Identification of a Novel miR-122-5p/CDC25A Axis and Potential Therapeutic Targets for Chronic Myeloid Leukemia
Abstract
1. Introduction
2. Results
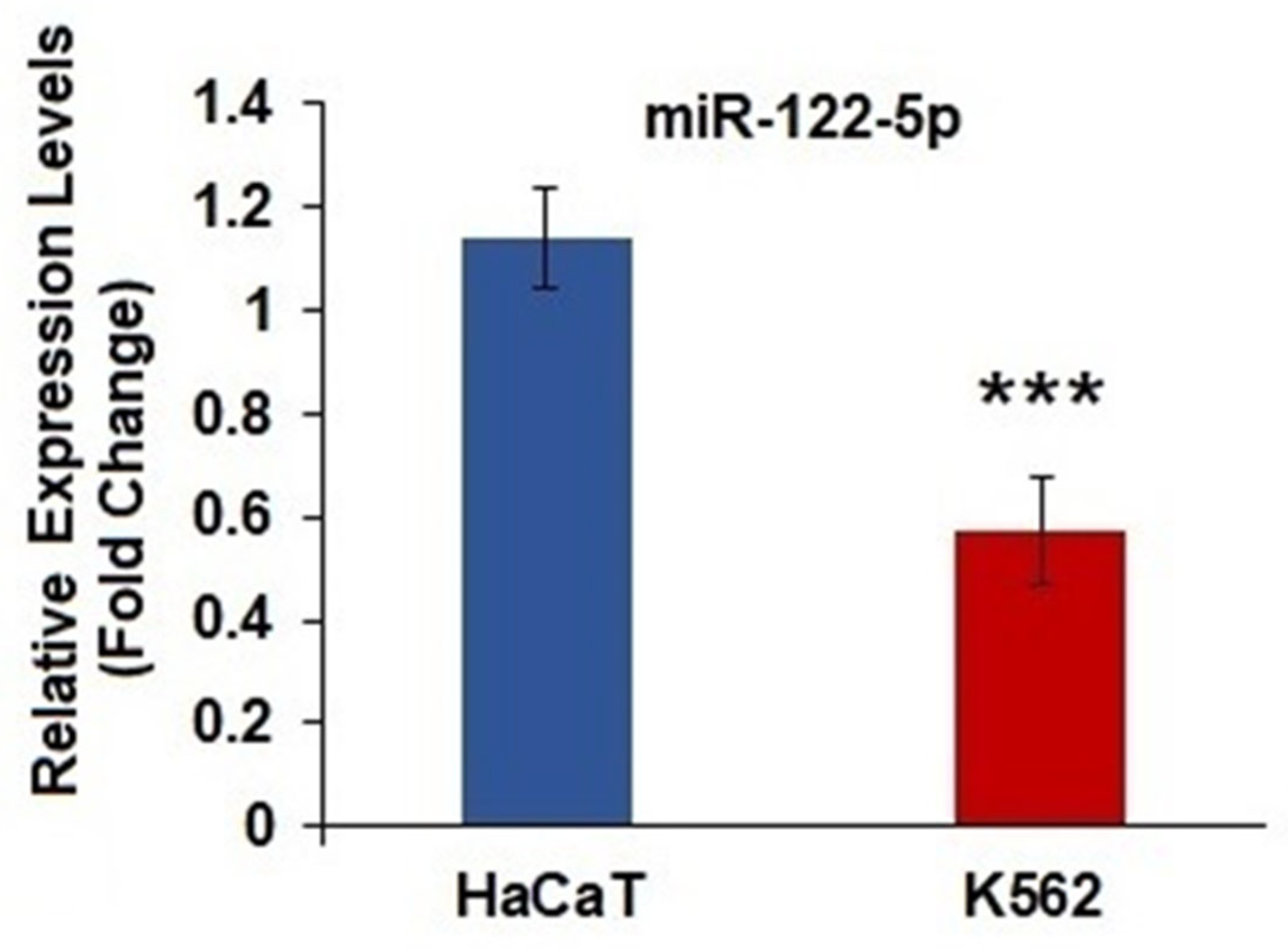
2.1. miR-122-5p Was Underexpressed in K562 Cells
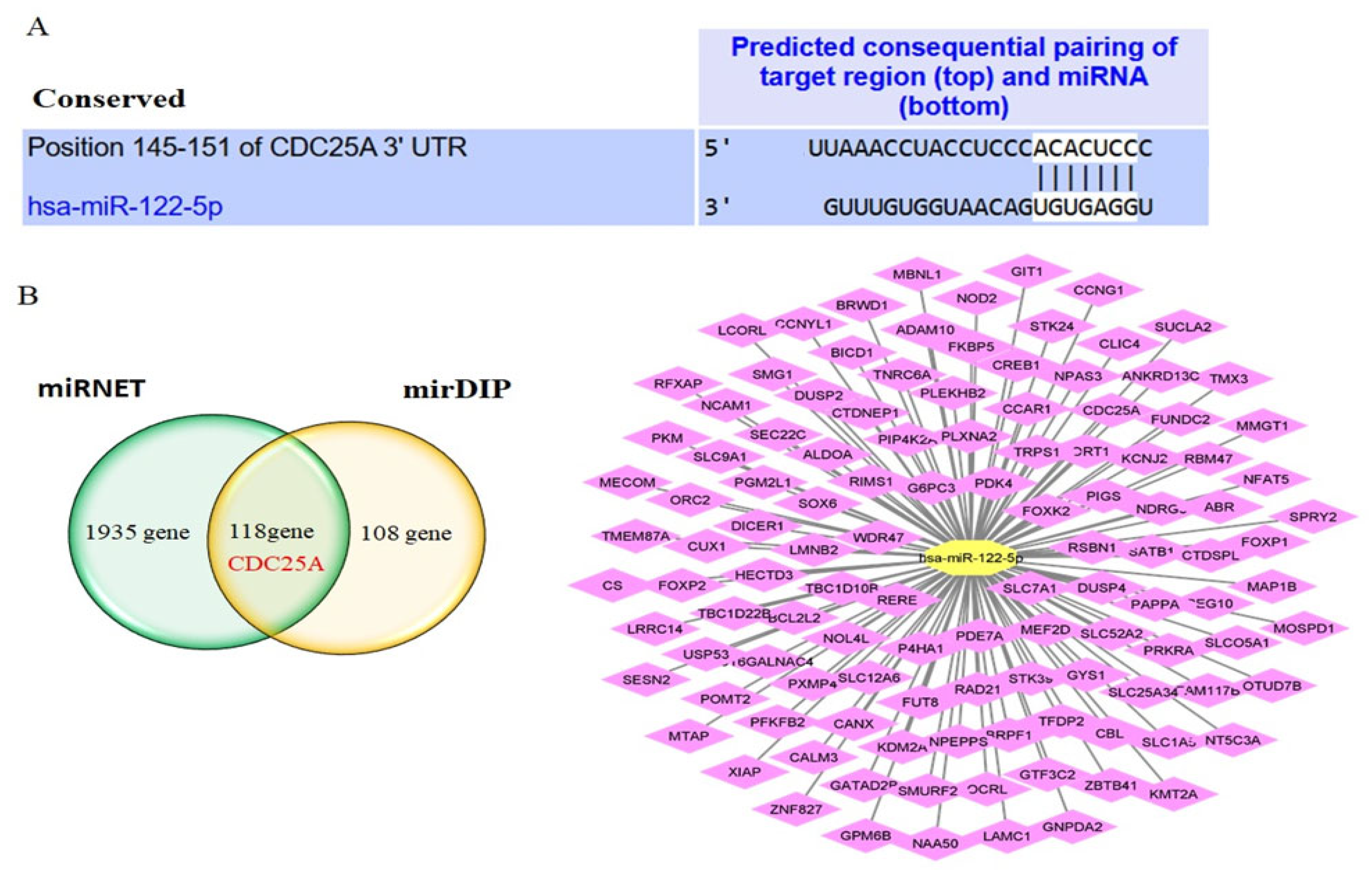
2.2. CDC25A Is a Direct Target of miR-122-5p
2.3. Expression Level of CDC25A and CDK4
2.4. CDC25A Expression and Functions in Chronic Myeloproliferative Disorder
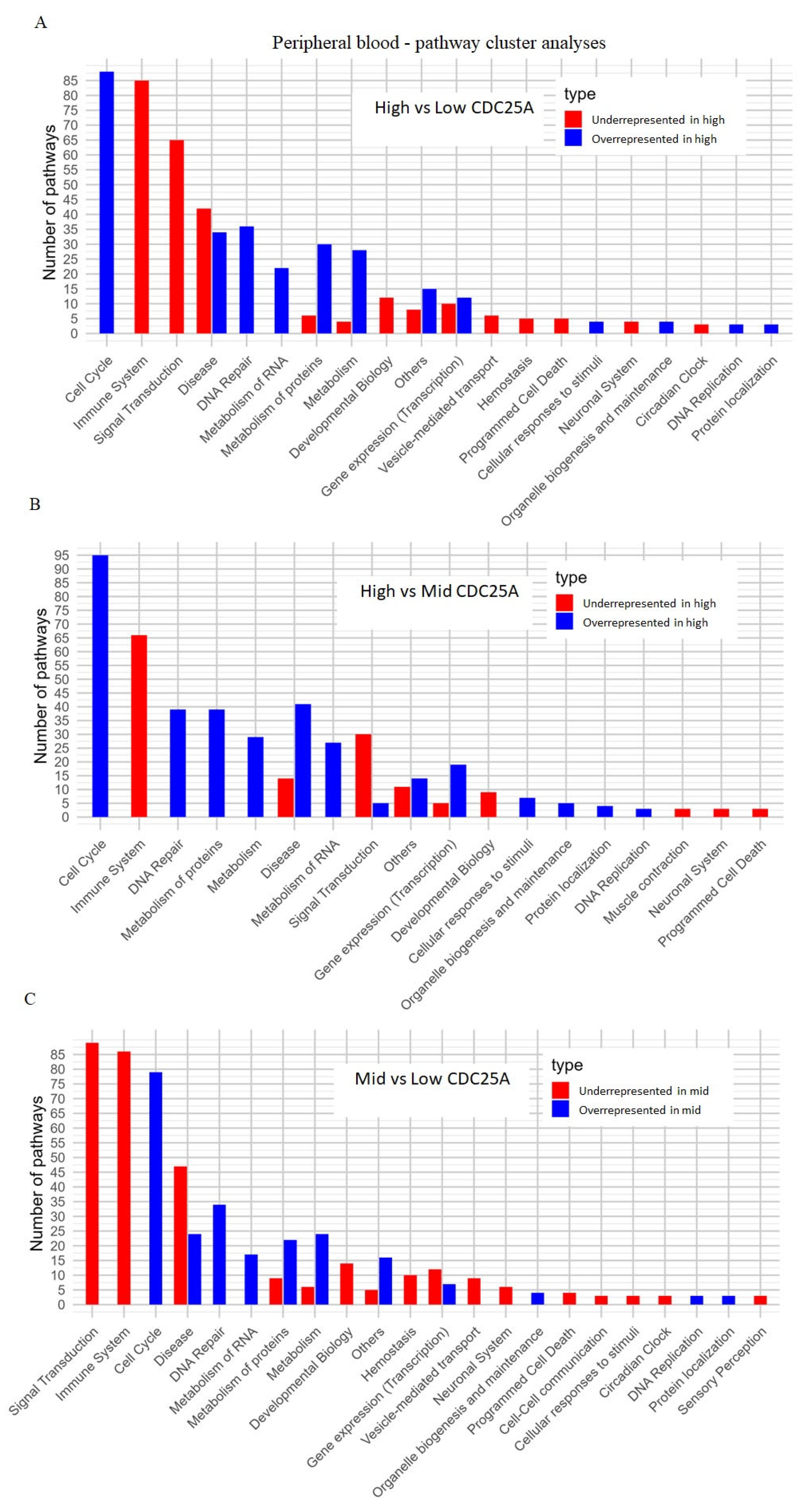
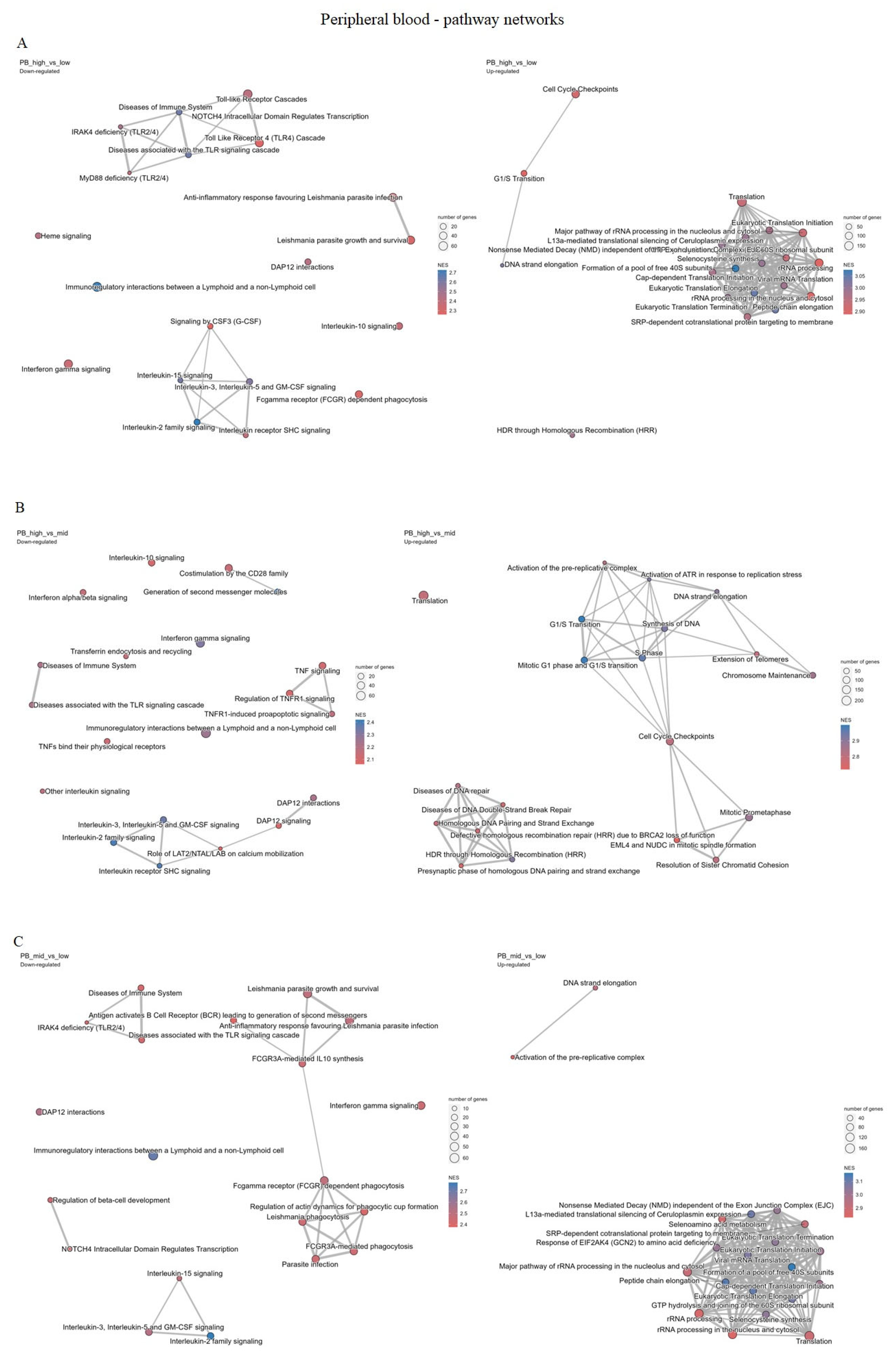
2.4.1. Peripheral Blood
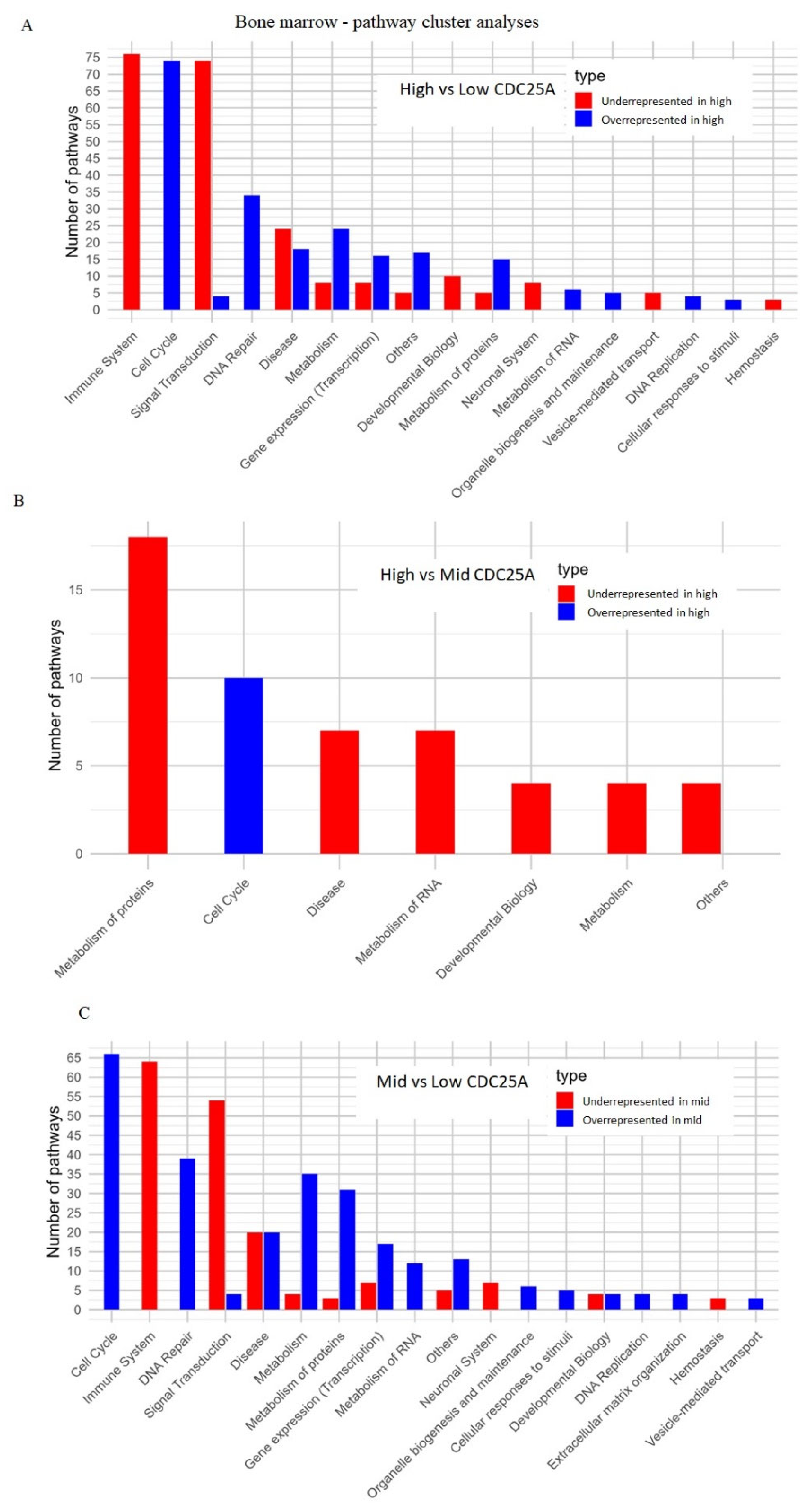
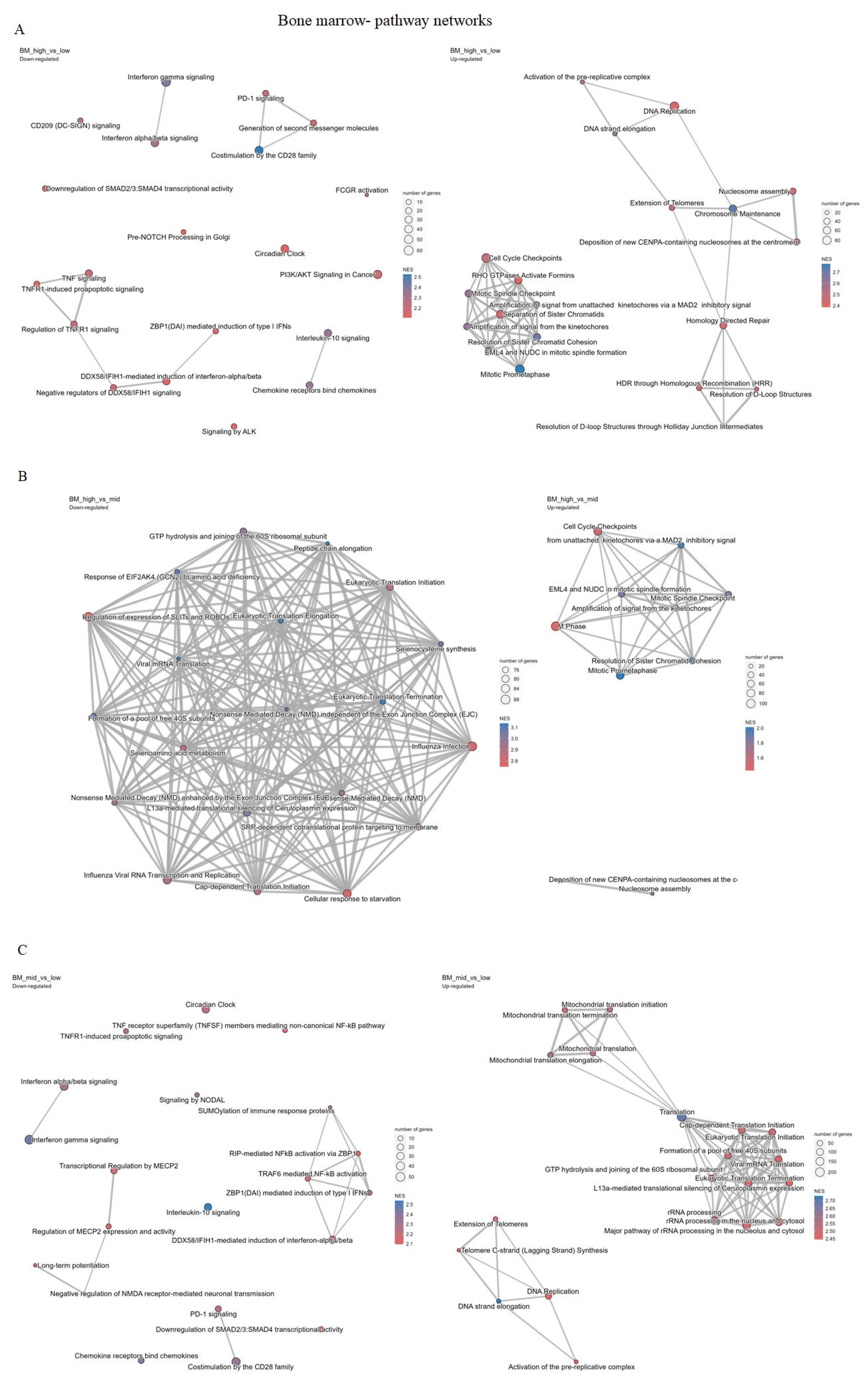
2.4.2. Bone Marrow
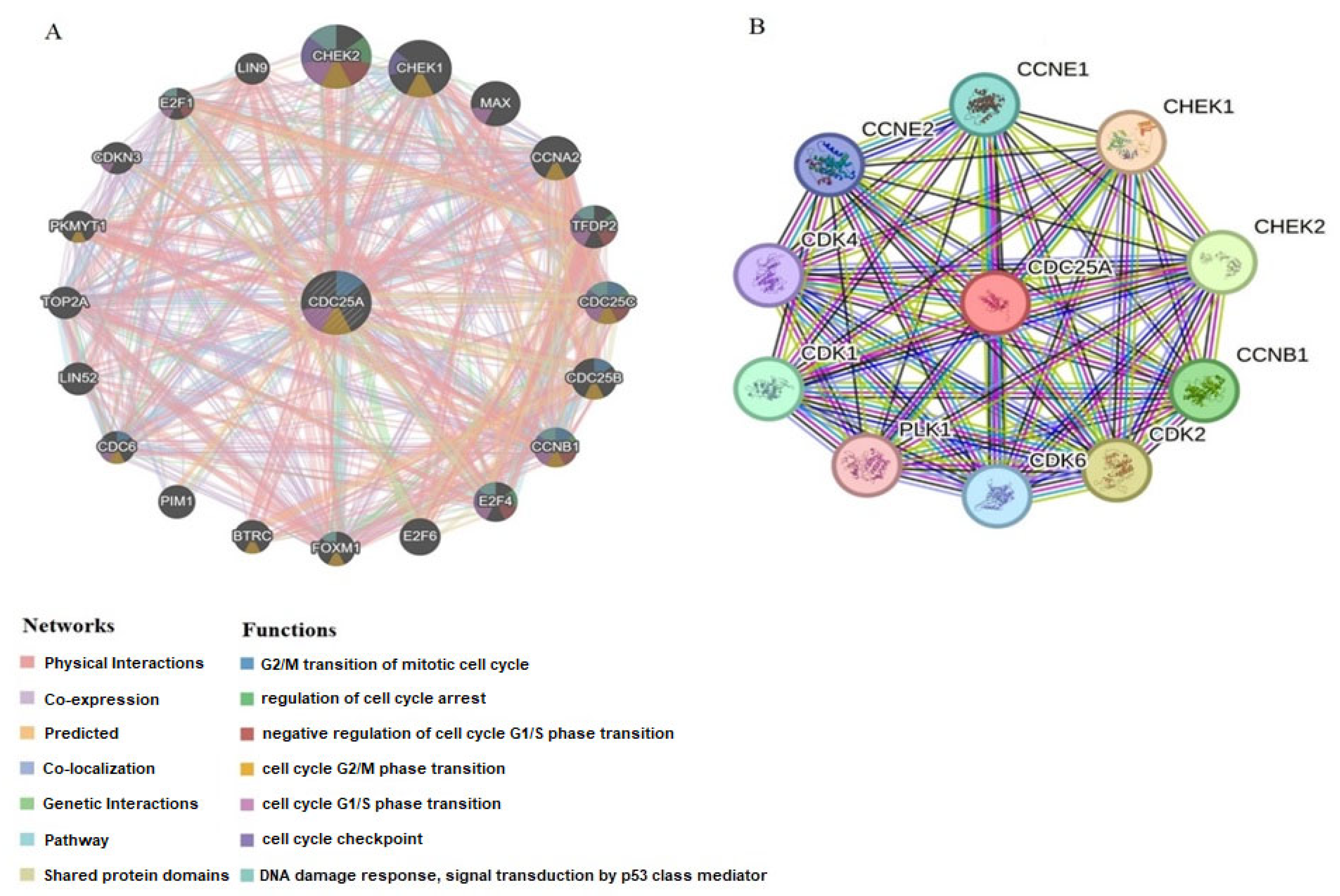
2.5. Interaction Network Analysis of CDC25A
3. Discussion
4. Materials and Methods
4.1. Differentially Expressed Genes (DEGs) and Data Processing
4.2. Prediction of miRNA Targets
4.3. CDC25A Gene Expression Analysis in K562
CDC25A Gene Expression Analysis and Functions in Chronic Myeloproliferative Disorder
4.4. Gene and Protein Network of CDC25A
4.5. Cell Culture
4.6. RNA Extraction and Quantitative Real-Time PCR (RT-qPCR) for CDC25A, CDK4
4.7. miRNA Extraction and RT-qPCR Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AML | Acute myeloid leukemia |
| CDC25A | Cell division cycle 25A |
| CML | Chronic myeloid leukemia |
| CDK | cyclin-cyclin-dependent kinase |
| CDK4 | Cyclin-dependent kinase 4 |
| CT | Cycle threshold |
| DGE | Differential gene expression |
| DMEM | Dulbecco’s Modified Eagle Medium |
| FBS | Fetal bovine serum |
| GEO | Gene Expression Omnibus |
| HaCaT | Human keratinocyte cells |
| IM | Imatinib mesylate |
| miRNAs | MicroRNAs |
| Ph | Philadelphia chromosome |
| PVCA | Principal Variance Component Analysis |
| RT-qPCR | Quantitative real-time PCR |
| STRING | Search Tool for the Retrieval of Interacting Gene |
| TKI | tyrosine kinase inhibitors |
References
- Yang, Y.; Zhong, F.; Jiang, J.; Li, M.; Yao, F.; Liu, J.; Cheng, Y.; Xu, S.; Chen, S.; Zhang, H.; et al. Bioinformatic analysis of the expression profile and identification of RhoGDI2 as a biomarker in imatinib-resistant K562 cells. Hematology 2023, 28, 2244856. [Google Scholar] [CrossRef] [PubMed]
- Ochi, Y. Genetic landscape of chronic myeloid leukemia. Int. J. Hematol. 2023, 117, 30–36. [Google Scholar] [CrossRef]
- El-Tanani, M.; Nsairat, H.; Matalka, I.I.; Lee, Y.F.; Rizzo, M.; Aljabali, A.A.; Mishra, V.; Mishra, Y.; Hromić-Jahjefendić, A.; Tambuwala, M.M. The impact of the BCR-ABL oncogene in the pathology and treatment of chronic myeloid leukemia. Pathol. Res. Pract. 2024, 254, 155161. [Google Scholar] [CrossRef]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2025 update on diagnosis, therapy, and monitoring. Am. J. Hematol. 2024, 99, 2191–2212. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, W.; Xue, W.; Wang, Y.; Chen, P.; Li, Q.; Li, Y.; Hu, X.; Zhao, Y.; Zhou, H. miR-181a plays the tumor-suppressor role in chronic myeloid leukemia CD34+ cells partially via SERPINE1. Cell Mol. Life Sci. 2023, 81, 10. [Google Scholar] [CrossRef]
- Laganà, A.; Scalzulli, E.; Bisegna, M.L.; Ielo, C.; Martelli, M.; Breccia, M. Understanding and overcoming resistance to tyrosine kinase inhibitors (TKIs) in Chronic myeloid leukemia (CML). Expert. Rev. Hematol. 2025, 18, 65–79. [Google Scholar] [CrossRef]
- Litwińska, Z.; Machaliński, B. miRNAs in chronic myeloid leukemia: Small molecules, essential function. Leuk. Lymphoma 2017, 58, 1297–1305. [Google Scholar] [CrossRef]
- Yang, H.; Liu, Y.; Chen, L.; Zhao, J.; Guo, M.; Zhao, X.; Wen, Z.; He, Z.; Chen, C.; Xu, L. MiRNA-Based Therapies for Lung Cancer: Opportunities and Challenges? Biomolecules 2023, 13, 877. [Google Scholar] [CrossRef] [PubMed]
- Suri, C.; Swarnkar, S.; Bhaskar, L.V.K.S.; Verma, H.K. Non-Coding RNA as a Biomarker in Lung Cancer. Noncoding RNA 2024, 10, 50. [Google Scholar] [CrossRef]
- Navabi, A.; Akbari, B.; Abdalsamadi, M.; Naseri, S. The role of microRNAs in the development, progression and drug resistance of chronic myeloid leukemia and their potential clinical significance. Life Sci. 2022, 296, 120437. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.Y.; Liu, X.J.; Zhan, Y.; Liu, M.H.; Zhang, X.Y.; Li, Z.Y.; Lu, Y.-Q.; Luo, J.-M.; Yang, L. MiR-140-5p induces cell apoptosis and decreases Warburg effect in chronic myeloid leukemia by targeting SIX1. Biosci. Rep. 2019, 39, BSR20190150. [Google Scholar] [CrossRef]
- Faramin, L.M.; Hashemipour, N.; Niaraki, N.; Soghala, S.; Moradi, A.; Sarhangi, S.; Hatami, M.; Aghaei-Zarch, F.; Khosravifar, M.; Mohammadzadeh, A.; et al. MicroRNA-122 in human cancers: From mechanistic to clinical perspectives. Cancer Cell Int. 2023, 23, 29. [Google Scholar] [CrossRef]
- Lara-Chica, M.; Correa-Sáez, A.; Jiménez-Izquierdo, R.; Garrido-Rodríguez, M.; Ponce, F.J.; Moreno, R.; Morrison, K.; Di Vona, C.; Arató, K.; Jiménez-Jiménez, C.; et al. A novel CDC25A/DYRK2 regulatory switch modulates cell cycle and survival. Cell Death Differ. 2022, 29, 105–117. [Google Scholar] [CrossRef]
- Li, H.; Jiang, M.; Cui, M.; Feng, G.; Dong, J.; Li, Y.; Xiao, H.; Fan, S. MiR-365 enhances the radiosensitivity of non-small cell lung cancer cells through targeting CDC25A. Biochem. Biophys. Res. Commun. 2019, 512, 392–398. [Google Scholar] [CrossRef]
- Qin, H.; Liu, W. MicroRNA-99a-5p suppresses breast cancer progression and cell-cycle pathway through downregulating CDC25A. J. Cell Physiol. 2019, 234, 3526–3537. [Google Scholar] [CrossRef]
- Fernandez-Vidal, A.; Ysebaert, L.; Didier, C.; Betous, R.; De-Toni, F.; Prade-Houdellier, N.; Demur, C.; Contour-GalcérA, M.-O.; PrévOst, G.P.; Ducommun, B.; et al. Cell adhesion regulates CDC25A expression and proliferation in acute myeloid leukemia. Cancer Res. 2006, 66, 7128–7135. [Google Scholar] [CrossRef]
- Chen, S.; Tang, Y.; Yang, C.; Li, K.; Huang, X.; Cao, J. Silencing CDC25A inhibits the proliferation of liver cancer cells by downregulating IL-6 in vitro and in vivo. Int. J. Mol. Med. 2020, 45, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Huang, S. The role of Cdc25A in the regulation of cell proliferation and apoptosis. Anticancer Agents Med. Chem. 2012, 12, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Dakilah, I.; Harb, A.; Abu-Gharbieh, E.; El-Huneidi, W.; Taneera, J.; Hamoudi, R.; Semreen, M.H.; Bustanji, Y. Potential of CDC25 phosphatases in cancer research and treatment: Key to precision medicine. Front. Pharmacol. 2024, 15, 1324001. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Li, Q.; Xu, A.; Hu, Y.; Sun, C. Ferroptosis in hematological malignancies and its potential network with abnormal tumor metabolism. Biomed. Pharmacother. 2022, 148, 112747. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Wu, J.; Wang, Q.; Liang, Y.; Li, X.; Chen, G.; Ma, L.; Liu, X.; Zhou, F. Global burden of hematologic malignancies and evolution patterns over the past 30 years. Blood Cancer J. 2023, 13, 82. [Google Scholar] [CrossRef]
- Li, S.Q.; Liu, J.; Zhang, J.; Wang, X.L.; Chen, D.; Wang, Y.; Xu, Y.-M.; Huang, B.; Lin, J.; Li, J.; et al. Transcriptome profiling reveals the high incidence of hnRNPA1 exon 8 inclusion in chronic myeloid leukemia. J. Adv. Res. 2020, 24, 301–310. [Google Scholar] [CrossRef]
- Chakrabortty, A.; Patton, D.J.; Smith, B.F.; Agarwal, P. miRNAs: Potential as biomarkers and therapeutic targets for cancer. Genes 2023, 14, 1375. [Google Scholar] [CrossRef]
- Xishan, Z.; Xianjun, L.; Ziying, L.; Guangxin, C.; Gang, L. The malignancy suppression role of miR-23a by targeting the BCR/ABL oncogene in chromic myeloid leukemia. Cancer Gene Ther. 2014, 21, 397–404. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Tao, K.; Xiao, Q.; Huang, Z.; Zhong, L.; Cao, W.; Wen, J.; Feng, W. MiR-29b suppresses CML cell proliferation and induces apoptosis via regulation of BCR/ABL1 protein. Exp. Cell Res. 2013, 319, 1094–1101. [Google Scholar] [CrossRef]
- Chen, Z.; Fu, S.; Shan, Y.; He, Z.; Gu, J.; Wu, H.; Lin, J.; Huang, Y.; Wang, H.; Lu, Y.; et al. Circ_0001047 inhibits prostate cancer progression and enhances abiraterone sensitivity via miR-122-5p/FKBP5/PHLPP1/AKT axis in vitro. Discov. Oncol. 2024, 15, 569. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xu, C.; Liu, C.; Wu, X.; Chen, X.; Hou, J.; Wang, L. TGF-β1-Induced LINC01094 promotes epithelial-mesenchymal transition in hepatocellular carcinoma through the miR-122-5p/TGFBR2–SAMD2–SMAD3 Axis. Funct. Integr. Genomics. 2024, 24, 123. [Google Scholar] [CrossRef] [PubMed]
- Salzmann, R.J.; Garbin, A.; Gaffo, E.; Elia, C.; Martire, G.; Bortoluzzi, S.; Tondo, A.; Muggeo, P.; Sala, A.; Pizzi, M.; et al. Extracellular Vesicle miR-122-5p as a Prognostic Biomarker in Pediatric Classical Hodgkin Lymphoma. Int. J. Mol. Sci. 2024, 25, 13243. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yuan, Y.; Yang, X.; Hong, Z.; Yang, L. Decreased expression of microRNA-122 is associated with an unfavorable prognosis in childhood acute myeloid leukemia and function analysis indicates a therapeutic potential. Pathol. Res. Pract. 2017, 213, 1166–1172. [Google Scholar] [CrossRef]
- Sadeghi, H.; Golalipour, M.; Yamchi, A.; Farazmandfar, T.; Shahbazi, M. CDC25A pathway toward tumorigenesis: Molecular targets of CDC25A in cell-cycle regulation. J. Cell Biochem. 2019, 120, 2919–2928. [Google Scholar] [CrossRef]
- Ding, F.N.; Gao, B.H.; Wu, X.; Gong, C.W.; Wang, W.Q.; Zhang, S.M. miR-122-5p modulates the radiosensitivity of cervical cancer cells by regulating cell division cycle 25A (CDC25A). FEBS Open Bio. 2019, 9, 1869–1879. [Google Scholar] [CrossRef]
- Sørensen, C.S.; Syljuåsen, R.G.; Falck, J.; Schroeder, T.; Rönnstrand, L.; Khanna, K.K.; Zhou, B.-B.; Bartek, J.; Lukas, J. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell 2003, 3, 247–258. [Google Scholar] [CrossRef]
- Li, J.; Qiao, H.; Wu, F.; Sun, S.; Feng, C.; Li, C.; Yan, W.; Lv, W.; Wu, H.; Liu, M.; et al. A novel hypoxia- and lactate metabolism-related signature to predict prognosis and immunotherapy responses for breast cancer by integrating machine learning and bioinformatic analyses. Front. Immunol. 2022, 13, 998140. [Google Scholar] [CrossRef]
- Tian, Y.; Xing, Y.; Zhang, Z.; Peng, R.; Zhang, L.; Sun, Y. Bioinformatics Analysis of Key Genes and circRNA-miRNA-mRNA Regulatory Network in Gastric Cancer. Biomed. Res. Int. 2020, 2020, 2862701. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Lim, B. Targeting apoptosis in cancer. Curr. Oncol. Rep. 2022, 24, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Gong, Q.; Chen, F. CDC25A inhibition suppresses cell proliferation and induces G 1/S-phase cell cycle arrest in nasopharyngeal carcinoma. Mol. Med. Rep. 2023, 27, 109. [Google Scholar] [CrossRef]
- Kristjá, K.; Rudolph, J. Cdc25 Phosphatases and Cancer. Chem. Biol. 2004, 11, 1043–1051. [Google Scholar] [CrossRef]
- Rissland, O.S.; Hong, S.J.; Bartel, D.P. MicroRNA Destabilization Enables Dynamic Regulation of the miR-16 Family in Response to Cell Cycle Changes. Mol. Cell 2011, 43, 993–1004. [Google Scholar] [CrossRef]
- Johnson, C.D.; Esquela-Kerscher, A.; Stefani, G.; Byrom, M.; Kelnar, K.; Ovcharenko, D.; Wilson, M.; Wang, X.; Shelton, J.; Shingara, J.; et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007, 67, 7713–7722. [Google Scholar] [CrossRef]
- Lee, S.O.; Masyuk, T.; Splinter, P.; Banales, J.M.; Masyuk, A.; Stroope, A.; LaRusso, N. MicroRNA15a modulates expression of the cell-cycle regulator Cdc25A and affects hepatic cystogenesis in a rat model of polycystic kidney disease. J. Clin. Investig. 2008, 118, 3714–3724. [Google Scholar] [CrossRef]
- Yang, X.; Feng, M.; Jiang, X.; Wu, Z.; Li, Z.; Aau, M.; Yu, Q. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb–E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009, 23, 2388–2393. [Google Scholar] [CrossRef]
- Wan, P.; Zou, F.; Zhang, X.; Li, H.; Dulak, A.; Robert, J.; John, S.L.; Wang, Z.; Zhang, L.; Yu, L.; et al. miR21 negatively regulates CDC25A and cell cylce progression in colpon cancer cells. Cancer Res. 2009, 69, 8157–8165. [Google Scholar]
- Shi, L.; Zhang, J.; Pan, T.; Zhou, J.; Gong, W.; Liu, N.; Fu, Z.; You, Y. MiR-125b is critical for the suppression of human U251 glioma stem cell proliferation. Brain Res. 2010, 1312, 120–126. [Google Scholar] [CrossRef]
- Sarkar, S.; Dey, B.K.; Dutta, A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol. Biol. Cell 2010, 21, 2138–2149. [Google Scholar] [CrossRef]
- Marcet, B.; Chevalier, B.; Luxardi, G.; Coraux, C.; Zaragosi, L.E.; Cibois, M.; Robbe-Sermesant, K.; Jolly, T.; Moreilhon, C.; Cardinaud, B. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat. Cell Biol. 2011, 13, 693–701, Erratum in Nat. Cell Biol. 2011, 13, 1280. [Google Scholar] [CrossRef] [PubMed]
- Bertero, T.; Gastaldi, C.; Bourget-Ponzio, I.; Mari, B.; Meneguzzi, G.; Barbry, P.; Ponzio, G.; Rezzonico, R. CDC25A targeting by miR-483-3p decreases CCND-CDK4/6 assembly and contributes to cell cycle arrest. Cell Death Differ. 2013, 20, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zheng, M.; Lu, R.; Du, J.; Zhao, Q.; Li, Z.; Li, Y.; Zhang, S. The role of CDC25C in cell cycle regulation and clinical cancer therapy: A systematic review. Cancer Cell Int. 2020, 20, 213. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.; Jiang, G. Differential expression and functional analysis of lung cancer gene expression datasets: A systems biology perspective. Oncol. Lett. 2019, 18, 776–782. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef] [PubMed]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.C.; Tsay, M.; Lu, R.; Jurisica, I. MirDIP 4.1—Integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef]
- Shirdel, E.A.; Xie, W.; Mak, T.W.; Jurisica, I. NAViGaTing the micronome using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS ONE 2011, 6, e17429. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Wu, C.; Jin, X.; Tsueng, G.; Afrasiabi, C.; Su, A.I. BioGPS: Building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016, 44, D313–D316. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Orozco, C.; Boyer, J.; Leglise, M.; Goodale, J.; Batalov, S.; Hodge, C.L.; Haase, J.; Janes, J.; Huss, J.W.; et al. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome. Biol. 2009, 10, R130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wilmot, B.; Bottomly, D.; Dao, K.H.T.; Stevens, E.; Eide, C.A.; Hodge, C.L.; Haase, J.; Janes, J.; Huss, J.W.; et al. Genomic landscape of neutrophilic leukemias of ambiguous diagnosis. Blood 2019, 134, 867–879. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein—Protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612, Correction in Nucleic Acids Res. 2021, 49, 10800. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, 214–220. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ozer Yaman, S.; Petrović, N.; Yaman, S.; Akidan, O.; Cimbek, A.; Baycelebi, G.; Srdić-Rajić, T.; Šami, A.; Misir, S. Identification of a Novel miR-122-5p/CDC25A Axis and Potential Therapeutic Targets for Chronic Myeloid Leukemia. Int. J. Mol. Sci. 2025, 26, 11401. https://doi.org/10.3390/ijms262311401
Ozer Yaman S, Petrović N, Yaman S, Akidan O, Cimbek A, Baycelebi G, Srdić-Rajić T, Šami A, Misir S. Identification of a Novel miR-122-5p/CDC25A Axis and Potential Therapeutic Targets for Chronic Myeloid Leukemia. International Journal of Molecular Sciences. 2025; 26(23):11401. https://doi.org/10.3390/ijms262311401
Chicago/Turabian StyleOzer Yaman, Serap, Nina Petrović, Selcuk Yaman, Osman Akidan, Ahmet Cimbek, Gulsah Baycelebi, Tatjana Srdić-Rajić, Ahmad Šami, and Sema Misir. 2025. "Identification of a Novel miR-122-5p/CDC25A Axis and Potential Therapeutic Targets for Chronic Myeloid Leukemia" International Journal of Molecular Sciences 26, no. 23: 11401. https://doi.org/10.3390/ijms262311401
APA StyleOzer Yaman, S., Petrović, N., Yaman, S., Akidan, O., Cimbek, A., Baycelebi, G., Srdić-Rajić, T., Šami, A., & Misir, S. (2025). Identification of a Novel miR-122-5p/CDC25A Axis and Potential Therapeutic Targets for Chronic Myeloid Leukemia. International Journal of Molecular Sciences, 26(23), 11401. https://doi.org/10.3390/ijms262311401




