Interaction Between Enhancers and Promoters in Chicken Genome
Abstract
1. Introduction
2. Results
2.1. Similarity Between Erythrocyte and Fibroblast TADs and the Consensus TAD Set
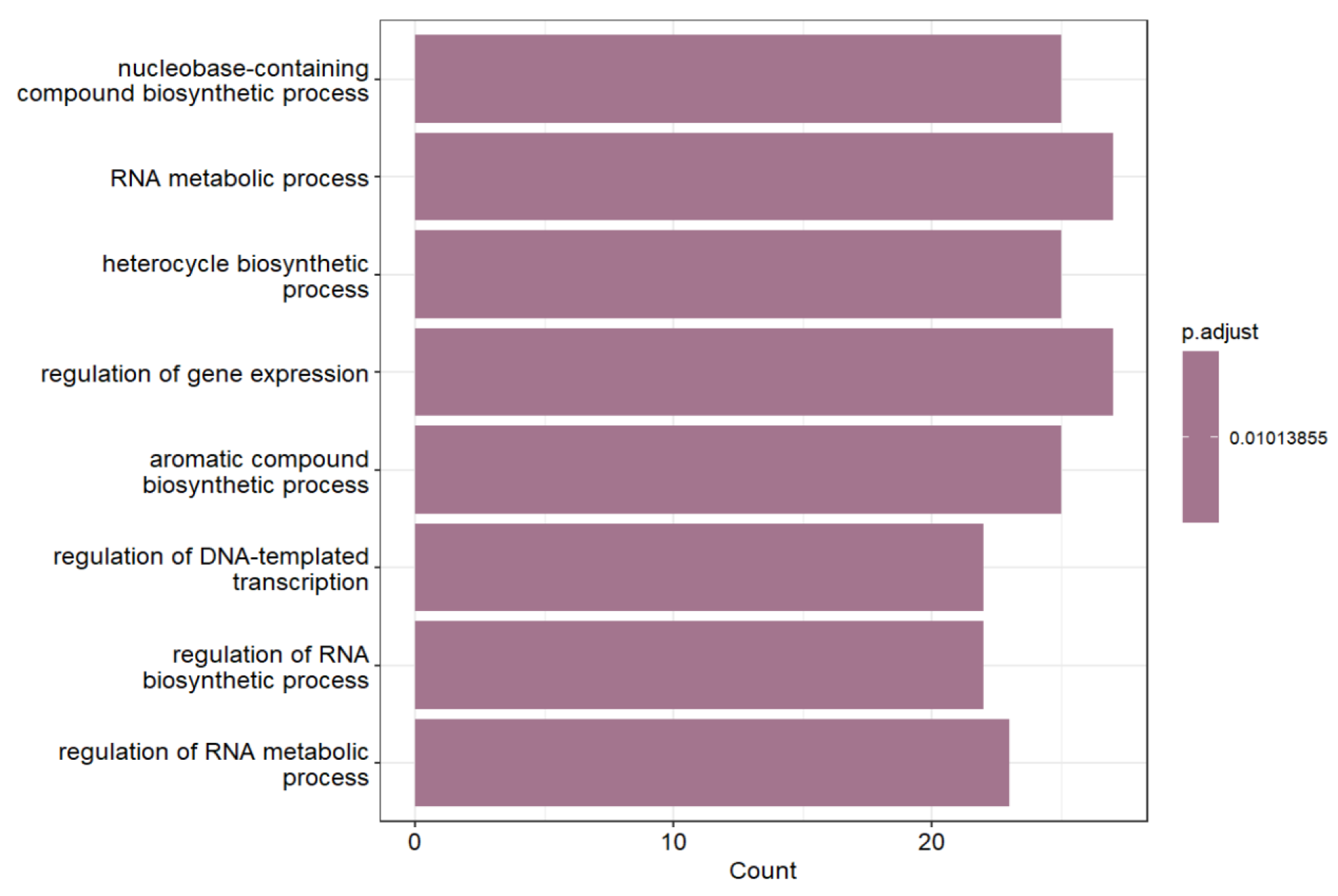
2.2. Enhancer-Mediated Epigenetic Regulation of Promoter Activity
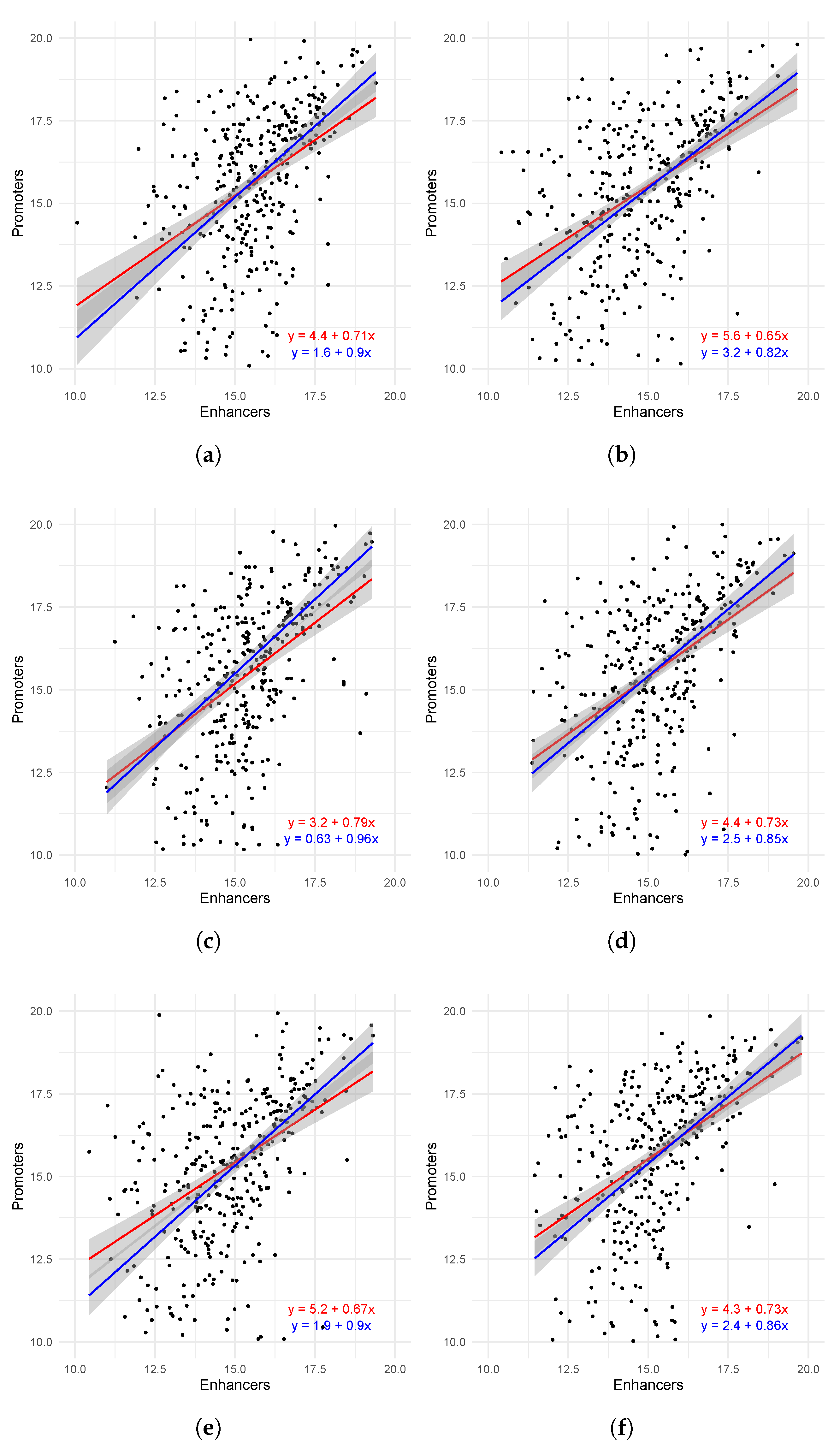
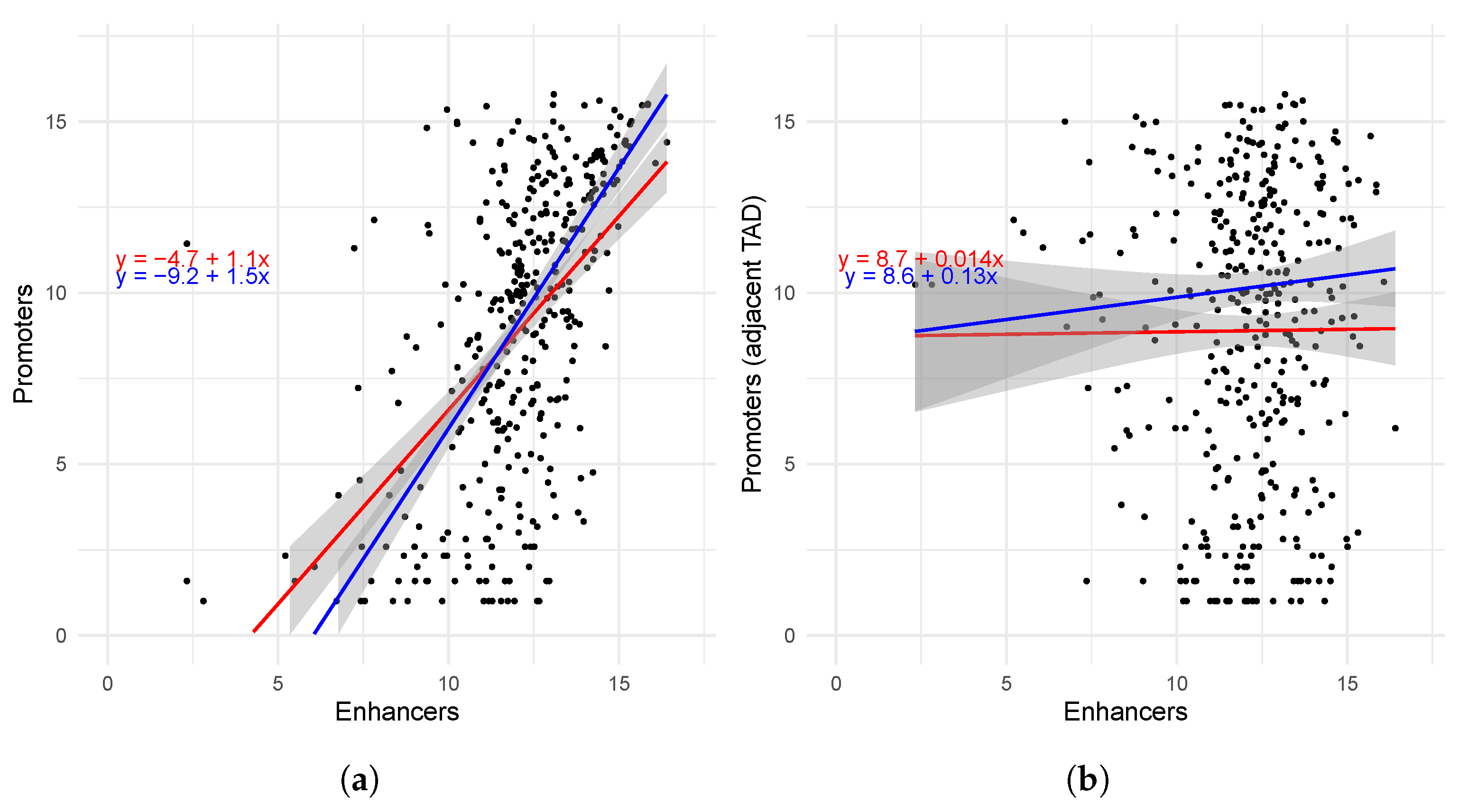
2.3. Interaction Between Promoters and Enhancers on the TAD Level
2.4. Interaction Between Enhancers and Promoters on the TAD Level in the IsoSeq Experiment on Piao Chickens
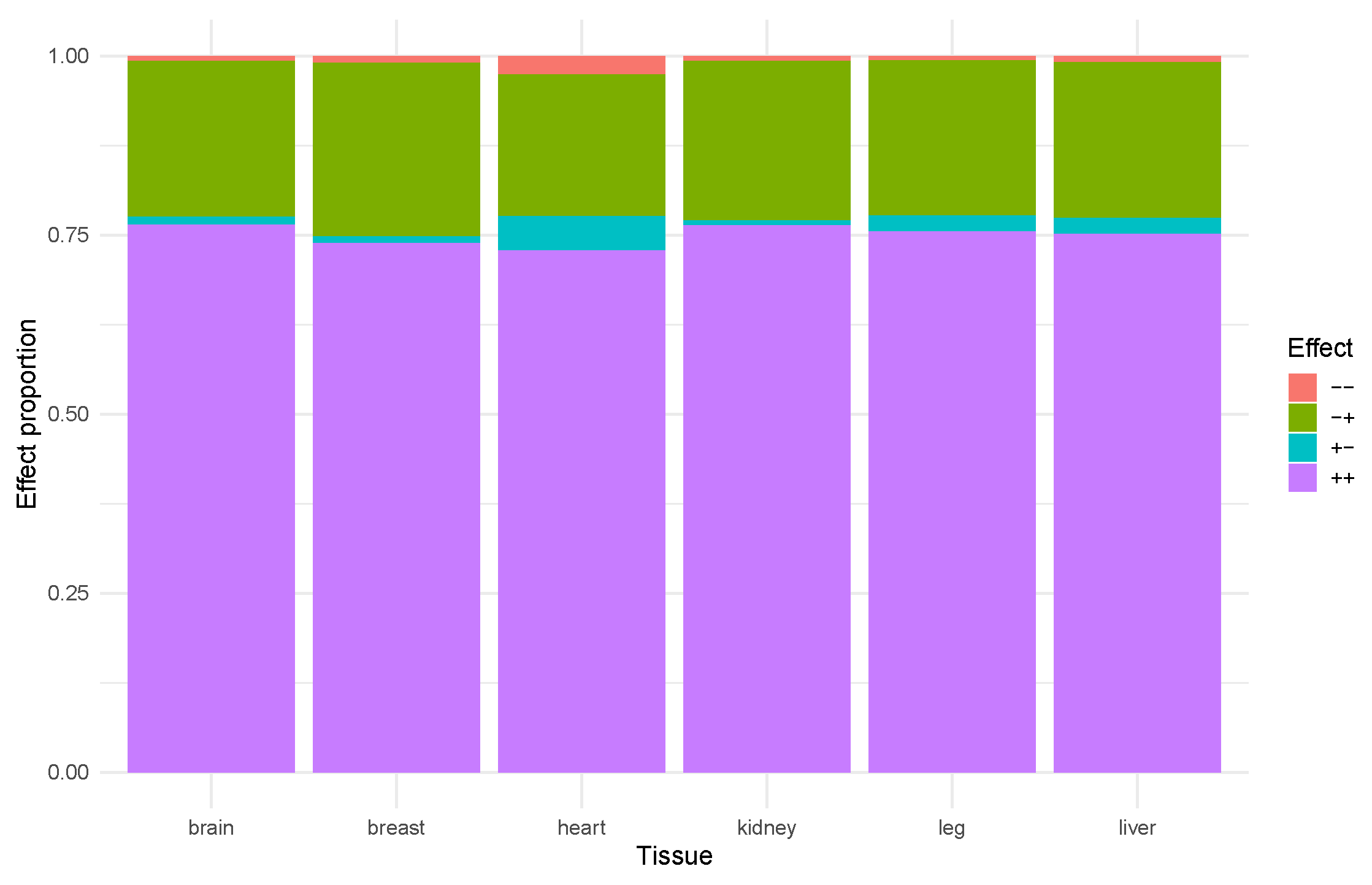
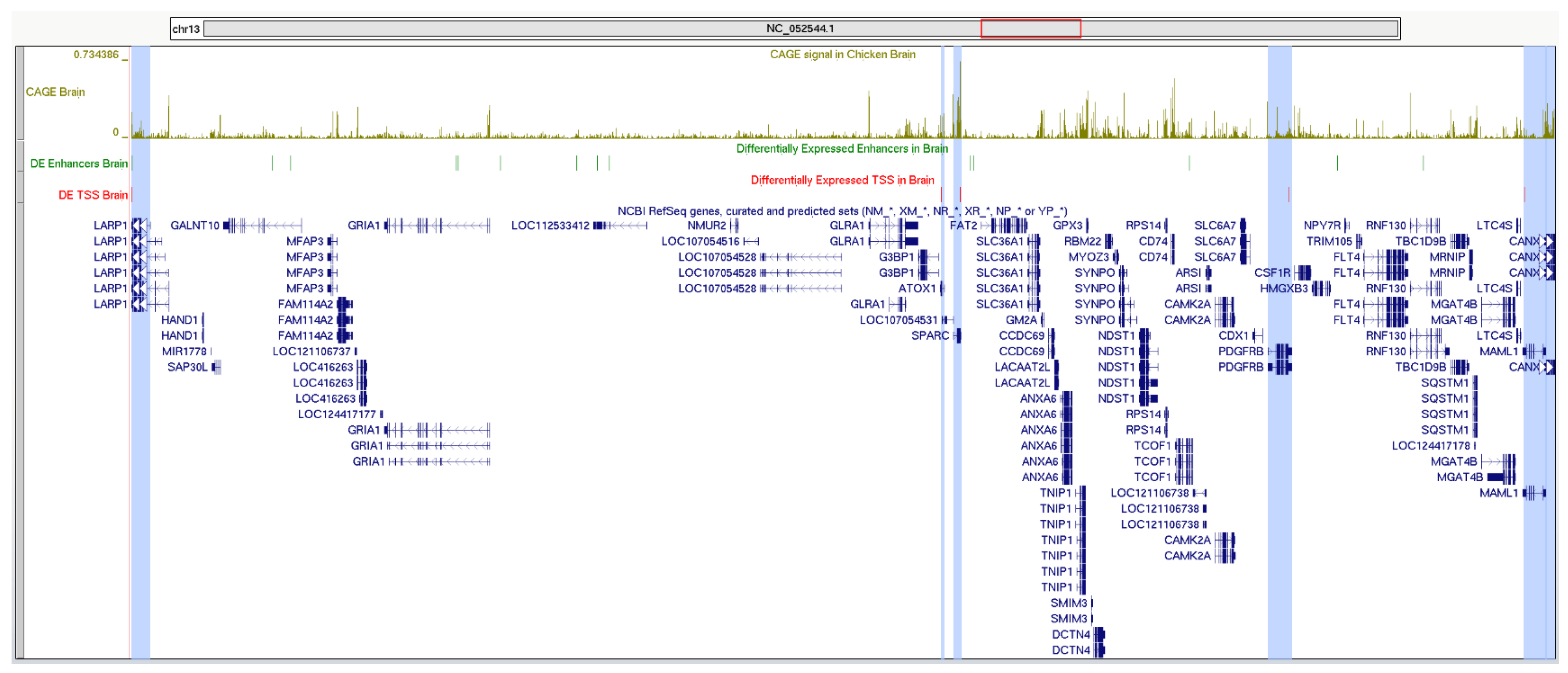
2.5. Interactions Between Enhancers and Promoters in Chicken Brain
3. Discussion
4. Materials and Methods
4.1. Public CAGE and IsoSeq Data
4.2. Expression of Promoters and Enhancers
4.3. Co-Localization of Promoters and Enhancers in Topologically Associating Domains
- Linear Proximity: Identifying enhancer–TSS/gene pairs located within 500,000 base pairs (bp) of each other.
- Co-Localization Within the Same TAD: Extracting all regulatory element–TSS/gene pairs that co-localize within the same TAD domain.
- Functional Filtering: From the set of interactions, only pairs in which both the enhancer and the corresponding TSS or gene were differentially expressed were retained.
4.4. Tissue-Wise Quantification of Interactions Between Promoters and Enhancers
4.5. Correlation Between Aggregated Expression of Promoters and Enhancers in TADs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CAGE | Cap Analysis of Gene Expression; |
| TAD | Topologically Associating Domain; |
| TSS | Transcription Start Site. |
Appendix A
References
- Rahnamoun, H.; Orozco, P.; Lauberth, S.M. The role of enhancer RNAs in epigenetic regulation of gene expression. Transcription 2020, 11, 19–25. [Google Scholar] [CrossRef]
- Calo, E.; Wysocka, J. Modification of enhancer chromatin: What, how, and why? Mol. Cell 2013, 49, 825–837. [Google Scholar] [CrossRef]
- Chepelev, I.; Wei, G.; Wangsa, D.; Tang, Q.; Zhao, K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012, 22, 490–503. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Wang, Y.; Wang, M.; Wang, Y.; Zhu, X.; Gu, S.; Zhong, C.; An, L.; Shan, M.; Damas, J.; et al. An atlas of regulatory elements in chicken: A resource for chicken genetics and genomics. Sci. Adv. 2023, 9, eade1204. [Google Scholar] [CrossRef] [PubMed]
- Kawaji, H.; Kasukawa, T.; Forrest, A.; Carninci, P.; Hayashizaki, Y. The FANTOM5 collection, a data series underpinning mammalian transcriptome atlases in diverse cell types. Sci. Data 2017, 4, 170113. [Google Scholar] [CrossRef]
- Lizio, M.; Deviatiiarov, R.; Nagai, H.; Galan, L.; Arner, E.; Itoh, M.; Lassmann, T.; Kasukawa, T.; Hasegawa, A.; Ros, M.A.; et al. Systematic analysis of transcription start sites in avian development. PLoS Biol. 2017, 15, e2002887. [Google Scholar] [CrossRef]
- Fishman, V.; Battulin, N.; Nuriddinov, M.; Maslova, A.; Zlotina, A.; Strunov, A.; Chervyakova, D.; Korablev, A.; Serov, O.; Krasikova, A. 3D organization of chicken genome demonstrates evolutionary conservation of topologically associated domains and highlights unique architecture of erythrocytes’ chromatin. Nucleic Acids Res. 2019, 47, 648–665. [Google Scholar] [CrossRef]
- McArthur, E.; Capra, J.A. Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability. Am. J. Hum. Genet. 2021, 108, 269–283. [Google Scholar] [CrossRef]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Sexton, T.; Yaffe, E.; Kenigsberg, E.; Bantignies, F.; Leblanc, B.; Hoichman, M.; Parrinello, H.; Tanay, A.; Cavalli, G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 2012, 148, 458–472. [Google Scholar] [CrossRef]
- Ray-Jones, H.; Spivakov, M. Transcriptional enhancers and their communication with gene promoters. Cell. Mol. Life Sci. 2021, 78, 6453–6485. [Google Scholar] [CrossRef]
- Andersson, R.; Gebhard, C.; Miguel-Escalada, I.; Hoof, I.; Bornholdt, J.; Boyd, M.; Chen, Y.; Zhao, X.; Schmidl, C.; Suzuki, T.; et al. An atlas of active enhancers across human cell types and tissues. Nature 2014, 507, 455–461. [Google Scholar] [CrossRef]
- Bonev, B.; Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 2016, 17, 661–678. [Google Scholar] [CrossRef]
- Oudelaar, A.M.; Higgs, D.R. The relationship between genome structure and function. Nat. Rev. Genet. 2020, 22, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.K.; Chen, S.Y.; Gan, Z.Q.; Zhang, Y.Z.; Yue, T.; Chen, M.M.; Xue, Y.; Hu, H.; Guo, A.Y. AnimalTFDB 4.0: A comprehensive animal transcription factor database updated with variation and expression annotations. Nucleic Acids Res. 2023, 51, D39–D45. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Acosta, R.; Addleman, N.J.; Adrian, J.; Afzal, V.; Ai, R.; Aken, B.; Akiyama, J.A.; Jammal, O.A.; Amrhein, H.; et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Schmidt, C.J.; Lamont, S.J. Transcriptome analysis reveals potential mechanisms underlying differential heart development in fast- and slow-growing broilers under heat stress. BMC Genom. 2017, 18, 295. [Google Scholar] [CrossRef]
- Nora, E.P.; Goloborodko, A.; Valton, A.L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017, 169, 930–944.e22. [Google Scholar] [CrossRef]
- Dixon, J.; Gorkin, D.; Ren, B. Chromatin Domains: The Unit of Chromosome Organization. Mol. Cell 2016, 62, 668–680. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, T.; Wang, Z. Functional similarities of protein-coding genes in topologically associating domains and spatially-proximate genomic regions. Genes 2022, 13, 480. [Google Scholar] [CrossRef]
- Zinani, O.Q.H.; Keseroğlu, K.; Özbudak, E.M. Regulatory mechanisms ensuring coordinated expression of functionally related genes. Trends Genet. 2022, 38, 73–81. [Google Scholar] [CrossRef]
- Wolf, S.; Melo, D.; Garske, K.M.; Pallares, L.F.; Lea, A.J.; Ayroles, J.F. Characterizing the landscape of gene expression variance in humans. PLoS Genet. 2023, 19, e1010833. [Google Scholar] [CrossRef]
- van Duin, L.; Krautz, R.; Rennie, S.; Andersson, R. Transcription factor expression is the main determinant of variability in gene co-activity. Mol. Syst. Biol. 2023, 19, e11392. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, J.; Sun, Q.; Xue, Z.; Tang, Z.; Liu, W.; Liu, J.; Miao, B.; Su, N.; He, Y.; et al. Calnexin promotes glioblastoma progression by inducing protective mitophagy through the MEK/ERK/BNIP3 pathway. Theranostics 2025, 15, 2624–2648. [Google Scholar] [CrossRef]
- Grushina, V.A.; Yevshin, I.S.; Gusev, O.A.; Kolpakov, F.A.; Stanishevskaya, O.I.; Fedorova, E.S.; Zinovieva, N.A.; Pintus, S.S. Prediction and annotation of alternative transcription starts and promoter shift in the chicken genome. J. Bioinform. Comput. Biol. 2025, 23, 2550004. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Haberle, V.; Forrest, A.R.R.; Hayashizaki, Y.; Carninci, P.; Lenhard, B. CAGEr: Precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res. 2015, 43, e51. [Google Scholar] [CrossRef]
- Thodberg, M.; Thieffry, A.; Vitting-Seerup, K.; Andersson, R.; Sandelin, A. CAGEfightR: Analysis of 5’-end data using R/Bioconductor. BMC Bioinform. 2019, 20, 487. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Hinrichs, A.S.; Karolchik, D.; Baertsch, R.; Barber, G.P.; Bejerano, G.; Clawson, H.; Diekhans, M.; Furey, T.S.; Harte, R.A.; Hsu, F.; et al. The UCSC Genome Browser Database: Update 2006. Nucleic Acids Res. 2006, 34, D590–D598. [Google Scholar] [CrossRef]


| TADs | Genome | #Intervals | Jaccard |
|---|---|---|---|
| Erythrocyte | galGal5 | 507 | |
| Erythrocyte | GRCg7b | 515 | |
| Fibroblast | galGal5 | 403 | |
| Fibroblast | GRCg7b | 420 | |
| Erythrocyte × Fibroblast | GRCg7b | 769 | 0.780 |
| Erythrocyte only | GRCg7b | 9 | |
| Fibroblast only | GRCg7b | 14 |
| Tissue | Effect | Count | ++ | −+ | +− | − − |
|---|---|---|---|---|---|---|
| Brain | TFs | 31 | 108 | 2 | ||
| Cofactors | 23 | 51 | 4 | |||
| Breast | TFs | 9 | 15 | |||
| Cofactors | 7 | 7 | 1 | |||
| Heart | TFs | 20 | 42 | 3 | ||
| Cofactors | 19 | 31 | 2 | 2 | 1 | |
| Kidney | TFs | 26 | 70 | 1 | ||
| Cofactors | 17 | 29 | 2 | |||
| Liver | TFs | 30 | 91 | |||
| Cofactors | 23 | 41 | 4 | 1 | ||
| Leg | TFs | 17 | 30 | |||
| Cofactors | 13 | 19 |
| Tissue | r | |||||
|---|---|---|---|---|---|---|
| Brain | 0.437 | 3.38 × 10−18 | 0.489 | 5.93 × 10−23 | 0.343 | 2.88 × 10−22 |
| Breast | 0.457 | 5.84 × 10−20 | 0.506 | 8.8 × 10−25 | 0.360 | 2.52 × 10−24 |
| Heart | 0.482 | 2.73 × 10−22 | 0.515 | 0.0 | 0.362 | 1.47 × 10−24 |
| Kidney | 0.473 | 2.16 × 10−21 | 0.513 | 1.68 × 10−25 | 0.365 | 5.17 × 10−25 |
| Leg | 0.461 | 2.84 × 10−20 | 0.509 | 0.0 | 0.363 | 9.85 × 10−25 |
| Liver | 0.489 | 6.12 × 10−23 | 0.532 | 0.0 | 0.377 | 1.32 × 10−26 |
| TADs | r | |||||
|---|---|---|---|---|---|---|
| Same TAD | 0.534 | 9.13 × 10−29 | 0.551 | 6.43 × 10−31 | 0.400 | 1.66 × 10−30 |
| Adjacent TADs | 0.00662 | 0.899 | 0.0487 | 0.35 | 0.0323 | 0.354 |
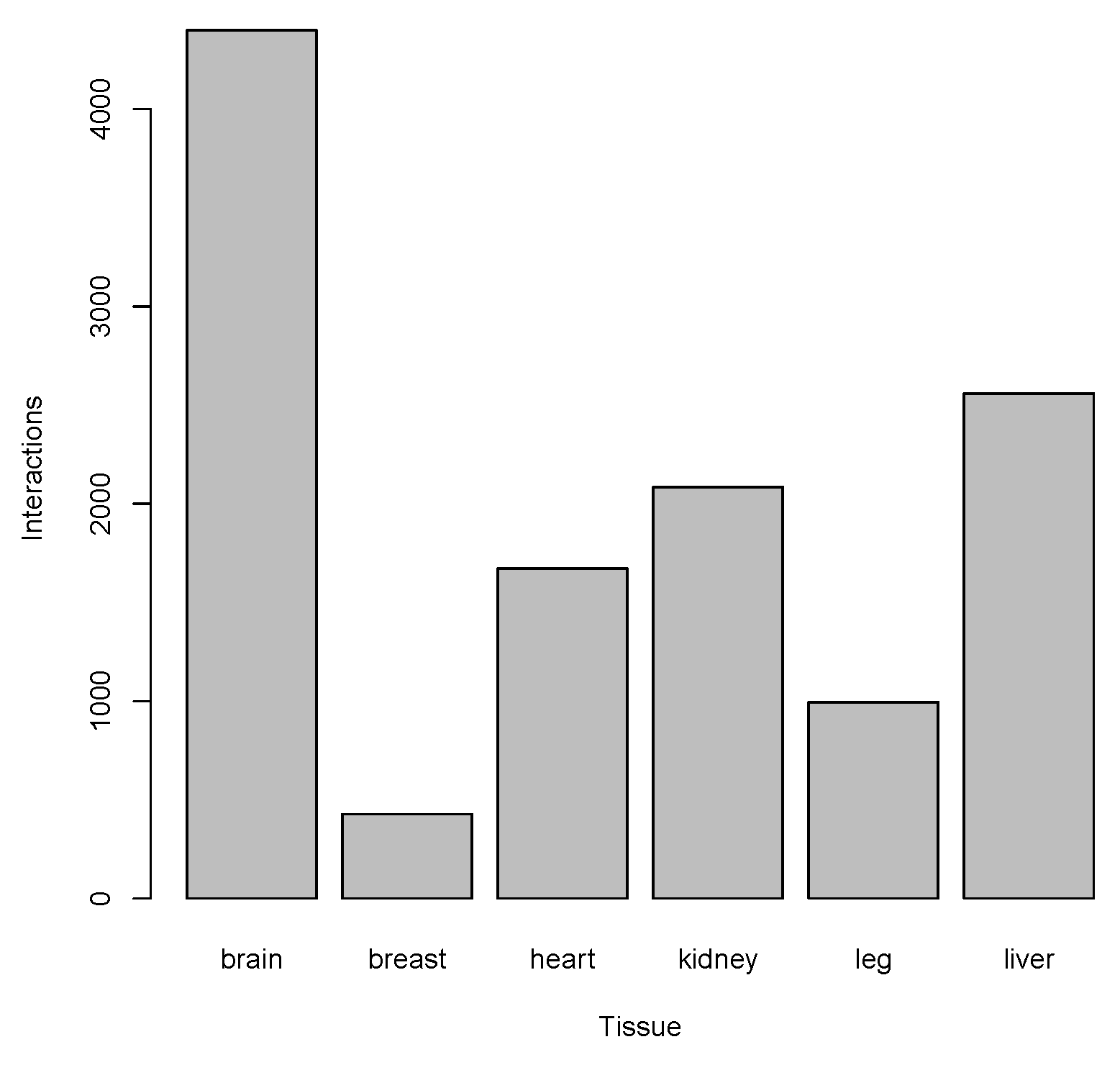
| Tissue | #Interactions |
|---|---|
| Brain | 4400 |
| Breast | 428 |
| Heart | 1672 |
| Kidney | 2085 |
| Leg | 995 |
| Liver | 2559 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grushina, V.A.; Filatova, A.P.; Gagarina, V.S.; Prasolov, D.E.; Kolpakov, F.A.; Gusev, O.A.; Pintus, S.S. Interaction Between Enhancers and Promoters in Chicken Genome. Int. J. Mol. Sci. 2025, 26, 11407. https://doi.org/10.3390/ijms262311407
Grushina VA, Filatova AP, Gagarina VS, Prasolov DE, Kolpakov FA, Gusev OA, Pintus SS. Interaction Between Enhancers and Promoters in Chicken Genome. International Journal of Molecular Sciences. 2025; 26(23):11407. https://doi.org/10.3390/ijms262311407
Chicago/Turabian StyleGrushina, Valentina A., Anastasia P. Filatova, Valeria S. Gagarina, Danila E. Prasolov, Fedor A. Kolpakov, Oleg A. Gusev, and Sergey S. Pintus. 2025. "Interaction Between Enhancers and Promoters in Chicken Genome" International Journal of Molecular Sciences 26, no. 23: 11407. https://doi.org/10.3390/ijms262311407
APA StyleGrushina, V. A., Filatova, A. P., Gagarina, V. S., Prasolov, D. E., Kolpakov, F. A., Gusev, O. A., & Pintus, S. S. (2025). Interaction Between Enhancers and Promoters in Chicken Genome. International Journal of Molecular Sciences, 26(23), 11407. https://doi.org/10.3390/ijms262311407







