Single-Cell Sequencing Reveals Novel Tumor Populations and Their Interplay with the Immune Microenvironment in a Pleomorphic Rhabdomyosarcoma
Abstract
1. Introduction
2. Results
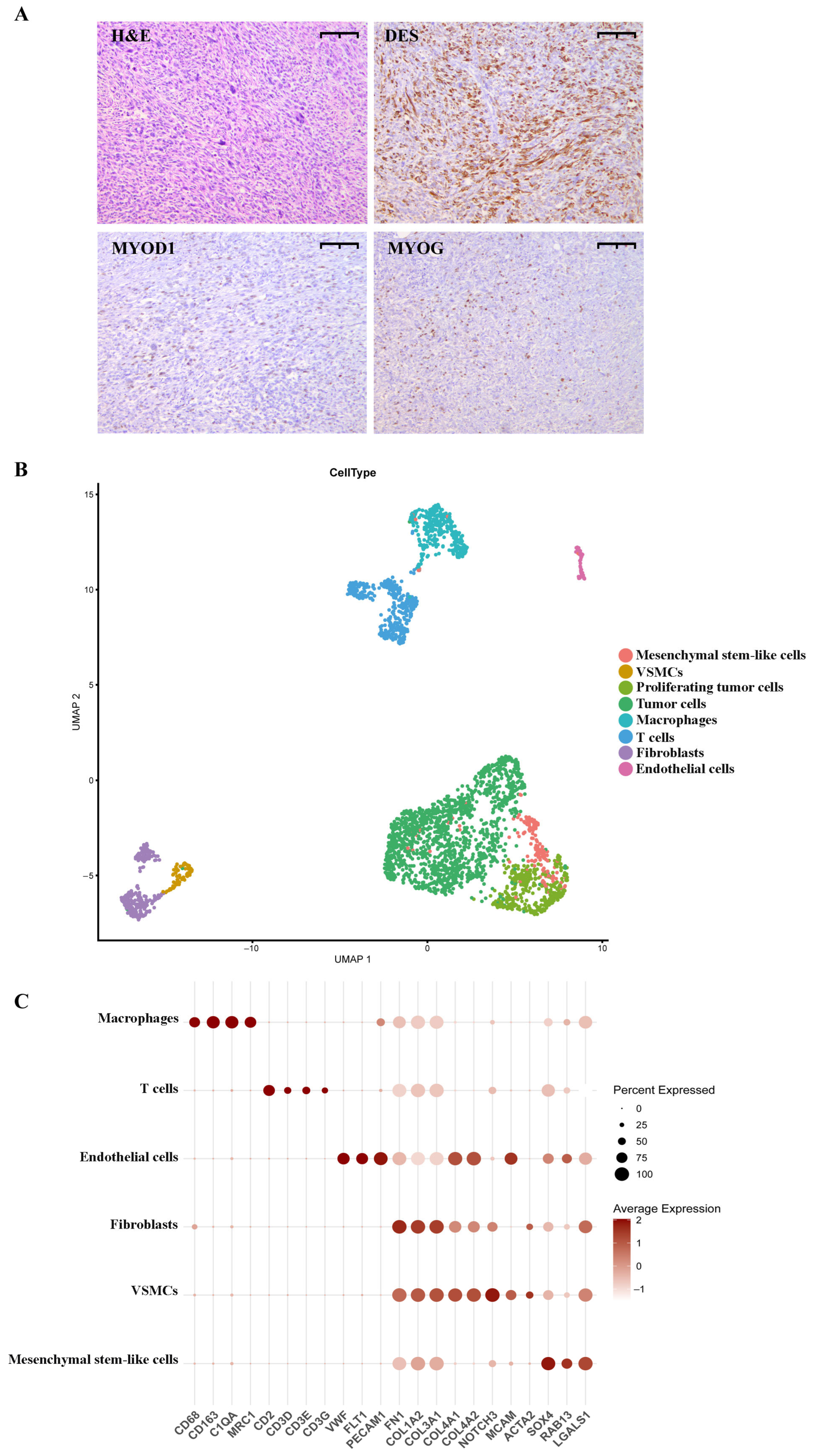
2.1. pRMS Case Presentation
2.2. Annotation of pRMS Cell Clusters
2.3. pRMS Tumor Cell Heterogeneity
2.3.1. Cell Ploidy and Differential Gene Expression
2.3.2. Functional Characterization
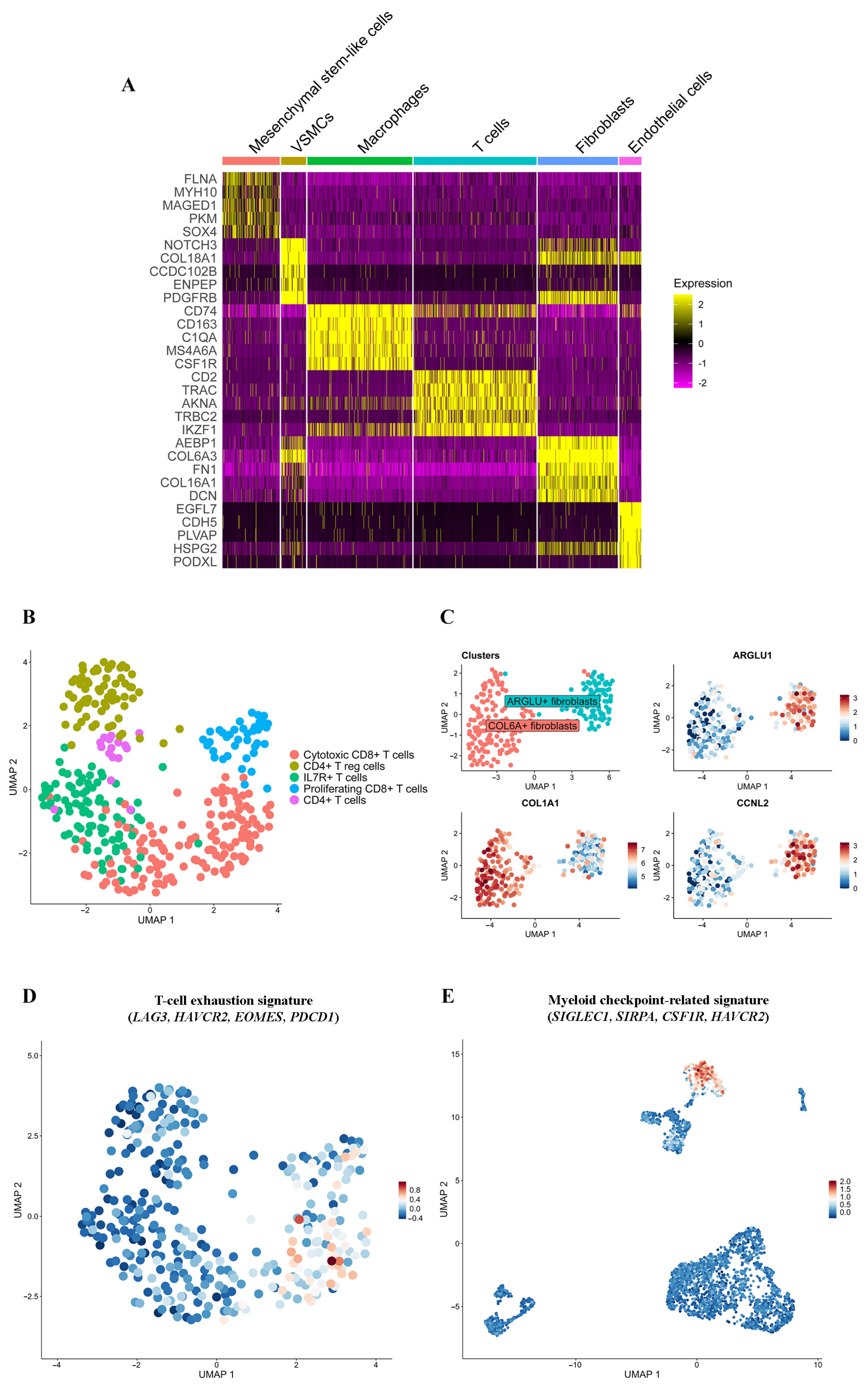
2.4. pRMS Tumor Microenvironment
2.4.1. T Lymphocytes
2.4.2. Macrophages
2.4.3. Fibroblasts
2.4.4. Mesenchymal Stem-like Cells
2.4.5. Endothelial Cells and VSMCs
2.5. Tumor–Immune Cell Communications in pRMS
3. Discussion
4. Methods
4.1. Consent and Approval
4.2. Histological Analysis
4.3. Immunohistochemistry
4.4. Sample Preparation for Single-Cell RNA Sequencing
4.5. Single-Cell RNA Library Construction and Sequencing
4.6. Single-Cell RNA Sequencing Quality Control and Data Processing
4.7. Analysis of Differentially Expressed Genes
4.8. Copy Number Karyotyping of Aneuploid Cells
4.9. Cell–Cell Interaction Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fan, R.; Parham, D.M.; Wang, L.L. An Integrative Morphologic and Molecular Approach for Diagnosis and Subclassification of Rhabdomyosarcoma. Arch. Pathol. Lab. Med. 2022, 146, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Agaram, N.P. Evolving classification of rhabdomyosarcoma. Histopathology 2022, 80, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Furlong, M.A.; Mentzel, T.; Fanburg-Smith, J.C. Pleomorphic rhabdomyosarcoma in adults: A clinicopathologic study of 38 cases with emphasis on morphologic variants and recent skeletal muscle-specific markers. Mod. Pathol. 2001, 14, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, S.D.; Pissaloux, D.; Crombe, A.; Coindre, J.M.; Le Loarer, F. Pleomorphic Sarcomas: The State of the Art. Surg. Pathol. Clin. 2019, 12, 63–105. [Google Scholar] [CrossRef]
- Kobayashi, H.; Okajima, K.; Zhang, L.; Hirai, T.; Ishibashi, Y.; Tsuda, Y.; Ikegami, M.; Kawai, A.; Tanaka, S. Prognostic factors and treatment outcomes in patients with pleomorphic rhabdomyosarcoma: A population-based cohort study. Jpn. J. Clin. Oncol. 2024, 54, 471–478. [Google Scholar] [CrossRef]
- Noujaim, J.; Thway, K.; Jones, R.L.; Miah, A.; Khabra, K.; Langer, R.; Kasper, B.; Judson, I.; Benson, C.; Kollar, A. Adult Pleomorphic Rhabdomyosarcoma: A Multicentre Retrospective Study. Anticancer Res. 2015, 35, 6213–6217. [Google Scholar]
- Liu, J.; Liu, P.; Gong, F.; Tian, Y.; Zhao, X. Case Report: A PD-L1-Positive Patient with Pleomorphic Rhabdomyosarcoma Achieving an Impressive Response to Immunotherapy. Front. Immunol. 2022, 13, 815598. [Google Scholar] [CrossRef]
- Torabi, A.; Amaya, C.N.; Wians, F.H., Jr.; Bryan, B.A. PD-1 and PD-L1 expression in bone and soft tissue sarcomas. Pathology 2017, 49, 506–513. [Google Scholar] [CrossRef]
- Kournoutas, I.; Monga, V.; Davick, J.; Rieth, J. Objective Responses in Metastatic Pleomorphic Rhabdomyosarcoma Treated with Combination of Doxorubicin and Pembrolizumab: A Case Series. Case Rep. Oncol. 2024, 17, 344–351. [Google Scholar] [CrossRef]
- Beird, H.C.; Wu, C.C.; Nakazawa, M.; Ingram, D.; Daniele, J.R.; Lazcano, R.; Little, L.; Davies, C.; Daw, N.C.; Wani, K.; et al. Complete loss of TP53 and RB1 is associated with complex genome and low immune infiltrate in pleomorphic rhabdomyosarcoma. HGG Adv. 2023, 4, 100224. [Google Scholar] [CrossRef]
- Saoud, C.; Dermawan, J.K.; Sharma, A.E.; Tap, W.; Wexler, L.H.; Antonescu, C.R. Genomic profiling of pleomorphic rhabdomyosarcoma reveals a genomic signature distinct from that of embryonal rhabdomyosarcoma. Genes Chromosomes Cancer 2024, 63, 23238. [Google Scholar] [CrossRef]
- Chelsky, Z.L.; Paulson, V.A.; Chen, E.Y. Molecular analysis of 10 pleomorphic rhabdomyosarcomas reveals potential prognostic markers and druggable targets. Genes Chromosomes Cancer 2022, 61, 138–147. [Google Scholar] [CrossRef]
- Poumeaud, F.; Valentin, T.; Fares, N.; Segier, B.; Watson, S.; Verret, B.; Tlemsani, C.; Penel, N.; Lejeune, S.; Firmin, N.; et al. Sarcomas developed in patients with Lynch Syndrome are enriched in pleomorphic soft-tissue sarcomas and are sensitive to immunotherapy. Eur. J. Cancer 2025, 216, 115196. [Google Scholar] [CrossRef]
- Machado, E.R.; van de Vlekkert, D.; Sheppard, H.S.; Perry, S.; Downing, S.M.; Laxton, J.; Ashmun, R.; Finkelstein, D.B.; Neale, G.A.; Hu, H.; et al. Haploinsufficiency of the lysosomal sialidase NEU1 results in a model of pleomorphic rhabdomyosarcoma in mice. Commun. Biol. 2022, 5, 992. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chen, D.; Wang, B.; Chai, W.; Yan, M.; Chen, Y.; Zhan, Y.; Yang, R.; Zhou, E.; Dai, S.; et al. Single-cell landscape of undifferentiated pleomorphic sarcoma. Oncogene 2024, 43, 1353–1368. [Google Scholar] [CrossRef]
- Yuan, L.L.; Chen, Z.; Qin, J.; Qin, C.J.; Bian, J.; Dong, R.F.; Yuan, T.B.; Xu, Y.T.; Kong, L.Y.; Xia, Y.Z. Single-cell sequencing reveals the landscape of the tumor microenvironment in a skeletal undifferentiated pleomorphic sarcoma patient. Front. Immunol. 2022, 13, 1019870. [Google Scholar] [CrossRef]
- Knouse, K.A.; Wu, J.; Whittaker, C.A.; Amon, A. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc. Natl. Acad. Sci. USA 2014, 111, 13409–13414. [Google Scholar] [CrossRef] [PubMed]
- Prakash, J.; Shaked, Y. The Interplay between Extracellular Matrix Remodeling and Cancer Therapeutics. Cancer Discov. 2024, 14, 1375–1388. [Google Scholar] [CrossRef]
- Huang, H.; Mu, Y.; Li, S. The biological function of Serpinb9 and Serpinb9-based therapy. Front. Immunol. 2024, 15, 1422113. [Google Scholar] [CrossRef] [PubMed]
- Andreatta, M.; Corria-Osorio, J.; Muller, S.; Cubas, R.; Coukos, G.; Carmona, S.J. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat. Commun. 2021, 12, 2965. [Google Scholar] [CrossRef]
- Ambhore, N.S.; Kalidhindi, R.S.R.; Pabelick, C.M.; Hawse, J.R.; Prakash, Y.S.; Sathish, V. Differential estrogen-receptor activation regulates extracellular matrix deposition in human airway smooth muscle remodeling via NF-kappaB pathway. FASEB J. 2019, 33, 13935–13950. [Google Scholar] [CrossRef]
- Li, D.Y.; Xiong, X.Z. ICOS(+) Tregs: A Functional Subset of Tregs in Immune Diseases. Front. Immunol. 2020, 11, 2104. [Google Scholar] [CrossRef]
- Abbas, H.A.; Hao, D.; Tomczak, K.; Barrodia, P.; Im, J.S.; Reville, P.K.; Alaniz, Z.; Wang, W.; Wang, R.; Wang, F.; et al. Single cell T cell landscape and T cell receptor repertoire profiling of AML in context of PD-1 blockade therapy. Nat. Commun. 2021, 12, 6071. [Google Scholar] [CrossRef]
- Li, J.; He, Y.; Hao, J.; Ni, L.; Dong, C. High Levels of Eomes Promote Exhaustion of Anti-tumor CD8(+) T Cells. Front. Immunol. 2018, 9, 2981. [Google Scholar] [CrossRef]
- Gonzalez, N.M.; Zou, D.; Gu, A.; Chen, W. Schrödinger’s T Cells: Molecular Insights Into Stemness and Exhaustion. Front. Immunol. 2021, 12, 725618. [Google Scholar] [CrossRef]
- Levring, T.B.; Kongsbak-Wismann, M.; Rode, A.K.O.; Al-Jaberi, F.A.H.; Lopez, D.V.; Met, O.; Woetmann, A.; Bonefeld, C.M.; Odum, N.; Geisler, C. Tumor necrosis factor induces rapid down-regulation of TXNIP in human T cells. Sci. Rep. 2019, 9, 16725. [Google Scholar] [CrossRef] [PubMed]
- Petkau, G.; Mitchell, T.J.; Chakraborty, K.; Bell, S.E.; D Angeli, V.; Matheson, L.; Turner, D.J.; Saveliev, A.; Gizlenci, O.; Salerno, F.; et al. The timing of differentiation and potency of CD8 effector function is set by RNA binding proteins. Nat. Commun. 2022, 13, 2274. [Google Scholar] [CrossRef]
- Wei, J.; Yu, W.; Chen, J.; Huang, G.; Zhang, L.; Chen, Z.; Hu, M.; Gong, X.; Du, H. Single-cell and spatial analyses reveal the association between gene expression of glutamine synthetase with the immunosuppressive phenotype of APOE+CTSZ+TAM in cancers. Mol. Oncol. 2023, 17, 611–628. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Yang, T.; Liang, H.; Deng, M. Myeloid checkpoints for cancer immunotherapy. Chin. J. Cancer Res. 2022, 34, 460–482. [Google Scholar] [CrossRef]
- Zhao, L.; Cheng, S.; Fan, L.; Zhang, B.; Xu, S. TIM-3: An update on immunotherapy. Int. Immunopharmacol. 2021, 99, 107933. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, Y.; Weeraratna, A.T. Fibroblasts in cancer: Unity in heterogeneity. Cell 2023, 186, 1580–1609. [Google Scholar] [CrossRef]
- Liu, J.; Qiu, J.; Zhang, Z.; Zhou, L.; Li, Y.; Ding, D.; Zhang, Y.; Zou, D.; Wang, D.; Zhou, Q.; et al. SOX4 maintains the stemness of cancer cells via transcriptionally enhancing HDAC1 revealed by comparative proteomics study. Cell Biosci. 2021, 11, 23. [Google Scholar] [CrossRef]
- Wang, H.; Xu, H.; Chen, W.; Cheng, M.; Zou, L.; Yang, Q.; Chan, C.B.; Zhu, H.; Chen, C.; Nie, J.; et al. Rab13 Sustains Breast Cancer Stem Cells by Supporting Tumor-Stroma Cross-talk. Cancer Res. 2022, 82, 2124–2140. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Cai, J.; Hou, Y.; Huang, Z.; Wang, Z. Role of EZH2 in cancer stem cells: From biological insight to a therapeutic target. Oncotarget 2017, 8, 37974–37990. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Niu, X.; Cheng, R. The pan-cancer landscape of crosstalk between leukocyte transendothelial migration-related genes and tumor microenvironment relevant to prognosis and immunotherapy response. Transl. Cancer Res. 2024, 13, 5247–5264. [Google Scholar] [CrossRef]
- He, F.; Xu, J.; Zeng, F.; Wang, B.; Yang, Y.; Xu, J.; Sun, X.; Ren, T.; Tang, X. Integrative analysis of Ewing’s sarcoma reveals that the MIF-CD74 axis is a target for immunotherapy. Cell Commun. Signal. 2025, 23, 23. [Google Scholar] [CrossRef]
- Yin, T.; Guoping, W.; Ma, Z.; Wang, L.; Chen, R.; Xiang, K.; Tan, L.; Wang, Y.; Chong, M.; Liang, Y.; et al. Amyloid-β precursor protein promotes tumor growth by establishing an immune-exclusive tumor microenvironment. bioRxiv 2025. [Google Scholar] [CrossRef]
- Ganguly, D.; Schmidt, M.O.; Coleman, M.; Ngo, T.C.; Sorrelle, N.; Dominguez, A.T.A.; Murimwa, G.Z.; Toombs, J.E.; Lewis, C.; Fang, Y.V.; et al. Pleiotrophin drives a prometastatic immune niche in breast cancer. J. Exp. Med. 2023, 220, e20220610. [Google Scholar] [CrossRef]
- Wang, R.; Feng, W.; Wang, H.; Wang, L.; Yang, X.; Yang, F.; Zhang, Y.; Liu, X.; Zhang, D.; Ren, Q.; et al. Blocking migration of regulatory T cells to leukemic hematopoietic microenvironment delays disease progression in mouse leukemia model. Cancer Lett. 2020, 469, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Rhost, S.; Hughes, E.; Harrison, H.; Rafnsdottir, S.; Jacobsson, H.; Gregersson, P.; Magnusson, Y.; Fitzpatrick, P.; Andersson, D.; Berger, K.; et al. Sortilin inhibition limits secretion-induced progranulin-dependent breast cancer progression and cancer stem cell expansion. Breast Cancer Res. 2018, 20, 137. [Google Scholar] [CrossRef]
- Danielli, S.G.; Porpiglia, E.; De Micheli, A.J.; Navarro, N.; Zellinger, M.J.; Bechtold, I.; Kisele, S.; Volken, L.; Marques, J.G.; Kasper, S.; et al. Single-cell profiling of alveolar rhabdomyosarcoma reveals RAS pathway inhibitors as cell-fate hijackers with therapeutic relevance. Sci. Adv. 2023, 9, eade9238. [Google Scholar] [CrossRef] [PubMed]
- Jakos, T.; Pislar, A.; Jewett, A.; Kos, J. Cysteine Cathepsins in Tumor-Associated Immune Cells. Front. Immunol. 2019, 10, 2037. [Google Scholar] [CrossRef]
- Tessaro, F.H.G.; Ko, E.Y.; De Simone, M.; Piras, R.; Broz, M.T.; Goodridge, H.S.; Balzer, B.; Shiao, S.L.; Guarnerio, J. Single-cell RNA-seq of a soft-tissue sarcoma model reveals the critical role of tumor-expressed MIF in shaping macrophage heterogeneity. Cell Rep. 2022, 39, 110977. [Google Scholar] [CrossRef]
- Luo, W.R. Rethinking cancer. Zhonghua Zhong Liu Za Zhi 2025, 47, 463–467. [Google Scholar]
- Hao, Y.; Stuart, T.; Kowalski, M.H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C.; et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2024, 42, 293–304. [Google Scholar] [CrossRef]
- Xi, N.M.; Li, J.J. Protocol for executing and benchmarking eight computational doublet-detection methods in single-cell RNA sequencing data analysis. STAR Protoc. 2021, 2, 100699. [Google Scholar] [CrossRef]
- McGinnis, C.S.; Murrow, L.M.; Gartner, Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019, 8, 329–337.e324. [Google Scholar] [CrossRef]
- Bais, A.S.; Kostka, D. scds: Computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics 2020, 36, 1150–1158. [Google Scholar] [CrossRef]
- Germain, P.L.; Lun, A.; Garcia Meixide, C.; Macnair, W.; Robinson, M.D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Research 2021, 10, 979. [Google Scholar] [CrossRef] [PubMed]
- Wolock, S.L.; Lopez, R.; Klein, A.M. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 2019, 8, 281–291.e289. [Google Scholar] [CrossRef] [PubMed]
- Finak, G.; McDavid, A.; Yajima, M.; Deng, J.; Gersuk, V.; Shalek, A.K.; Slichter, C.K.; Miller, H.W.; McElrath, M.J.; Prlic, M.; et al. MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015, 16, 278. [Google Scholar] [CrossRef] [PubMed]
- De Falco, A.; Caruso, F.; Su, X.D.; Iavarone, A.; Ceccarelli, M. A variational algorithm to detect the clonal copy number substructure of tumors from scRNA-seq data. Nat. Commun. 2023, 14, 1074. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Plikus, M.V.; Nie, Q. CellChat for systematic analysis of cell-cell communication from single-cell transcriptomics. Nat. Protoc. 2025, 20, 180–219. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopantseva, E.E.; Ikonnikov, A.V.; Menyailo, M.E.; Fetisov, T.I.; Korobeynikova, A.A.; Kirsanov, K.I.; Tararykova, A.A.; Bokhyan, B.Y.; Kozlov, N.A.; Yakubovskaya, M.G.; et al. Single-Cell Sequencing Reveals Novel Tumor Populations and Their Interplay with the Immune Microenvironment in a Pleomorphic Rhabdomyosarcoma. Int. J. Mol. Sci. 2025, 26, 11420. https://doi.org/10.3390/ijms262311420
Kopantseva EE, Ikonnikov AV, Menyailo ME, Fetisov TI, Korobeynikova AA, Kirsanov KI, Tararykova AA, Bokhyan BY, Kozlov NA, Yakubovskaya MG, et al. Single-Cell Sequencing Reveals Novel Tumor Populations and Their Interplay with the Immune Microenvironment in a Pleomorphic Rhabdomyosarcoma. International Journal of Molecular Sciences. 2025; 26(23):11420. https://doi.org/10.3390/ijms262311420
Chicago/Turabian StyleKopantseva, Elena E., Alexander V. Ikonnikov, Maxim E. Menyailo, Timur I. Fetisov, Anastasia A. Korobeynikova, Kirill I. Kirsanov, Anastasia A. Tararykova, Beniamin Yu. Bokhyan, Nikolay A. Kozlov, Marianna G. Yakubovskaya, and et al. 2025. "Single-Cell Sequencing Reveals Novel Tumor Populations and Their Interplay with the Immune Microenvironment in a Pleomorphic Rhabdomyosarcoma" International Journal of Molecular Sciences 26, no. 23: 11420. https://doi.org/10.3390/ijms262311420
APA StyleKopantseva, E. E., Ikonnikov, A. V., Menyailo, M. E., Fetisov, T. I., Korobeynikova, A. A., Kirsanov, K. I., Tararykova, A. A., Bokhyan, B. Y., Kozlov, N. A., Yakubovskaya, M. G., & Denisov, E. V. (2025). Single-Cell Sequencing Reveals Novel Tumor Populations and Their Interplay with the Immune Microenvironment in a Pleomorphic Rhabdomyosarcoma. International Journal of Molecular Sciences, 26(23), 11420. https://doi.org/10.3390/ijms262311420




