shaba, E.; Rossi, D.; Sorrentino, V.; Bini, L. Protein Extraction Methods Suitable for Muscle Tissue Proteomic Analysis. Proteomes 2024, 12, 27. [Google Scholar] [CrossRef]
- Plotkin, D.L.; Mattingly, M.L.; Anglin, D.A.; Michel, J.M.; Godwin, J.S.; McIntosh, M.C.; Bergamasco, J.G.A.; Scarpelli, M.C.; Angleri, V.; Taylor, L.W.; et al. Skeletal Muscle Myosin Heavy Chain Protein Fragmentation as a Potential Marker of Protein Degradation in Response to Resistance Training and Disuse Atrophy. bioRxiv 2024, 2024.05.24.595789. [Google Scholar] [CrossRef] [PubMed]
- Boncompagni, S.; Protasi, F.; Franzini-Armstrong, C. Sequential Stages in the Age-Dependent Gradual Formation and Accumulation of Tubular Aggregates in Fast Twitch Muscle Fibers: SERCA and Calsequestrin Involvement. Age 2012, 34, 27–41. [Google Scholar] [CrossRef]
- Wadmore, K.; Azad, A.J.; Gehmlich, K. The Role of Z-Disc Proteins in Myopathy and Cardiomyopathy. Int. J. Mol. Sci. 2021, 22, 3058. [Google Scholar] [CrossRef] [PubMed]
- Takada, F.; Vander Woude, D.L.; Tong, H.Q.; Thompson, T.G.; Watkins, S.C.; Kunkel, L.M.; Beggs, A.H. Myozenin: An α-Actinin- and γ-Filamin-Binding Protein of Skeletal Muscle Z Lines. Proc. Natl. Acad. Sci. USA 2001, 98, 1595–1600. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Shinohara, H.; Kurobe, N.; Inaguma, Y.; Shimizu, K.; Ohshima, K. Tissue Distribution and Developmental Profiles of Immunoreactive αB Crystallin in the Rat Determined with a Sensitive Immunoassay System. Biochim. Biophys. Acta (BBA)-Gen. Subj. 1991, 1074, 201–208. [Google Scholar] [CrossRef]
- Der Perng, M.; Wen, S.F.; van den IJssel, P.; Prescott, A.R.; Quinlan, R.A. Desmin Aggregate Formation by R120G αB-Crystallin Is Caused by Altered Filament Interactions and Is Dependent upon Network Status in Cells. Mol. Biol. Cell 2004, 15, 2335–2346. [Google Scholar] [CrossRef]
- Spriet, L.L.; Heigenhauser, G.J.F. Regulation of Pyruvate Dehydrogenase (PDH) Activity in Human Skeletal Muscle During Exercise. Exerc. Sport Sci. Rev. 2002, 30, 91–95. [Google Scholar] [CrossRef]
- Adeva-Andany, M.; López-Ojén, M.; Funcasta-Calderón, R.; Ameneiros-Rodríguez, E.; Donapetry-García, C.; Vila-Altesor, M.; Rodríguez-Seijas, J. Comprehensive Review on Lactate Metabolism in Human Health. Mitochondrion 2014, 17, 76–100. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Hagras, M.A.; Konstantopoulou, V.; Mayr, J.A.; Stuchebrukhov, A.A.; Meierhofer, D. Mutations in NDUFS1 Cause Metabolic Reprogramming and Disruption of the Electron Transfer. Cells 2019, 8, 1149. [Google Scholar] [CrossRef]
- Qi, B.; Song, L.; Hu, L.; Guo, D.; Ren, G.; Peng, T.; Liu, M.; Fang, Y.; Li, C.; Zhang, M.; et al. Cardiac-Specific Overexpression of Ndufs1 Ameliorates Cardiac Dysfunction after Myocardial Infarction by Alleviating Mitochondrial Dysfunction and Apoptosis. Exp. Mol. Med. 2022, 54, 946–960. [Google Scholar] [CrossRef]
- Dong, T.; Zhang, Z.; Sun, L.; Zhang, W.; Zhu, Z.; Lin, L.; Yang, L.; Lv, A.; Liu, C.; Li, Q.; et al. Mic60 Is Essential to Maintain Mitochondrial Integrity and to Prevent Encephalomyopathy. Brain Pathol. 2023, 33, e13157. [Google Scholar] [CrossRef]
- Poppelreuther, M.; Lundsgaard, A.-M.; Mensberg, P.; Sjøberg, K.; Vilsbøll, T.; Kiens, B.; Füllekrug, J. Acyl-CoA Synthetase Expression in Human Skeletal Muscle Is Reduced in Obesity and Insulin Resistance. Physiol. Rep. 2023, 11, e15817. [Google Scholar] [CrossRef]
- Ling, R.; Chen, G.; Tang, X.; Liu, N.; Zhou, Y.; Chen, D. Acetyl-CoA Synthetase 2(ACSS2): A Review with a Focus on Metabolism and Tumor Development. Discov. Oncol. 2022, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Kusuhara, K.; Madsen, K.; Jensen, L.; Hellsten, Y.; Pilegaard, H. Calcium Signalling in the Regulation of PGC-1α, PDK4 and HKII mRNA Expression. Biol. Chem. 2007, 388, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Roberts-Wilson, T.K.; Reddy, R.N.; Bailey, J.L.; Zheng, B.; Ordas, R.; Gooch, J.L.; Price, S.R. Calcineurin Signaling and PGC-1α Expression Are Suppressed during Muscle Atrophy Due to Diabetes. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2010, 1803, 960–967. [Google Scholar] [CrossRef]
- Tiao, G.; Lieberman, M.; Fischer, J.E.; Hasselgren, P.O. Intracellular Regulation of Protein Degradation during Sepsis Is Different in Fast- and Slow-Twitch Muscle. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 1997, 272, R849–R856. [Google Scholar] [CrossRef]
- Sacheck, J.M.; Hyatt, J.-P.K.; Raffaello, A.; Jagoe, R.T.; Roy, R.R.; Edgerton, V.R.; Lecker, S.H.; Goldberg, A.L. Rapid Disuse and Denervation Atrophy Involve Transcriptional Changes Similar to Those of Muscle Wasting During Systemic Diseases. FASEB J. 2007, 21, 140–155. [Google Scholar] [CrossRef] [PubMed]
- Durham, W.J.; Aracena-Parks, P.; Long, C.; Rossi, A.E.; Goonasekera, S.A.; Boncompagni, S.; Galvan, D.L.; Gilman, C.P.; Baker, M.; Shirokova, N.; et al. RyR1 S-Nitrosylation Underlies Environmental Heat Stroke and Sudden Death in Y522S RyR1 Knock-in Mice. Cell 2008, 133, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-B.; Wei, Y.-H. AMPK-Mediated Increase of Glycolysis as an Adaptive Response to Oxidative Stress in Human Cells: Implication of the Cell Survival in Mitochondrial Diseases. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2012, 1822, 233–247. [Google Scholar] [CrossRef]
- de Sousa Neto, I.V.; Carvalho, M.M.; de Cassia Marqueti, R.; Almeida, J.A.; de Sousa Oliveira, K.; Barin, F.R.; Petriz, B.; de Araújo, H.S.S.; Franco, O.L.; Durigan, J.L.Q. Proteomic Changes in Skeletal Muscle of Aged Rats in Response to Resistance Training. Cell Biochem. Funct. 2020, 38, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, A.; Monti, D.M.; Scaloni, A.; De Simone, G.; Monti, S.M. Protective Role of Carbonic Anhydrases III and VII in Cellular Defense Mechanisms upon Redox Unbalance. Oxidative Med. Cell Longev. 2018, 2018, 2018306. [Google Scholar] [CrossRef]
- Brancaccio, P.; Lippi, G.; Maffulli, N. Biochemical Markers of Muscular Damage. Clin. Chem. Lab. Med. 2010, 48, 757–767. [Google Scholar] [CrossRef]
- Carter, N.D.; Heath, R.; Jeffery, S.; Jackson, M.J.; Newham, D.J.; Edwards, R.H.T. Carbonic Anhydrase III in Duchenne Muscular Dystrophy. Clin. Chim. Acta 1983, 133, 201–208. [Google Scholar] [CrossRef]
- Dowling, P.; Gargan, S.; Zweyer, M.; Sabir, H.; Swandulla, D.; Ohlendieck, K. Proteomic Profiling of Carbonic Anhydrase CA3 in Skeletal Muscle. Expert Rev. Proteom. 2021, 18, 1073–1086. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Vantaggiato, L.; Cameli, P.; Bergantini, L.; d’Alessandro, M.; Shaba, E.; Carleo, A.; Di Giuseppe, F.; Angelucci, S.; Sebastiani, G.; Dotta, F.; et al. Serum Proteomic Profile of Asthmatic Patients After Six Months of Benralizumab and Mepolizumab Treatment. Biomedicines 2022, 10, 761. [Google Scholar] [CrossRef]
- Oakley, B.R.; Kirsch, D.R.; Morris, N.R. A Simplified Ultrasensitive Silver Stain for Detecting Proteins in Polyacrylamide Gels. Anal. Biochem. 1980, 105, 361–363. [Google Scholar] [CrossRef]
- Gharahdaghi, F.; Weinberg, C.R.; Meagher, D.A.; Imai, B.S.; Mische, S.M. Mass Spectrometric Identification of Proteins from Silver-Stained Polyacrylamide Gel: A Method for the Removal of Silver Ions to Enhance Sensitivity. Electrophoresis 1999, 20, 601–605. [Google Scholar] [CrossRef]
- Landi, C.; Bargagli, E.; Carleo, A.; Refini, R.M.; Bennett, D.; Bianchi, L.; Cillis, G.; Prasse, A.; Bini, L.; Rottoli, P. Bronchoalveolar Lavage Proteomic Analysis in Pulmonary Fibrosis Associated with Systemic Sclerosis: S100A6 and 14-3-3ε as Potential Biomarkers. Rheumatology 2019, 58, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
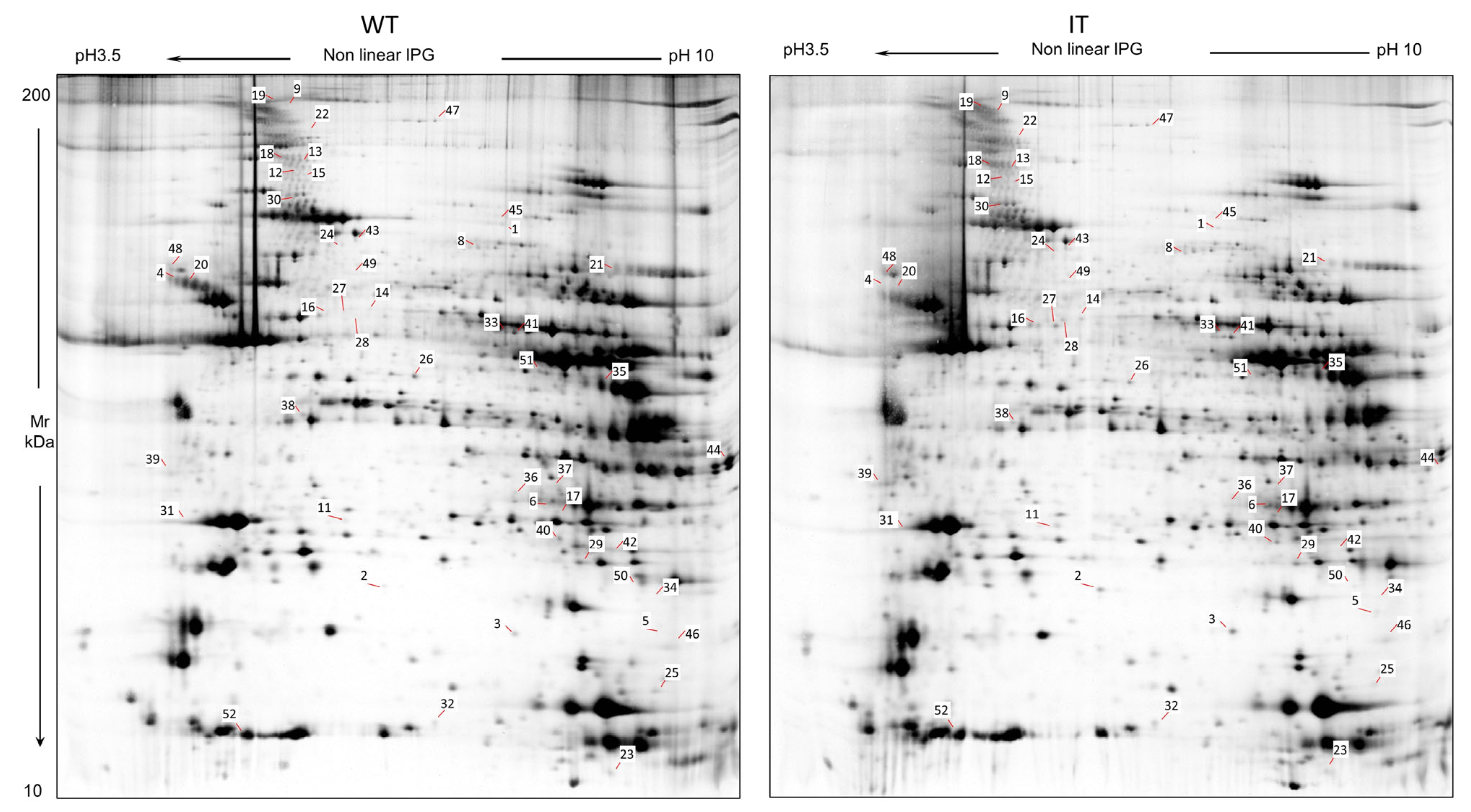
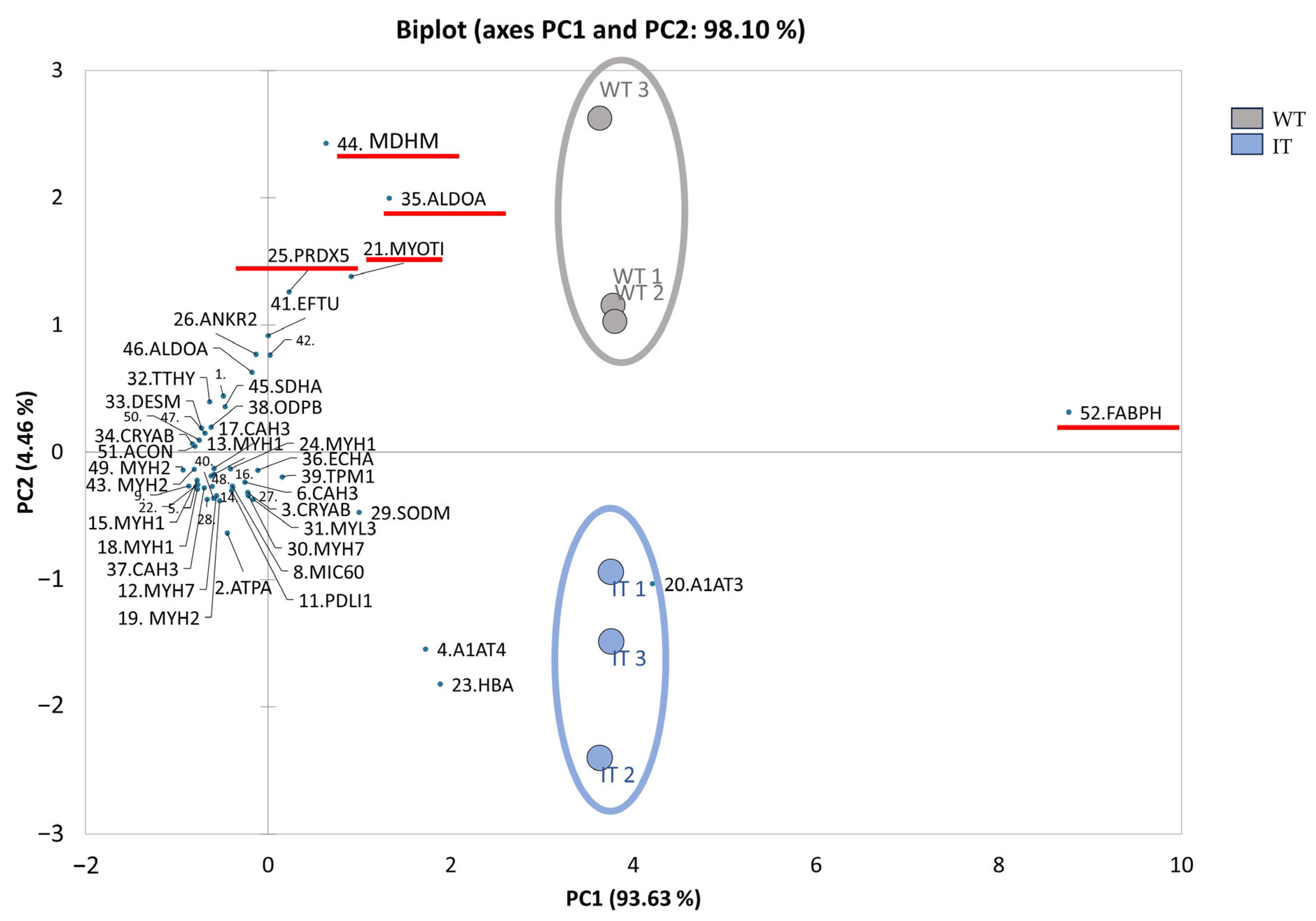
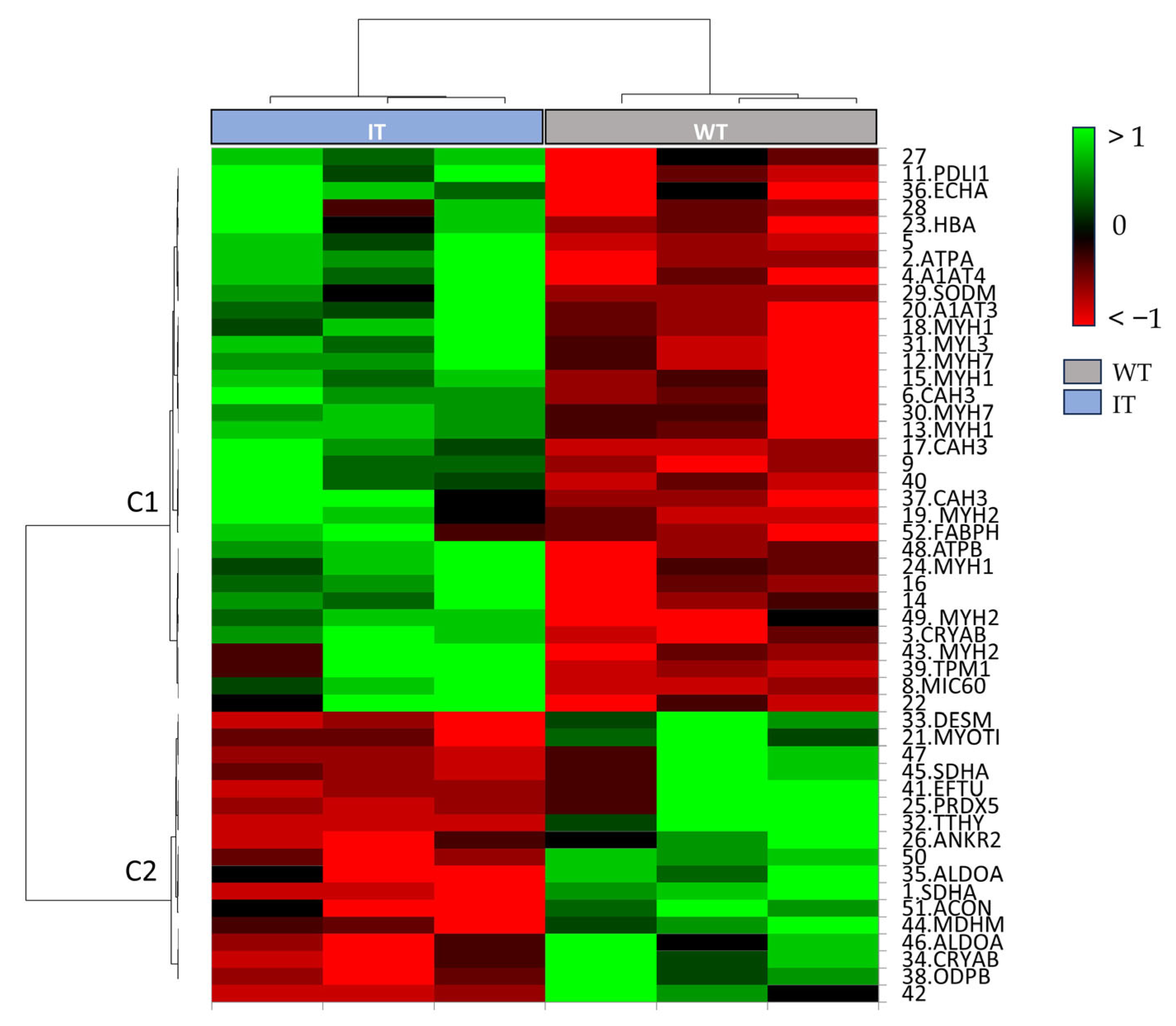
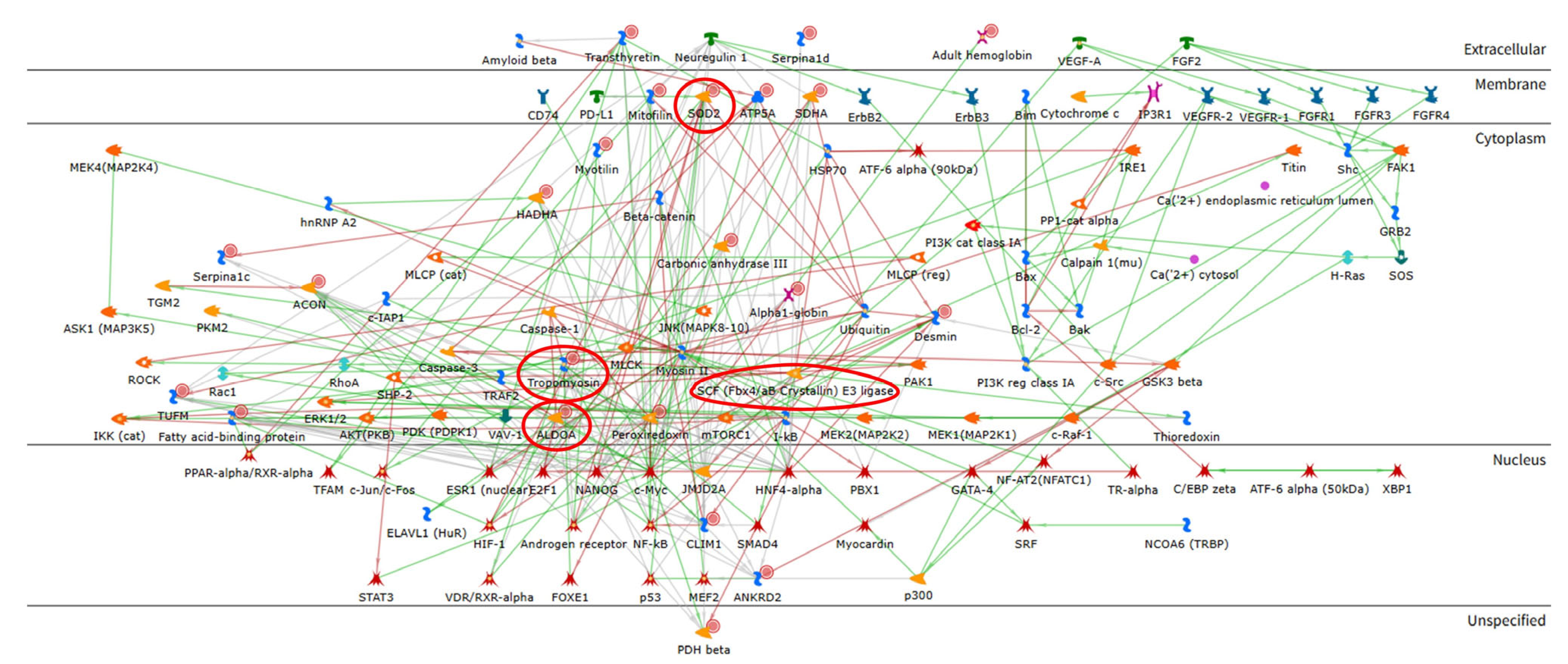

| Spot N. | Protein Name | Entry Name | Gene Name | AC | ANOVA Test | FC | Theoretical pI–MW | MASCOT Search Results | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Anova (p) | Adjusted (p) (FDR) | WT | IT | Score | No. of Matched Peptides | Sequence Coverage (%) | |||||||
| 1 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit. mitochondrial | SDHA_MOUSE | Sdha | Q8K2B3 | 0.0002 | 0.011 | 0.0228 | 0.0113 | 2.01 | 7.06–73,623 | 150 | 12/16 | 18 |
| 2 | ATP synthase subunit alpha. mitochondrial Fragment C | ATPA_MOUSE | Atp5f1a | Q03265 | 0.002 | 0.0361 | 0.0096 | 0.0349 | 3.64 | 9.22–59,830 | 65 | 6/10 | 14 |
| 3 | Alpha-crystallin B chain | CRYAB_MOUSE | Cryab | P23927 | 0.0035 | 0.0361 | 0.0191 | 0.0398 | 2.09 | 6.76–20,056 | 189 | 12/18 | 62 |
| 4 | Alpha-1-antitrypsin 1–4 | A1AT4_MOUSE | Serpina1d | Q00897 | 0.0042 | 0.0361 | 0.0619 | 0.1419 | 2.29 | 5.24–46,140 | 125 | 8/12 | 24 |
| 5 | 0.0045 | 0.0361 | 0.0051 | 0.0124 | 2.43 | ||||||||
| 6 | Carbonic anhydrase 3 | CAH3_MOUSE | Ca3 | P16015 | 0.0073 | 0.0387 | 0.0214 | 0.0342 | 1.6 | 6.89–29,633 | 204 | 12/17 | 69 |
| 8 | MICOS complex subunit Mic60 | MIC60_MOUSE | Immt | Q8CAQ8 | 0.0097 | 0.0387 | 0.0162 | 0.0299 | 1.85 | 6.18–84,247 | 98 | 7/8 | 15 |
| 9 | 0.0102 | 0.0387 | 0.0027 | 0.0098 | 3.68 | ||||||||
| 11 | PDZ and LIM domain protein 1 | PDLI1_MOUSE | Pdlim1 | O70400 | 0.0136 | 0.0387 | 0.0157 | 0.0299 | 1.9 | 6.38–36,208 | 101 | 9/17 | 34 |
| 12 | Myosin-7 Fragment C | MYH7_MOUSE | Myh7 | Q91Z83 | 0.0154 | 0.0387 | 0.0114 | 0.0227 | 2 | 5.59–223,539 | 124 | 23/38 | 13 |
| 13 | Myosin-1 Fragment C | MYH1_MOUSE | Myh1 | Q5SX40 | 0.0162 | 0.0387 | 0.0113 | 0.0176 | 1.55 | 5.60–224,116 | 148 | 21/29 | 11 |
| 14 | Actin. alpha skeletal muscle | ACTS_MOUSE | Acta1 | P68134 | 0.0179 | 0.0387 | 0.0095 | 0.0207 | 2.18 | 5.23–42,366 | 221 | 17/29 | 58 |
| 15 | Myosin-1 Fragment C | MYH1_MOUSE | Myh1 | Q5SX40 | 0.0191 | 0.0387 | 0.0065 | 0.0116 | 1.78 | 5.60–224,116 | 72 | 13/22 | 7 |
| 16 | Actin. alpha skeletal muscle | ACTS_MOUSE | Acta1 | P68134 | 0.0192 | 0.0387 | 0.0163 | 0.0299 | 1.83 | 5.23–42,366 | 75 | 6/14 | 19 |
| 17 | Carbonic anhydrase 3 | CAH3_MOUSE | Ca3 | P16015 | 0.0213 | 0.0401 | 0.0121 | 0.019 | 1.57 | 6.89–29,633 | 189 | 11/15 | 61 |
| 18 | Myosin-1 Fragment C | MYH1_MOUSE | Myh1 | Q5SX40 | 0.0276 | 0.047 | 0.0059 | 0.0131 | 2.21 | 5.60–224,116 | 210 | 26/32 | 14 |
| 19 | Myosin-2 | MYH2_MOUSE | Myh2 | G3UW82 | 0.0324 | 0.049 | 0.0104 | 0.0274 | 2.63 | 5.61–224,050 | 105 | 14/19 | 10 |
| 20 | Alpha-1-antitrypsin 1–3 | A1AT3_MOUSE | Serpina1c | Q00896 | 0.0359 | 0.049 | 0.145 | 0.2292 | 1.58 | 5.25–45,966 | 202 | 14/18 | 40 |
| 21 | Myotilin | MYOTI_MOUSE | Myot | Q9JIF9 | 0.0372 | 0.049 | 0.0765 | 0.0506 | 1.51 | 9.02–55,738 | 204 | 16/22 | 36 |
| 22 | 0.0382 | 0.049 | 0.006 | 0.0131 | 2.2 | ||||||||
| 23 | Hemoglobin subunit alpha | HBA_MOUSE | Hba | P01942 | 0.0383 | 0.049 | 0.0651 | 0.151 | 2.32 | 7.96–15,133 | 104 | 7/12 | 39 |
| 24 | Myosin-1 Fragment C | MYH1_MOUSE | Myh1 | Q5SX40 | 0.0384 | 0.049 | 0.017 | 0.0267 | 1.56 | 5.60–224,116 | 106 | 14/17 | 8 |
| 25 | Peroxiredoxin-5. mitochondrial | PRDX5_MOUSE | Prdx5 | P99029 | 0.0392 | 0.049 | 0.0526 | 0.0268 | 1.96 | 9.10–22226 | 116 | 6/7 | 40 |
| 26 | Ankyrin repeat domain–containing protein 2 | ANKR2_MOUSE | Ankrd2 | Q9WV06 | 0.0415 | 0.0496 | 0.0363 | 0.0208 | 1.75 | 5.95–36,855 | 273 | 18/23 | 67 |
| 27 | Actin. alpha skeletal muscle | ACTS_MOUSE | Acta1 | P68134 | 0.0444 | 0.0496 | 0.0196 | 0.0388 | 1.98 | 5.23–42,366 | 139 | 12/23 | 35 |
| 28 | Actin. alpha skeletal muscle | ACTS_MOUSE | Acta1 | P68134 | 0.0473 | 0.0497 | 0.0073 | 0.0195 | 2.66 | 5.23–42,366 | 109 | 9/15 | 28 |
| 29 | Superoxide dismutase [Mn]. mitochondrial | SODM_MOUSE | Sod2 | P09671 | 0.0497 | 0.0497 | 0.0543 | 0.0901 | 1.66 | 5.57–33,118 | 94 | 7/19 | 37 |
| 30 | Myosin-7 Fragment C | MYH7_MOUSE | Myh7 | Q91Z83 | 0.0497 | 0.0497 | 0.0223 | 0.0367 | 1.65 | 5.59–223,539 | 199 | 25/29 | 14 |
| 31 | Myosin light chain 3 | MYL3_MOUSE | Myl3 | P09542 | 0.0072 | 0.0387 | 0.0224 | 0.0403 | 1.8 | 5.03–22,521 | 154 | 10/14 | 51 |
| 32 | Transthyretin | TTHY_MOUSE | Ttr | P07309 | 0.0087 | 0.0387 | 0.0179 | 0.0058 | 3.07 | 5.77–15,880 | 96 | 5/8 | 43 |
| 33 | Desmin | DESM_MOUSE | Des | P31001 | 0.0134 | 0.0387 | 0.0131 | 0.0057 | 2.29 | 5.21–53,465 | 180 | 13/16 | 32 |
| 34 | Alpha–crystallin B chain | CRYAB_MOUSE | Cryab | P23927 | 0.0139 | 0.0387 | 0.0085 | 0.004 | 2.14 | 6.76–20,056 | 120 | 8/13 | 49 |
| 35 | Fructose–bisphosphate aldolase A | ALDOA_MOUSE | Aldoa | P05064 | 0.0173 | 0.0387 | 0.0945 | 0.0576 | 1.64 | 8.31–39787 | 120 | 9/11 | 30 |
| 36 | Trifunctional enzyme subunit alpha. mitochondrial | ECHA_MOUSE | Hadha | Q8BMS1 | 0.0168 | 0.0387 | 0.026 | 0.039 | 1.5 | 9.24–83,302 | 101 | 7/7 | 9 |
| 37 | Carbonic anhydrase 3 | CAH3_MOUSE | Ca3 | P16015 | 0.0194 | 0.0387 | 0.0074 | 0.0172 | 2.33 | 6.89–29,633 | 255 | 14/17 | 70 |
| 38 | Pyruvate dehydrogenase E1 component subunit beta. mitochondrial | ODPB_MOUSE | Pdhb | Q9D051 | 0.0216 | 0.0401 | 0.0163 | 0.0099 | 1.65 | 6.41–39,254 | 56 | 5/9 | 16 |
| 39 | Tropomyosin alpha-1 chain | TPM1_MOUSE | Tpm1 | P58771 | 0.028 | 0.047 | 0.0328 | 0.0516 | 1.57 | 4.69–32,718 | 112 | 9/14 | 19 |
| 40 | 0.0327 | 0.049 | 0.0092 | 0.0233 | 2.53 | ||||||||
| 41 | Elongation factor Tu. mitochondrial | EFTU_MOUSE | Tufm | Q8BFR5 | 0.0366 | 0.049 | 0.0421 | 0.0234 | 1.8 | 7.23–49,876 | 236 | 17/21 | 37 |
| 42 | 0.0396 | 0.049 | 0.0445 | 0.0231 | 1.92 | ||||||||
| 43 | Myosin-2 Fragment C | MYH2_MOUSE | Myh2 | G3UW82 | 0.0386 | 0.049 | 0.0062 | 0.0093 | 1.5 | 5.61–224,050 | 82 | 9/12 | 6 |
| 44 | Malate dehydrogenase. mitochondrial | MDHM_MOUSE | Mdh2 | P08249 | 0.042 | 0.0496 | 0.0756 | 0.0265 | 2.86 | 8.93–36,053 | 104 | 7/12 | 30 |
| 45 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit. mitochondrial | SDHA_MOUSE | Sdha | Q8K2B3 | 0.0429 | 0.0496 | 0.0224 | 0.0136 | 1.64 | 7.06–73,623 | 114 | 8/9 | 14 |
| 46 | Fructose-bisphosphate aldolase A | ALDOA_MOUSE | Aldoa | P05064 | 0.0457 | 0.0497 | 0.035 | 0.0196 | 1.78 | 8.31–39,787 | 155 | 8/10 | 24 |
| 47 | 0.0461 | 0.0497 | 0.0134 | 0.0085 | 1.57 | ||||||||
| 48 | ATP synthase F(1) complex catalytic subunit beta. mitochondrial | ATPB_MOUSE | Atp5f1b | P56480 | 0.0049 | 0.0361 | 0.0115 | 0.0199 | 1.73 | 5.19–56,265 | 210 | 14/17 | 39 |
| 49 | Myosin–2 Fragment C | MYH2_MOUSE | Myh2 | G3UW82 | 0.0139 | 0.0387 | 0.0026 | 0.0043 | 1.64 | 5.61– 224050 | 113 | 14/17 | 8 |
| 50 | 0.0032 | 0.0361 | 0.011 | 0.0067 | 1.64 | ||||||||
| 51 | Aconitate hydratase. mitochondrial Fragment C | ACON_MOUSE | Aco2 | Q99KI0 | 0.0274 | 0.047 | 0.009 | 0.0058 | 1.54 | 8.08–86151 | 145 | 12/17 | 21 |
| 52 | Fatty acid-binding protein. heart | FABPH_MOUSE | Fabp3 | P11404 | 0.0494 | 0.0497 | 0.2797 | 0.4205 | 1.5 | 6.11–14810 | 183 | 12/23 | 73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vantaggiato, L.; Shaba, E.; Fiore, F.; Rossi, D.; Sorrentino, V.; Bini, L.; Landi, C. Molecular Insights into Central Core Disease: Proteomic Signatures and Potential Therapeutic Biomarkers in RYR1 I4895T Mice. Int. J. Mol. Sci. 2025, 26, 11451. https://doi.org/10.3390/ijms262311451
Vantaggiato L, Shaba E, Fiore F, Rossi D, Sorrentino V, Bini L, Landi C. Molecular Insights into Central Core Disease: Proteomic Signatures and Potential Therapeutic Biomarkers in RYR1 I4895T Mice. International Journal of Molecular Sciences. 2025; 26(23):11451. https://doi.org/10.3390/ijms262311451
Chicago/Turabian StyleVantaggiato, Lorenza, Enxhi Shaba, Federica Fiore, Daniela Rossi, Vincenzo Sorrentino, Luca Bini, and Claudia Landi. 2025. "Molecular Insights into Central Core Disease: Proteomic Signatures and Potential Therapeutic Biomarkers in RYR1 I4895T Mice" International Journal of Molecular Sciences 26, no. 23: 11451. https://doi.org/10.3390/ijms262311451
APA StyleVantaggiato, L., Shaba, E., Fiore, F., Rossi, D., Sorrentino, V., Bini, L., & Landi, C. (2025). Molecular Insights into Central Core Disease: Proteomic Signatures and Potential Therapeutic Biomarkers in RYR1 I4895T Mice. International Journal of Molecular Sciences, 26(23), 11451. https://doi.org/10.3390/ijms262311451









