Identification of Novel TAT-I24-Related Peptides with Antiviral Activities
Abstract
1. Introduction
2. Results
2.1. Identification of Novel Sequences Conferring Antiviral Activity
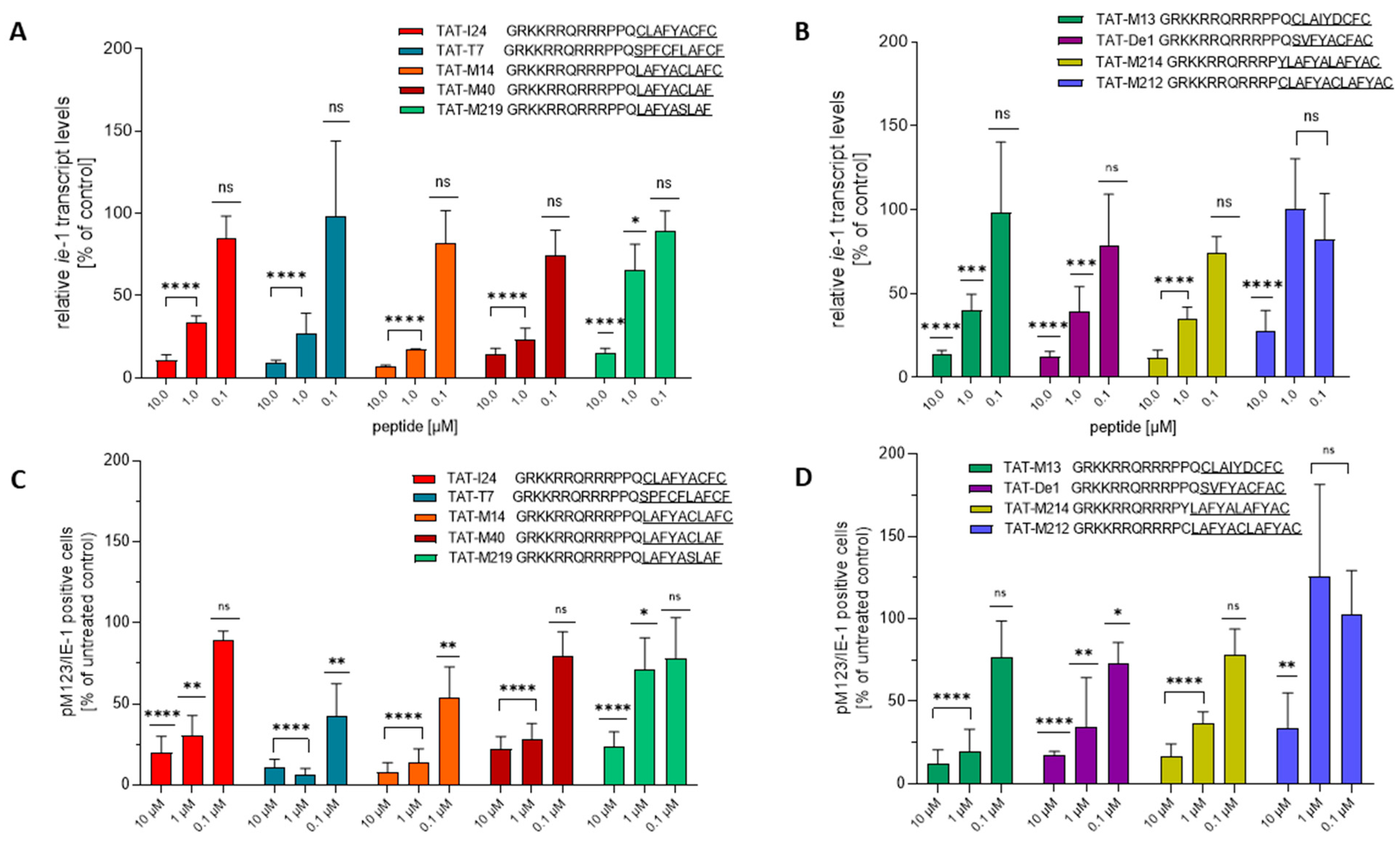
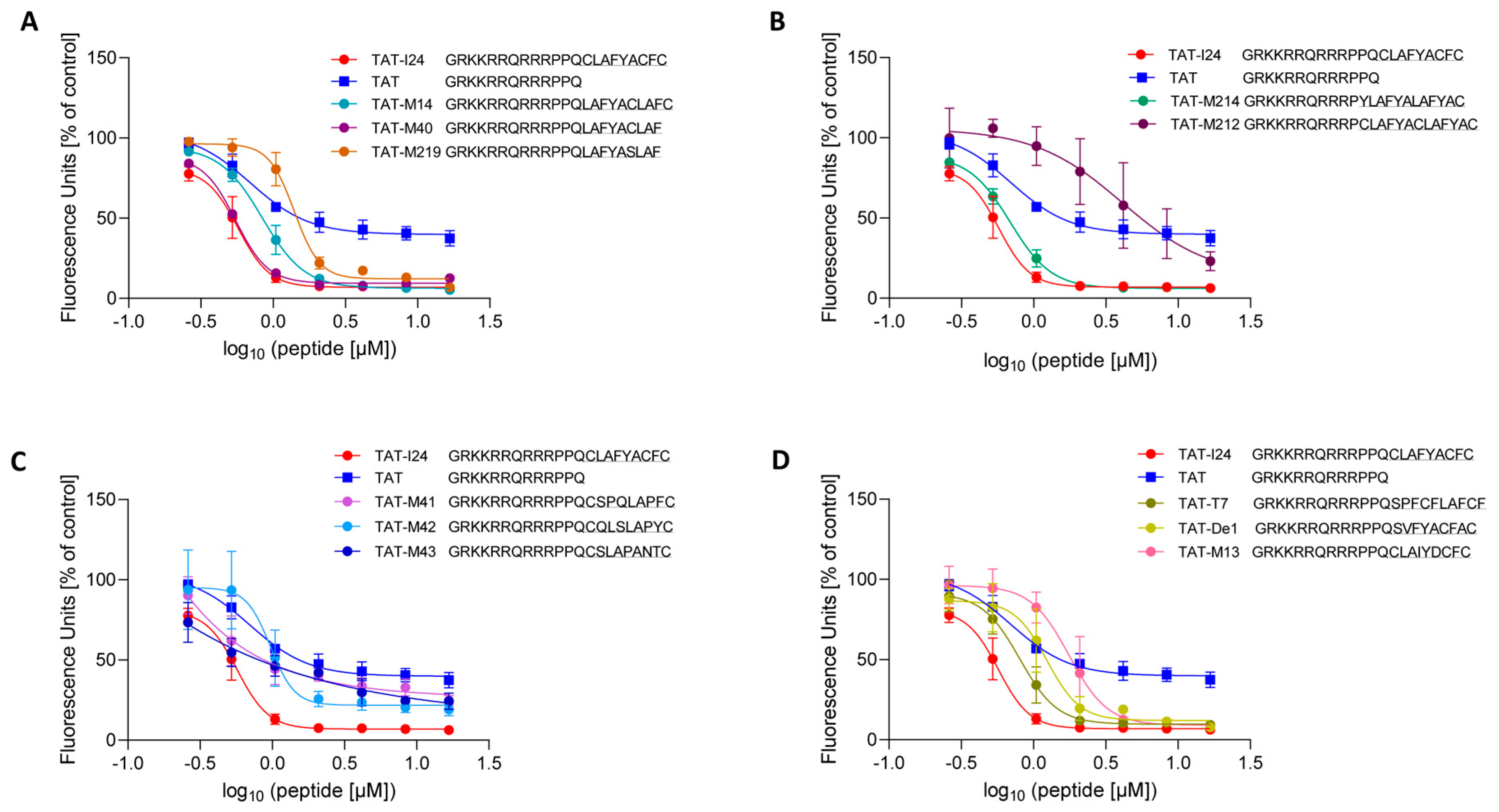
2.2. Antiviral Activities of TAT-Fusion Peptides
2.3. Effect of TAT-Fusion Peptides on m123/ie-1 Transcript Levels and pM123/IE1 Protein Expression in NIH/3T3 Cells
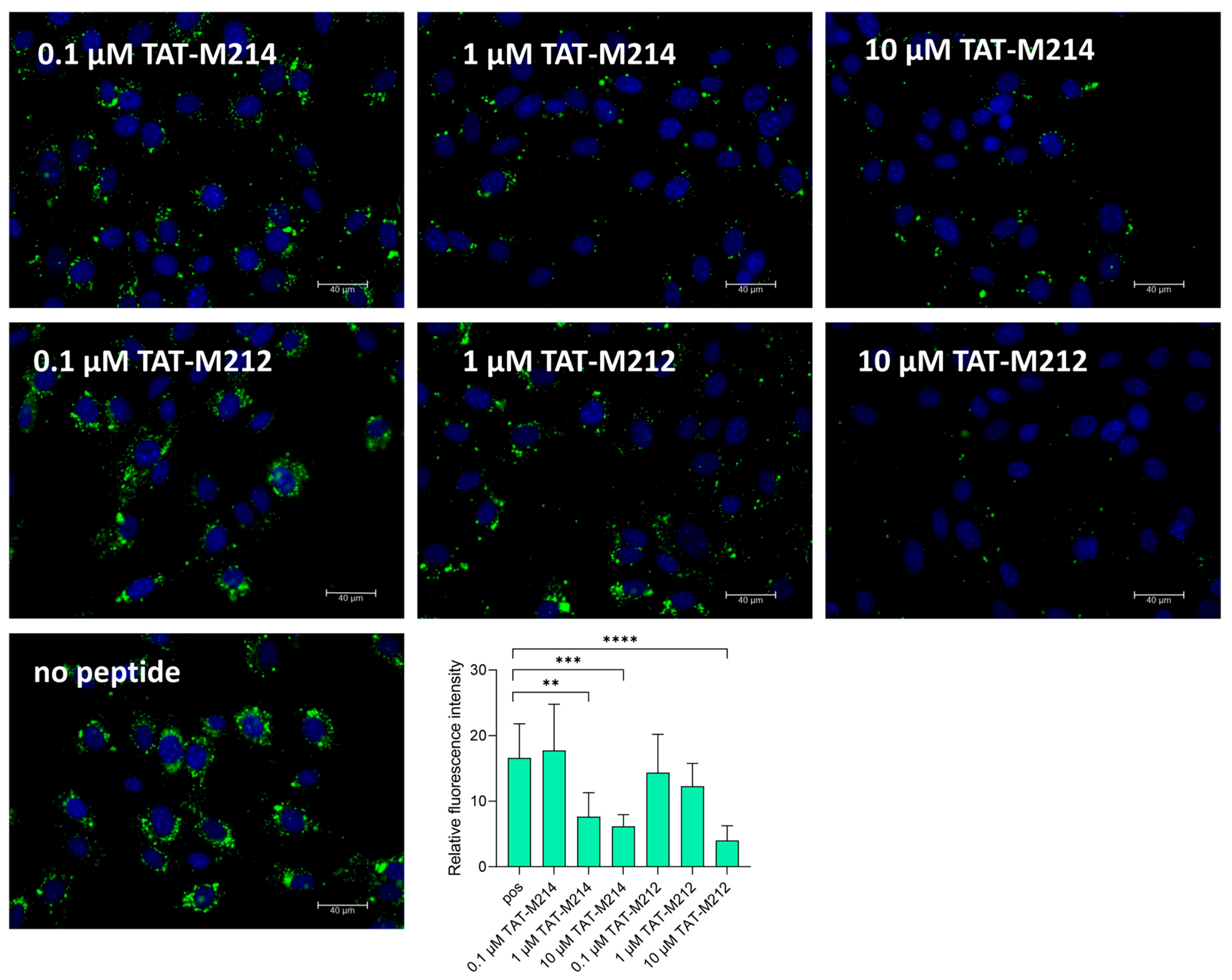
2.4. Effect of TAT-Fusion Peptides on Entry of a DiO-Labelled MCMV
2.5. Effect of TAT-Fusion Peptides on Red Blood Cells
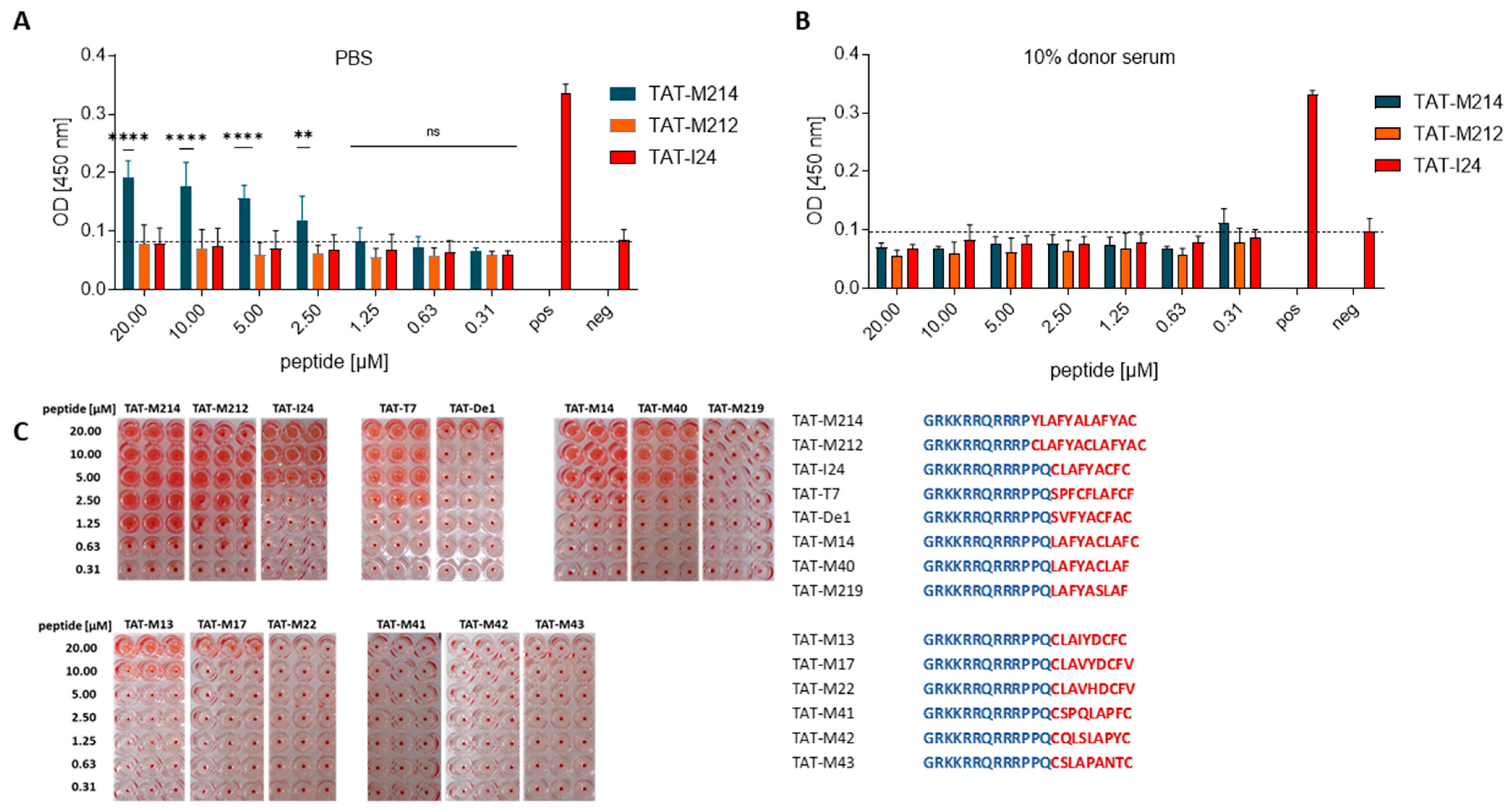
2.6. DNA-Binding of TAT-Fusion Peptides
2.7. Structural Features of the TAT-Fusion Peptides
3. Discussion
4. Materials and Methods
4.1. Peptides
4.2. Cell Culture
4.3. Viruses
4.3.1. Baculovirus
4.3.2. Mouse Cytomegalovirus
4.4. Cell Viability Assay
4.5. Staining and Microscopy
4.6. RNA Isolation, cDNA Synthesis and Quantitative Real-Time PCR
4.7. Isolation of Red Blood Cells and Haemolysis Assay
4.8. DNA-Binding Assay
4.9. Structure Prediction
4.10. Database Searches
4.11. Data Analysis
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMP | Antimicrobial peptide |
| RBC | Red blood cells |
| T7RNAP | T7 DNA-dependent RNA polymerase |
| PBS | phosphate-buffered saline |
References
- Ho, C.S.; Wong, C.T.H.; Aung, T.T.; Lakshminarayanan, R.; Mehta, J.S.; Rauz, S.; McNally, A.; Kintses, B.; Peacock, S.J.; de la Fuente-Nunez, C.; et al. Antimicrobial Resistance: A Concise Update. Lancet Microbe 2025, 6, 100947. [Google Scholar] [CrossRef]
- Han, J.J.; Song, H.A.; Pierson, S.L.; Shen-Gunther, J.; Xia, Q. Emerging Infectious Diseases Are Virulent Viruses—Are We Prepared? An Overview. Microorganisms 2023, 11, 2618. [Google Scholar] [CrossRef]
- Erdem Buckram, M.; Kesmen, Z. Antimicrobial Peptides (AMPs): A Promising Class of Antimicrobial Compounds. J. Appl. Microbiol. 2022, 132, 1573–1596. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Siman-Tov, G.; Hall, G.; Bhalla, N.; Narayanan, A. Human Antimicrobial Peptides as Therapeutics for Viral Infections. Viruses 2019, 11, 704. [Google Scholar] [CrossRef]
- Cresti, L.; Cappello, G.; Pini, A. Antimicrobial Peptides towards Clinical Application—A Long History to Be Concluded. Int. J. Mol. Sci. 2024, 25, 4870. [Google Scholar] [CrossRef]
- Vanzolini, T.; Bruschi, M.; Rinaldi, A.C.; Magnani, M.; Fraternale, A. Multitalented Synthetic Antimicrobial Peptides and Their Antibacterial, Antifungal and Antiviral Mechanisms. Int. J. Mol. Sci. 2022, 23, 545. [Google Scholar] [CrossRef]
- Al Tahan, M.A.; Al-Khattawi, A.; Russell, C. Oral Peptide Delivery Systems: Synergistic Approaches Using Polymers, Lipids, Nanotechnology, and Needle-Based Carriers. J. Drug Deliv. Sci. Technol. 2025, 112, 107205. [Google Scholar] [CrossRef]
- Odunitan, T.T.; Apanisile, B.T.; Afolabi, J.A.; Adeniwura, P.O.; Akinboade, M.W.; Ibrahim, N.O.; Alare, K.P.; Saibu, O.A.; Adeosun, O.A.; Opeyemi, H.S.; et al. Beyond Conventional Drug Design: Exploring the Broad-Spectrum Efficacy of Antimicrobial Peptides. Chem. Biodivers. 2025, 22, e202401349. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wang, Q.; Ren, K.; Xu, T.; Zhang, Z.; Xu, M.; Rao, Z.; Zhang, X. A Review of Antimicrobial Peptides: Structure, Mechanism of Action, and Molecular Optimization Strategies. Fermentation 2024, 10, 540. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Harris, F.; Phoenix, D.A.; Dennison, S.R. Antimicrobial Peptides from Frogs of the Glandirana Genus. Biologics 2024, 4, 444–507. [Google Scholar] [CrossRef]
- Klotman, M.E.; Chang, T.L. Defensins in Innate Antiviral Immunity. Nat. Rev. Immunol. 2006, 6, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Chou, Y.Y.; Chang, T.L. Defensins in Viral Infections. J. Innate Immun. 2009, 1, 413–420. [Google Scholar] [CrossRef]
- Wilson, S.S.; Wiens, M.E.; Smith, J.G. Antiviral Mechanisms of Human Defensins. J. Mol. Biol. 2013, 425, 4965–4980. [Google Scholar] [CrossRef] [PubMed]
- Barlow, P.G.; Findlay, E.G.; Currie, S.M.; Davidson, D.J. Antiviral Potential of Cathelicidins. Future Microbiol. 2014, 9, 55–73. [Google Scholar] [CrossRef]
- Ahmed, A.; Siman-Tov, G.; Keck, F.; Kortchak, S.; Bakovic, A.; Risner, K.; Lu, T.K.; Bhalla, N.; de la Fuente-Nunez, C.; Narayanan, A. Human Cathelicidin Peptide LL-37 as a Therapeutic Antiviral Targeting Venezuelan Equine Encephalitis Virus Infections. Antivir. Res. 2019, 164, 61–69. [Google Scholar] [CrossRef]
- Soofiyani, S.R.; Soltani, E.; Nejad-Khelejani, M.K.; Ghanbari, R.; Memar, M.Y. Hepcidin: A Potent Antimicrobial Peptide Involved in Iron Homeostasis. Gene Rep. 2024, 37, 102082. [Google Scholar] [CrossRef]
- Benfield, A.H.; Henriques, S.T. Mode-of-Action of Antimicrobial Peptides: Membrane Disruption vs. Intracellular Mechanisms. Front. Med. Technol. 2020, 2, 610997. [Google Scholar] [CrossRef]
- Nicolas, P. Multifunctional Host Defense Peptides: Intracellular-Targeting Antimicrobial Peptides. FEBS J. 2009, 276, 6483–6496. [Google Scholar] [CrossRef] [PubMed]
- Park, C.B.; Kim, H.S.; Kim, S.C. Mechanism of Action of the Antimicrobial Peptide Buforin II: Buforin II Kills Microorganisms by Penetrating the Cell Membrane and Inhibiting Cellular Functions. Biochem. Biophys. Res. Commun. 1998, 244, 253–257. [Google Scholar] [CrossRef]
- Park, S.-C.; Kim, J.-Y.; Jeong, C.; Yoo, S.; Hahm, K.-S.; Park, Y. A Plausible Mode of Action of Pseudin-2, an Antimicrobial Peptide from Pseudis Paradoxa. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2011, 1808, 171–182. [Google Scholar] [CrossRef]
- Hsu, C.-H.; Chen, C.; Jou, M.-L.; Lee, A.Y.-L.; Lin, Y.-C.; Yu, Y.-P.; Huang, W.-T.; Wu, S.-H. Structural and DNA-Binding Studies on the Bovine Antimicrobial Peptide, Indolicidin: Evidence for Multiple Conformations Involved in Binding to Membranes and DNA. Nucleic Acids Res. 2005, 33, 4053–4064. [Google Scholar] [CrossRef]
- Hayden, R.M.; Goldberg, G.K.; Ferguson, B.M.; Schoeneck, M.W.; Libardo, M.D.J.; Mayeux, S.E.; Shrestha, A.; Bogardus, K.A.; Hammer, J.; Pryshchep, S.; et al. Complementary Effects of Host Defense Peptides Piscidin 1 and Piscidin 3 on DNA and Lipid Membranes: Biophysical Insights into Contrasting Biological Activities. J. Phys. Chem. B 2015, 119, 15235–15246. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.C.J.; Shirkey, J.D.; Park, J.; Bisht, K.; Cowan, A.J. An Overview of Antiviral Peptides and Rational Biodesign Considerations. BioDesign Res. 2022, 2022, 9898241. [Google Scholar] [CrossRef]
- Ruzsics, Z.; Hoffmann, K.; Riedl, A.; Krawczyk, A.; Widera, M.; Sertznig, H.; Schipper, L.; Kapper-Falcone, V.; Debreczeny, M.; Ernst, W.; et al. A Novel, Broad-Acting Peptide Inhibitor of Double-Stranded DNA Virus Gene Expression and Replication. Front. Microbiol. 2020, 11, 2934. [Google Scholar] [CrossRef] [PubMed]
- Harant, H.; Höfinger, S.; Kricek, F.; Ruf, C.; Ruzsics, Z.; Hengel, H.; Lindley, I.J.D. The Peptide TAT-I24 with Antiviral Activity against DNA Viruses Binds Double-Stranded DNA with High Affinity. Biologics 2021, 1, 41–60. [Google Scholar] [CrossRef]
- Kicker, E.; Kouros, A.; Zatloukal, K.; Harant, H. The Virus Entry Pathway Determines Sensitivity to the Antiviral Peptide TAT-I24. Viruses 2025, 17, 458. [Google Scholar] [CrossRef] [PubMed]
- Green, M.; Loewenstein, P.M. Autonomous Functional Domains of Chemically Synthesized Human Immunodeficiency Virus Tat Trans-Activator Protein. Cell 1988, 55, 1179–1188. [Google Scholar] [CrossRef]
- Vivès, E.; Brodin, P.; Lebleu, B. A Truncated HIV-1 Tat Protein Basic Domain Rapidly Translocates through the Plasma Membrane and Accumulates in the Cell Nucleus. J. Biol. Chem. 1997, 272, 16010–16017. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Brister, J.R.; Chan, J.; Connor, R.; Feldgarden, M.; Fine, A.M.; Funk, K.; Hoffman, J.; et al. Database Resources of the National Center for Biotechnology Information in 2025. Nucleic Acids Res. 2025, 53, D20–D29. [Google Scholar] [CrossRef]
- Basic Local Alignment Search Tool; National Library of Medicine (US), National Center for Biotechnology Information: Bethesda, MD, USA, 1988. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 7 July 2025).
- Deng, S.-J.; Pearce, K.H.; Dixon, E.P.; Hartley, K.A.; Stanley, T.B.; Lobe, D.C.; Garvey, E.P.; Kost, T.A.; Petty, R.L.; Rocque, W.J.; et al. Identification of Peptides That Inhibit the DNA Binding, Trans-Activator, and DNA Replication Functions of the Human Papillomavirus Type 11 E2 Protein. J. Virol. 2004, 78, 2637–2641. [Google Scholar] [CrossRef]
- Ziu, T.; Sambur, E.; Ruzsics, Z.; Hengel, H.; Grabherr, R.; Höfinger, S.; Harant, H. In Vitro Profiling of the Antiviral Peptide TAT-I24. Int. J. Mol. Sci. 2024, 25, 10463. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, A.; Seelig, J. High Affinity of the Cell-Penetrating Peptide HIV-1 Tat-PTD for DNA. Biochemistry 2007, 46, 8138–8145. [Google Scholar] [CrossRef] [PubMed]
- Ignatovich, I.A.; Dizhe, E.B.; Pavlotskaya, A.V.; Akifiev, B.N.; Burov, S.V.; Orlov, S.V.; Perevozchikov, A.P. Complexes of Plasmid DNA with Basic Domain 47-57 of the HIV-1 Tat Protein Are Transferred to Mammalian Cells by Endocytosis-Mediated Pathways. J. Biol. Chem. 2003, 278, 42625–42636. [Google Scholar] [CrossRef]
- Rey, J.; Murail, S.; De Vries, S.; Derreumaux, P.; Tuffery, P. PEP-FOLD4: A PH-Dependent Force Field for Peptide Structure Prediction in Aqueous Solution. Nucleic Acids Res. 2023, 51, W432–W437. [Google Scholar] [CrossRef]
- Tufféry, P.; Derreumaux, P. A Refined PH-Dependent Coarse-Grained Model for Peptide Structure Prediction in Aqueous Solution. Front. Bioinform. 2023, 3, 1113928. [Google Scholar] [CrossRef]
- Binette, V.; Mousseau, N.; Tuffery, P. A Generalized Attraction-Repulsion Potential and Revisited Fragment Library Improves PEP-FOLD Peptide Structure Prediction. J. Chem. Theory Comput. 2022, 18, 2720–2736. [Google Scholar] [CrossRef]
- Lamiable, A.; Thevenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tuffery, P. PEP-FOLD3: Faster Denovo Structure Prediction for Linear Peptides in Solution and in Complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef]
- PEP-FOLD4. Available online: http://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD4 (accessed on 19 August 2025).
- Bittrich, S.; Segura, J.; Duarte, J.M.; Burley, S.K.; Rose, Y. RCSB Protein Data Bank: Exploring Protein 3D Similarities via Comprehensive Structural Alignments. Bioinformatics 2024, 40, btae370. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhatt, R.; Bhikadiya, C.; Bi, C.; Biester, A.; Biswas, P.; Bittrich, S.; Blaumann, S.; Brown, R.; Chao, H.; et al. Updated Resources for Exploring Experimentally-Determined PDB Structures and Computed Structure Models at the RCSB Protein Data Bank. Nucleic Acids Res. 2025, 53, D564–D574. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.V.; Salguero, C.; Khan, S.N.; Meagher, J.L.; Brown, W.C.; Humbert, N.; de Rocquigny, H.; Smith, J.L.; D’Souza, V.M. HIV-1 Tat Interactions with Cellular 7SK and Viral TAR RNAs Identifies Dual Structural Mimicry. Nat. Commun. 2018, 9, 4266. [Google Scholar] [CrossRef]
- Torres, M.D.T.; Melo, M.C.R.; Flowers, L.; Crescenzi, O.; Notomista, E.; de la Fuente-Nunez, C. Mining for Encrypted Peptide Antibiotics in the Human Proteome. Nat. Biomed. Eng. 2022, 6, 67–75, Correction in Nat. Biomed. Eng. 2022, 6, 1451. https://doi.org/10.1038/s41551-022-00967-2. [Google Scholar] [CrossRef]
- Santos, M.A.; Silva, F.L.; Lira, B.O.V.; Cardozo Fh, J.L.; Vasconcelos, A.G.; Araujo, A.R.; Murad, A.M.; Garay, A.V.; Freitas, S.M.; Leite, J.R.S.A.; et al. Probing Human Proteins for Short Encrypted Antimicrobial Peptides Reveals Hs10, a Peptide with Selective Activity for Gram-Negative Bacteria. Biochim. Biophys. Acta Gen. Subj. 2023, 1867, 130265. [Google Scholar] [CrossRef]
- Ramada, M.H.S.; Brand, G.D.; Abrão, F.Y.; Oliveira, M.; Filho, J.L.C.; Galbieri, R.; Gramacho, K.P.; Prates, M.V.; Bloch, C. Encrypted Antimicrobial Peptides from Plant Proteins. Sci. Rep. 2017, 7, 13263. [Google Scholar] [CrossRef]
- Brand, G.D.; Ramada, M.H.S.; Genaro-Mattos, T.C.; Bloch, C. Towards an Experimental Classification System for Membrane Active Peptides. Sci. Rep. 2018, 8, 1194. [Google Scholar] [CrossRef]
- Brand, G.D.; Ramada, M.H.S.; Manickchand, J.R.; Correa, R.; Ribeiro, D.J.S.; Santos, M.A.; Vasconcelos, A.G.; Abrão, F.Y.; Prates, M.V.; Murad, A.M.; et al. Intragenic Antimicrobial Peptides (IAPs) from Human Proteins with Potent Antimicrobial and Anti-Inflammatory Activity. PLoS ONE 2019, 14, e0220656. [Google Scholar] [CrossRef]
- Hughes, D.J.; Kipar, A.; Milligan, S.G.; Cunningham, C.; Sanders, M.; Quail, M.A.; Rajandream, M.A.; Efstathiou, S.; Bowden, R.J.; Chastel, C.; et al. Characterization of a Novel Wood Mouse Virus Related to Murid Herpesvirus 4. J. Gen. Virol. 2010, 91, 867–879. [Google Scholar] [CrossRef]
- Virgin Iv, H.W.; Latreille, P.; Wamsley, P.; Hallsworth, K.; Weck, K.E.; Dal Canto, A.J.; Speck, S.H. Complete Sequence and Genomic Analysis of Murine Gammaherpesvirus 68. J. Virol. 1997, 71, 5894–5904. [Google Scholar] [CrossRef]
- Yokoyama, N.; Fujii, K.; Hirata, M.; Tamai, K.; Kiyono, T.; Kuzushima, K.; Nishiyama, Y.; Fujita, M.; Tsurumi, T. Assembly of the Epstein–Barr Virus BBLF4, BSLF1 and BBLF2/3 Proteins and Their Interactive Properties. J. Gen. Virol. 1999, 80, 2879–2887. [Google Scholar] [CrossRef]
- Fujii, K.; Yokoyama, N.; Kiyono, T.; Kuzushima, K.; Homma, M.; Nishiyama, Y.; Fujita, M.; Tsurumi, T. The Epstein-Barr Virus Pol Catalytic Subunit Physically Interacts with the BBLF4-BSLF1-BBLF2/3 Complex. J. Virol. 2000, 74, 2550–2557. [Google Scholar] [CrossRef]
- Devasahayam Arokia Balaya, R.; Kanekar, S.; Kumar, S.; Kandasamy, R.K. Role of DEAD/DEAH-Box Helicases in Immunity, Infection and Cancers. Cell Commun. Signal. 2025, 23, 292. [Google Scholar] [CrossRef]
- Redder, P.; Hausmann, S.; Khemici, V.; Yasrebi, H.; Linder, P. Bacterial Versatility Requires DEAD-Box RNA Helicases. FEMS Microbiol. Rev. 2015, 39, 392–412. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Yip, C.C.Y.; Huang, Y.; Tsoi, H.-W.; Chan, K.-H.; Yuen, K.-Y. Comparative Analysis of 22 Coronavirus HKU1 Genomes Reveals a Novel Genotype and Evidence of Natural Recombination in Coronavirus HKU1. J. Virol. 2006, 80, 7136–7145. [Google Scholar] [CrossRef]
- Tahir, M. Coronavirus Genomic Nsp14-ExoN, Structure, Role, Mechanism, and Potential Application as a Drug Target. J. Med. Virol. 2021, 93, 4258–4264. [Google Scholar] [CrossRef]
- Ogando, N.S.; Zevenhoven-Dobbe, J.C.; Van Der Meer, Y.; Bredenbeek, P.J.; Posthuma, C.C.; Snijder, E.J. The Enzymatic Activity of the Nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020, 94, e01246-20. [Google Scholar] [CrossRef]
- Fisher, S.Z. SARS CoV-2 Nsp10 in Complex with the ExoN Domain from Nsp14. Available online: https://www.rcsb.org/structure/9FW2 (accessed on 19 July 2025).
- Cheetham, G.M.T.; Jeruzalmi, D.; Steitz, T.A. Structural Basis for Initiation of Transcription from an RNA Polymerase-Promoter Complex. Nature 1999, 399, 80–83, Correction in Nature 1999, 400, 89. https://doi.org/10.1038/21929. [Google Scholar] [CrossRef] [PubMed]
- Jeruzalmi, D.; Steitz, T.A. Structure of T7 RNA Polymerase Complexed to the Transcriptional Inhibitor T7 Lysozyme. EMBO J. 1998, 17, 4101–4113. [Google Scholar] [CrossRef]
- Cheetham, G.M.T.; Steitz, T.A. Structure of a Transcribing T7 RNA Polymerase Initiation Complex. Science 1999, 286, 2305–2309. [Google Scholar] [CrossRef]
- Bultmann, H.; Teuton, J.; Brandt, C.R. Addition of a C-Terminal Cysteine Improves the Anti-Herpes Simplex Virus Activity of a Peptide Containing the Human Immunodeficiency Virus Type 1 TAT Protein Transduction Domain. Antimicrob. Agents Chemother. 2007, 51, 1596–1607. [Google Scholar] [CrossRef]
- Tang, W.H.; Wang, C.F.; Liao, Y. Di Fetal Bovine Serum Albumin Inhibits Antimicrobial Peptide Activity and Binds Drug Only in Complex with A1-Antitrypsin. Sci. Rep. 2021, 11, 1267. [Google Scholar] [CrossRef]
- Dennis, M.S.; Zhang, M.; Gloria Meng, Y.; Kadkhodayan, M.; Kirchhofer, D.; Combs, D.; Damico, L.A. Albumin Binding as a General Strategy for Improving the Pharmacokinetics of Proteins. J. Biol. Chem. 2002, 277, 35035–35043. [Google Scholar] [CrossRef]
- Bagińska, K.; Makowska, J.; Wiczk, W.; Kasprzykowski, F.; Chmurzyński, L. Conformational Studies of Alanine-Rich Peptide Using CD and FTIR Spectroscopy. J. Pept. Sci. 2008, 14, 283–289. [Google Scholar] [CrossRef]
- Sani, M.A.; Rajput, S.; Keizer, D.W.; Separovic, F. NMR Techniques for Investigating Antimicrobial Peptides in Model Membranes and Bacterial Cells. Methods 2024, 224, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Asselah, T.; Lampertico, P.; Aleman, S.; Bourlière, M.; Streinu-Cercel, A.; Bogomolov, P.; Morozov, V.; Stepanova, T.; Lazar, S.; Manuilov, D.; et al. Bulevirtide Monotherapy Is Safe and Well Tolerated in Chronic Hepatitis Delta: An Integrated Safety Analysis of Bulevirtide Clinical Trials at Week 48. Liver Int. 2025, 45, e16174. [Google Scholar] [CrossRef] [PubMed]
- Harant, H. Selective Inhibition of Murine Cytomegalovirus Viral Gene Expression by the Antiviral Peptide TAT-I24. Int. J. Mol. Sci. 2022, 23, 7246. [Google Scholar] [CrossRef]
- Trilling, M.; Le, V.T.K.; Fiedler, M.; Zimmermann, A.; Bleifuß, E.; Hengel, H. Identification of DNA-Damage DNA-Binding Protein 1 as a Conditional Essential Factor for Cytomegalovirus Replication in Interferon-γ-Stimulated Cells. PLoS Pathog. 2011, 7, e1002069. [Google Scholar] [CrossRef] [PubMed]
- Sæbø, I.P.; Bjørås, M.; Franzyk, H.; Helgesen, E.; Booth, J.A. Optimization of the Hemolysis Assay for the Assessment of Cytotoxicity. Int. J. Mol. Sci. 2023, 24, 2914. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
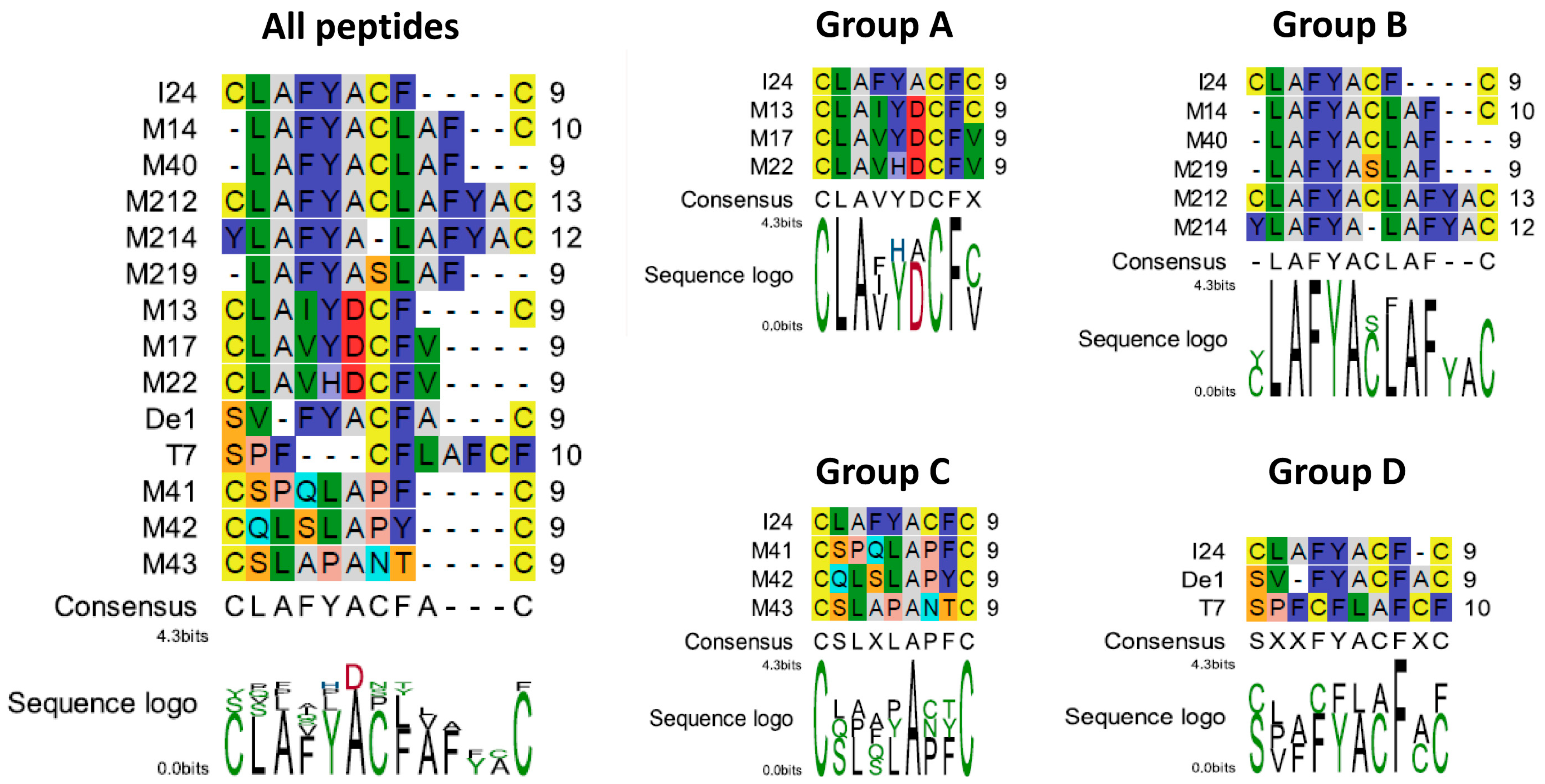


| Synthetic Peptide * | ||||||
|---|---|---|---|---|---|---|
| Sequence BLAST Hit | ID | Species | Amino Acids | Protein Name/Function | Name | Sequence |
| LAFYACF | YP_010085930.1 | Wood mouse herpesvirus (WMHV) | 279–285 | Helicase-primase primase subunit | I24 | CLAFYACFC |
| NP_044893.1 | Murid gammaherpesvirus 4 (MuHV4) | 279–285 | ||||
| CLAIYDCFC | ABD75543.1 | Human coronavirus HKU1 | 6325–6333 (ORF1ab) 276–284 (Nsp14) | Nonstructural protein 14/Guanine-N7 methyltransferase | M13 | CLAIYDCFC |
| CLAVYDCFV | UUT43647.1 | Betacoronavirus sp. | 6317–6325 (ORF1ab) 276–284 (Nsp14) | Nonstructural protein 14/Guanine-N7 methyltransferase | M17 | CLAVYDCFV |
| CLAVHDCFV | UPB65199.1 | SARS-CoV-2 | 6204–6212 (ORF1ab) 275–283 (Nsp14) | Nonstructural protein 14/Guanine-N7 methyltransferase | M22 | CLAVHDCFV |
| SVFYACFSC | MDI3365062.1 | Pantoea sp. V108_6 | 567–575 | DEAD/DEAH box helicase/RNA-binding proteins with ATPase activity | De1 ** | SVFYACFAC |
| SVFYACFA | WP_008932954 | Ectothiorhodospira sp. PHS-1 | 423–430 | ATP-binding protein | ||
| LAFYACLAFC | WP_240521350.1 | Pseudomonas | 140–149 | Hypothetical protein | M14 | LAFYACLAFC |
| SPQLAPFC | MFV1968217.1 | Pirellulaceae bacterium | 145–152 | SDR family oxido-reductase/NAD- or NADP-dependent oxidoreductases | M41 | CSPQLAPFC |
| QLSLAPYC | XP_060411117.1 | Colletotrichum navitas | 212–219 | LY79DRAFT_318317/uncharacterized protein | M42 | CQLSLAPYC |
| CSLAPANTC | MFA5187732.1 | Patescibacteria group bacterium | 819–827, 1015–1023 | Repeated sequence motifs in Ig-like domain-containing protein | M43 | CSLAPANTC |
| CSLAPTNTC | 524–532, 722–730, 1113–1121 | |||||
| SPFCFLAFCF | P00573 | bacteriophage T7 | 507–516 | T7 RNA Polymerase/DNA-directed RNA polymerase | T7 | SPFCFLAFCF |
| LAFYALAFY | MBK9337318.1 | Lewinellaceae bacterium | 128–136 | IPM98_12370/hypothetical protein | M214 | YLAFYALAFYAC |
| LAFFACLAFY | XP_013439911.1 | Eimeria necatrix | 43–52 | RER1 protein/Retrieval of endoplasmic reticulum membrane proteins from the early Golgi compartment | M212 *** | CLAFYACLAFYAC |
| Name | Sequence | Group | Baculovirus/ HEK293 IC50 [µM] | SI [HEK293] | MCMV/ NIH3T3 IC50 [µM] |
|---|---|---|---|---|---|
| TAT-I24 | GRKKRRQRRRPPQCLAFYACFC | 0.016 | 2480 [33] | 0.809 | |
| TAT-T7 | GRKKRRQRRRPPQSPFCFLAFCF | D | 0.017 | 1299 | 1.126 |
| TAT-M40 | GRKKRRQRRRPPQLAFYACLAF | B | 0.021 | 2585 | 1.846 |
| TAT-M214 | GRKKRRQRRRPYLAFYALAFYAC | B | 0.025 | 2306 | 3.668 |
| TAT-M13 | GRKKRRQRRRPPQCLAIYDCFC | A | 0.031 | 1731 | 1.219 |
| TAT-M14 | GRKKRRQRRRPPQLAFYACLAFC | B | 0.034 | 766 | 2.569 |
| TAT-De1 | GRKKRRQRRRPPQSVFYACFAC | D | 0.037 | 517 | 1.611 |
| TAT-M219 | GRKKRRQRRRPPQLAFYASLAF | B | 0.096 | 24 | 7.379 |
| TAT-M17 | GRKKRRQRRRPPQCLAVYDCFV | A | 0.102 | 792 | 5.849 |
| TAT-M22 | GRKKRRQRRRPPQCLAVHDCFV | A | 0.245 | 138 | 11.78 |
| TAT-M43 | GRKKRRQRRRPPQCSLAPANTC | C | 0.306 | n.d. | n.d. |
| TAT-M42 | GRKKRRQRRRPPQCQLSLAPYC | C | 0.378 | n.d. | n.d. |
| TAT-M41 | GRKKRRQRRRPPQCSPQLAPFC | C | 0.434 | n.d. | n.d. |
| TAT-M212 | GRKKRRQRRRPCLAFYACLAFYAC | B | 0.619 | >10,000 | 18.190 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harant, H.; Höfinger, S.; Grabherr, R.; Ruzsics, Z.; Hengel, H. Identification of Novel TAT-I24-Related Peptides with Antiviral Activities. Int. J. Mol. Sci. 2025, 26, 11433. https://doi.org/10.3390/ijms262311433
Harant H, Höfinger S, Grabherr R, Ruzsics Z, Hengel H. Identification of Novel TAT-I24-Related Peptides with Antiviral Activities. International Journal of Molecular Sciences. 2025; 26(23):11433. https://doi.org/10.3390/ijms262311433
Chicago/Turabian StyleHarant, Hanna, Siegfried Höfinger, Reingard Grabherr, Zsolt Ruzsics, and Hartmut Hengel. 2025. "Identification of Novel TAT-I24-Related Peptides with Antiviral Activities" International Journal of Molecular Sciences 26, no. 23: 11433. https://doi.org/10.3390/ijms262311433
APA StyleHarant, H., Höfinger, S., Grabherr, R., Ruzsics, Z., & Hengel, H. (2025). Identification of Novel TAT-I24-Related Peptides with Antiviral Activities. International Journal of Molecular Sciences, 26(23), 11433. https://doi.org/10.3390/ijms262311433







