Maternal Melatonin Supplementation Modulates Placental DNA Methylation and Gene Expression in Nutrient-Restricted Cattle
Abstract
1. Introduction
2. Results
2.1. Methylome and Transcriptome Profiles
2.2. DNA-Methylation Profiling Across Experimental Groups
2.2.1. Differential DNA Methylation by Nutrient Restriction
2.2.2. Melatonin Dependent Differential DNA Methylation
2.2.3. Sex-Specific DNA Methylation Differences
2.3. Functional Enrichment of DMGs
2.4. RNA Sequencing Profiles of Differentially Expressed Genes
2.5. Functional Enrichment Analysis of DEGs
2.6. Overlapping Analysis of DNA Methylation and Gene Expression
3. Discussion
3.1. Maternal Nutrient Restriction Affects DNA Methylation
3.2. Melatonin Alters the Epigenetic and Transcriptomic Profile
3.3. Sex-Specific Epigenetic and Transcriptomic Alterations
3.4. Sex-Specific Epigenetic and Transcriptomic Alterations in Response to Melatonin
3.5. Integration Analysis Between DMRs and DEGs
4. Materials and Methods
4.1. Experimental Design
4.2. Collection of Samples
4.3. DNA Methylation Analysis
4.3.1. Methyl MiniSeq Genome-Wide Bisulfite Sequencing and Library Preparation
4.3.2. Sequence Alignments and Data Analysis
4.3.3. Differential Methylation Analysis
4.4. Total RNA Extraction, Library Construction and RNA Sequencing
4.5. Bioinformatic Analysis
4.6. Analysis of Overlapping Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Velazquez, M.A. Impact of Maternal Malnutrition during the Periconceptional Period on Mammalian Preimplantation Embryo Development. Domest. Anim. Endocrinol. 2015, 51, 27–45. [Google Scholar] [CrossRef] [PubMed]
- King, J.C. A Summary of Pathways or Mechanisms Linking Preconception Maternal Nutrition with Birth Outcomes. J. Nutr. 2016, 146, 1437S–1444S. [Google Scholar] [CrossRef] [PubMed]
- Caton, J.S.; Crouse, M.S.; McLean, K.J.; Dahlen, C.R.; Ward, A.K.; Cushman, R.A.; Grazul-Bilska, A.T.; Neville, B.W.; Borowicz, P.P.; Reynolds, L.P. Maternal Periconceptual Nutrition, Early Pregnancy, and Developmental Outcomes in Beef Cattle. J. Anim. Sci. 2020, 98, skaa358. [Google Scholar] [CrossRef] [PubMed]
- Van Gronigen Case, G.; Storey, K.M.; Parmeley, L.E.; Schulz, L.C. Effects of Maternal Nutrient Restriction during the Periconceptional Period on Placental Development in the Mouse. PLoS ONE 2021, 16, e0244971. [Google Scholar] [CrossRef]
- Nwachukwu, C.U.; Woad, K.J.; Barnes, N.; Gardner, D.S.; Robinson, R.S. Maternal Protein Restriction Affects Fetal Ovary Development in Sheep. Reprod. Fertil. 2021, 2, 161. [Google Scholar] [CrossRef]
- Du, M.; Tong, J.; Zhao, J.; Underwood, K.R.; Zhu, M.; Ford, S.P.; Nathanielsz, P.W. Fetal Programming of Skeletal Muscle Development in Ruminant Animals. J. Anim. Sci. 2010, 88, E51–E60. [Google Scholar] [CrossRef]
- Wu, G.; Bazer, F.W.; Cudd, T.A.; Meininger, C.J.; Spencer, T.E. Maternal Nutrition and Fetal Development. J. Nutr. 2004, 134, 2169–2172. [Google Scholar] [CrossRef]
- Lemley, C.O.; Vonnahme, K.A. Physiology and Endocrinology Symposium: Alterations in Uteroplacental Hemodynamics during Melatonin Supplementation in Sheep and Cattle. J. Anim. Sci. 2017, 95, 2211–2221. [Google Scholar] [CrossRef]
- Mandruzzato, G.; Antsaklis, A.; Botet, F.; Chervenak, F.A.; Figueras, F.; Grunebaum, A.; Puerto, B.; Skupski, D.; Stanojevic, M. Intrauterine Restriction (IUGR). J. Perinat. Med. 2008, 36, 277–281. [Google Scholar] [CrossRef]
- Saleem, T.; Sajjad, N.; Fatima, S.; Habib, N.; Ali, S.R.; Qadir, M. Intrauterine Growth Retardation—Small Events, Big Consequences. Ital. J. Pediatr. 2011, 37, 41. [Google Scholar] [CrossRef]
- Sharma, D.; Shastri, S.; Sharma, P. Intrauterine Growth Restriction: Antenatal and Postnatal Aspects. Clin. Med. Insights Pediatr. 2016, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, L.; Vonnahme, K.; Lemley, C.; Redmer, D.; Grazul-Bilska, A.; Borowicz, P.; Caton, J. Maternal Stress and Placental Vascular Function and Remodeling. Curr. Vasc. Pharmacol. 2013, 11, 564–593. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Correa, Z.E.; Messman, R.D.; Sidelinger, D.R.; Heath King, E.; Sánchez-Rodríguez, H.L.; Burnett, D.D.; Lemley, C.O. Melatonin Alters Bovine Uterine Artery Hemodynamics, Vaginal Temperatures, and Fetal Morphometrics during Late Gestational Nutrient Restriction in a Season-Dependent Manner. J. Anim. Sci. 2021, 99, skab242. [Google Scholar] [CrossRef]
- Rueda-Clausen, C.F.; Morton, J.S.; Davidge, S.T. Effects of Hypoxia-Induced Intrauterine Growth Restriction on Cardiopulmonary Structure and Function during Adulthood. Cardiovasc. Res. 2009, 81, 713–722. [Google Scholar] [CrossRef]
- Mert, I.; Oruc, A.S.; Yuksel, S.; Cakar, E.S.; Buyukkagnici, U.; Karaer, A.; Danisman, N. Role of Oxidative Stress in Preeclampsia and Intrauterine Growth Restriction. J. Obstet. Gynaecol. Res. 2012, 38, 658–664. [Google Scholar] [CrossRef]
- Jiao, P.; Lu, H.; Hao, L.; Degen, A.A.; Cheng, J.; Yin, Z.; Mao, S.; Xue, Y. Nutrigenetic and Epigenetic Mechanisms of Maternal Nutrition–Induced Glucolipid Metabolism Changes in the Offspring. Nutr. Rev. 2025, 83, 728–748. [Google Scholar] [CrossRef]
- Chadio, S.; Kotsampasi, B.; Taka, S.; Liandris, E.; Papadopoulos, N.; Plakokefalos, E. Epigenetic Changes of Hepatic Glucocorticoid Receptor in Sheep Male Offspring Undernourished in Utero. Reprod. Fertil. Dev. 2017, 29, 1995–2004. [Google Scholar] [CrossRef]
- Rashidi, S.Y.; Rafiyan, M.; Asemi, R.; Asemi, Z.; Mohammadi, S. Effect of Melatonin as a Therapeutic Strategy against Intrauterine Growth Restriction: A Mini-Review of Current State. Ann. Med. Surg. 2024, 86, 5320. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Correa, Z.E.; Messman, R.D.; Swanson, R.M.; Lemley, C.O. Melatonin in Health and Disease: A Perspective for Livestock Production. Biomolecules 2023, 13, 490. [Google Scholar] [CrossRef]
- Tamura, H.; Nakamura, Y.; Terron, M.P.; Flores, L.J.; Manchester, L.C.; Tan, D.X.; Sugino, N.; Reiter, R.J. Melatonin and Pregnancy in the Human. Reprod. Toxicol. 2008, 25, 291–303. [Google Scholar] [CrossRef]
- Bates, K.; Herzog, E.D. Maternal-Fetal Circadian Communication During Pregnancy. Front. Endocrinol. 2020, 11, 519328. [Google Scholar] [CrossRef]
- Kamfar, W.W.; Khraiwesh, H.M.; Ibrahim, M.O.; Qadhi, A.H.; Azhar, W.F.; Ghafouri, K.J.; Alhussain, M.H.; Aldairi, A.F.; AlShahrani, A.M.; Alghannam, A.F.; et al. Comprehensive Review of Melatonin as a Promising Nutritional and Nutraceutical Supplement. Heliyon 2024, 10, e24266. [Google Scholar] [CrossRef]
- Reiter, R.J.; Tan, D.X.; Mayo, J.C.; Sainz, R.M.; Leon, J.; Czarnocki, Z. Melatonin as an Antioxidant: Biochemical Mechanisms and Pathophysiological Implications in Humans. Acta Biochim. Pol. 2003, 50, 1129–1146. [Google Scholar] [CrossRef]
- Brockus, K.E.; Hart, C.G.; Gilfeather, C.L.; Fleming, B.O.; Lemley, C.O. Dietary Melatonin Alters Uterine Artery Hemodynamics in Pregnant Holstein Heifers. Domest. Anim. Endocrinol. 2016, 55, 1–10. [Google Scholar] [CrossRef]
- Joo, E.H.; Kim, Y.R.; Kim, N.; Jung, J.E.; Han, S.H.; Cho, H.Y.; Joo, E.H.; Kim, Y.R.; Kim, N.; Jung, J.E.; et al. Effect of Endogenic and Exogenic Oxidative Stress Triggers on Adverse Pregnancy Outcomes: Preeclampsia, Fetal Growth Restriction, Gestational Diabetes Mellitus and Preterm Birth. Int. J. Mol. Sci. 2021, 22, 10122. [Google Scholar] [CrossRef]
- Keomanivong, F.E.; Lemley, C.O.; Camacho, L.E.; Yunusova, R.; Borowicz, P.P.; Caton, J.S.; Meyer, A.M.; Vonnahme, K.A.; Swanson, K.C. Influence of Nutrient Restriction and Melatonin Supplementation of Pregnant Ewes on Maternal and Fetal Pancreatic Digestive Enzymes and Insulin-Containing Clusters. Animal 2015, 10, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Amaral, H.B.S.; Silveira, M.M.; Nicolás, A.C.C.V.; Pimenta, L.K.L.; Chaves, J.E.V.; Caetano, A.R.; Franco, M.M.; Dode, M.A.N. Melatonin Improves Bovine Embryo Production and Quality via Antioxidant, Metabolic, and Epigenetic Pathways. Antioxidants 2025, 14, 1322. [Google Scholar] [CrossRef]
- Lee, H.S. Impact of Maternal Diet on the Epigenome during In Utero Life and the Developmental Programming of Diseases in Childhood and Adulthood. Nutrients 2015, 7, 9492. [Google Scholar] [CrossRef]
- Phomvisith, O.; Muroya, S.; Otomaru, K.; Oshima, K.; Oshima, I.; Nishino, D.; Haginouchi, T.; Gotoh, T. Maternal Undernutrition Affects Fetal Thymus DNA Methylation, Gene Expression, and, Thereby, Metabolism and Immunopoiesis in Wagyu (Japanese Black) Cattle. Int. J. Mol. Sci. 2024, 25, 9242. [Google Scholar] [CrossRef] [PubMed]
- de Mendoza, A.; Nguyen, T.V.; Ford, E.; Poppe, D.; Buckberry, S.; Pflueger, J.; Grimmer, M.R.; Stolzenburg, S.; Bogdanovic, O.; Oshlack, A.; et al. Large-Scale Manipulation of Promoter DNA Methylation Reveals Context-Specific Transcriptional Responses and Stability. Genome Biol. 2022, 23, 163. [Google Scholar] [CrossRef]
- Flores, K.B.; Wolschin, F.; Amdam, G.V. The Role of Methylation of DNA in Environmental Adaptation. Integr. Comp. Biol. 2013, 53, 359. [Google Scholar] [CrossRef]
- Sosa-Larios, T.C.; Cerbón, M.A.; Morimoto, S. Epigenetic Alterations Caused by Nutritional Stress during Fetal Programming of the Endocrine Pancreas. Arch. Med. Res. 2015, 46, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Namous, H.; Peñagaricano, F.; Del Corvo, M.; Capra, E.; Thomas, D.L.; Stella, A.; Williams, J.L.; Marsan, P.A.; Khatib, H. Integrative Analysis of Methylomic and Transcriptomic Data in Fetal Sheep Muscle Tissues in Response to Maternal Diet during Pregnancy. BMC Genom. 2018, 19, 123. [Google Scholar] [CrossRef]
- Peterson, M.; Gauvin, M.; Pillai, S.; Jones, A.; McFadden, K.; Cameron, K.; Reed, S.; Zinn, S.; Govoni, K. Maternal Under-and over-Nutrition during Gestation Causes Islet Hypertrophy and Sex-Specific Changes to Pancreas DNA Methylation in Fetal Sheep. Animals 2021, 11, 2531. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Z.; Lin, H.; Ma, D.; Tao, Q.; Liu, F. Epigenetic Regulation of Left–Right Asymmetry by DNA Methylation. EMBO J. 2017, 36, 2987–2997. [Google Scholar] [CrossRef] [PubMed]
- Goodman, L.D.; Cope, H.; Nil, Z.; Ravenscroft, T.A.; Charng, W.L.; Lu, S.; Tien, A.C.; Pfundt, R.; Koolen, D.A.; Haaxma, C.A.; et al. TNPO2 Variants Associate with Human Developmental Delays, Neurologic Deficits, and Dysmorphic Features and Alter TNPO2 Activity in Drosophila. Am. J. Hum. Genet. 2021, 108, 1669. [Google Scholar] [CrossRef]
- Leary, S.C.; Cobine, P.A.; Nishimura, T.; Verdijk, R.M.; De Krijger, R.; De Coo, R.; Tarnopolsky, M.A.; Winge, D.R.; Shoubridge, E.A. COX19 Mediates the Transduction of a Mitochondrial Redox Signal from SCO1 That Regulates ATP7A-Mediated Cellular Copper Efflux. Mol. Biol. Cell 2013, 24, 683. [Google Scholar] [CrossRef] [PubMed]
- Saijoh, Y.; Adachi, H.; Sakuma, R.; Yeo, C.Y.; Yashiro, K.; Watanabe, M.; Hashiguchi, H.; Mochida, K.; Ohishi, S.; Kawabata, M.; et al. Left–Right Asymmetric Expression of Lefty2 and Nodal Is Induced by a Signaling Pathway That Includes the Transcription Factor FAST2. Mol. Cell 2000, 5, 35–47. [Google Scholar] [CrossRef]
- Linowiecka, K.; Slominski, A.T.; Reiter, R.J.; Böhm, M.; Steinbrink, K.; Paus, R.; Kleszczyński, K. Melatonin: A Potential Regulator of DNA Methylation. Antioxidants 2023, 12, 1155. [Google Scholar] [CrossRef]
- Onaolapo, O.J.; Onaolapo, A.Y.; Olowe, O.A.; Udoh, M.O.; Udoh, D.O.; Nathaniel, I.T. Melatonin and Melatonergic Influence on Neuronal Transcription Factors: Implications for the Development of Novel Therapies for Neurodegenerative Disorders. Curr. Neuropharmacol. 2020, 18, 563. [Google Scholar] [CrossRef]
- Yan, Y.; Zhou, X.E.; Thomas, S.L.; Liu, M.; Lai, G.Q.; Worden, E.J.; Jones, P.A.; Xu, T.H. Mechanisms of DNMT3A–3L-Mediated de Novo DNA Methylation on Chromatin. Nat. Struct. Mol. Biol. 2025, 2025, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jahncke, J.N.; Wright, K.M. The Many Roles of Dystroglycan in Nervous System Development and Function. Dev. Dyn. 2023, 252, 61–80. [Google Scholar] [CrossRef]
- Hardeland, R. Melatonin and Chromatin. Melatonin Res. 2019, 2, 67–93. [Google Scholar] [CrossRef]
- Corral-Vazquez, C.; Blanco, J.; Aiese Cigliano, R.; Sarrate, Z.; Rivera-Egea, R.; Vidal, F.; Garrido, N.; Daub, C.; Anton, E. The RNA Content of Human Sperm Reflects Prior Events in Spermatogenesis and Potential Post-Fertilization Effects. Mol. Hum. Reprod. 2021, 27, gaab035. [Google Scholar] [CrossRef]
- Sakamoto, H.; Kawata, M. Gastrin-Releasing Peptide System in the Spinal Cord Controls Male Sexual Behaviour. J. Neuroendocrinol. 2009, 21, 432–435. [Google Scholar] [CrossRef]
- Mo, C.; Lv, C.; Zhu, G.; Zhang, J.; Li, Y.; Wang, Y.; Li, J. In Vitro Gastrin-Releasing Peptide Response in Chicken Ovarian Follicles. Poult. Sci. 2025, 104, 105220. [Google Scholar] [CrossRef]
- Freese, J.L.; Pino, D.; Pleasure, S.J. Wnt Signaling in Development and Disease. Neurobiol. Dis. 2009, 38, 148. [Google Scholar] [CrossRef] [PubMed]
- Sallam, A.M. Genomic Regions Underlying Variation in Wattles, Horns, and Supernumerary Teats Phenotypes in Egyptian Goats. Can. J. Anim. Sci. 2024, 104, 549–557. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, H.M.; Ju, Z.H.; Zhang, Y.; Huang, J.M.; Qi, C.; Hou, M.H.; An, L.G.; Zhong, J.F.; Wang, C.F. Association of Novel Single Nucleotide Polymorphisms of the CXCR1 Gene with the Milk Performance Traits of Chinese Native Cattle. Genet. Mol. Res. 2013, 12, 2725–2739. [Google Scholar] [CrossRef]
- Wróbel, T.M.; Jørgensen, F.S.; Pandey, A.V.; Grudzińska, A.; Sharma, K.; Yakubu, J.; Björkling, F. Non-Steroidal CYP17A1 Inhibitors: Discovery and Assessment. J. Med. Chem. 2023, 66, 6542–6566. [Google Scholar] [CrossRef]
- Zhai, G.; Shu, T.; Xia, Y.; Lu, Y.; Shang, G.; Jin, X.; He, J.; Nie, P.; Yin, Z. Characterization of Sexual Trait Development in Cyp17a1-Deficient Zebrafish. Endocrinology 2018, 159, 3549–3562. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Zhang, Y.; Wang, Z.; Guo, W.; Chen, J.; Ji, Q.; Wang, Y.; Li, R.; Xu, H.; Chen, X. Key Role of CYP17A1 in Leydig Cell Function and Testicular Development in Qianbei Ma Goats. Genomics 2025, 117, 110937. [Google Scholar] [CrossRef]
- Melko, M.; Douguet, D.; Bensaid, M.; Zongaro, S.; Verheggen, C.; Gecz, J.; Bardoni, B. Functional Characterization of the AFF (AF4/FMR2) Family of RNA-Binding Proteins: Insights into the Molecular Pathology of FRAXE Intellectual Disability. Hum. Mol. Genet. 2011, 20, 1873–1885. [Google Scholar] [CrossRef]
- Dong, Z.; Jiang, H.; Liang, S.; Wang, Y.; Jiang, W.; Zhu, C. Ribosomal Protein L15 Is Involved in Colon Carcinogenesis. Int. J. Med. Sci. 2019, 16, 1132–1141. [Google Scholar] [CrossRef]
- Pottmeier, P.; Nikolantonaki, D.; Lanner, F.; Peuckert, C.; Jazin, E. Sex-Biased Gene Expression during Neural Differentiation of Human Embryonic Stem Cells. Front. Cell Dev. Biol. 2024, 12, 1341373. [Google Scholar] [CrossRef]
- Ma, W.; Fang, H.; Pease, N.; Filippova, G.N.; Disteche, C.M.; Berletch, J.B. Sex-Biased and Parental Allele-Specific Gene Regulation by KDM6A. Biol. Sex Differ. 2022, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Yin, P.; Yan, H.; Zhong, X.; Ren, C.; Li, K.; Chin Heng, B.; Zhang, W.; Tong, G. Single-Cell Transcriptome Analysis of Human Oocyte Ageing. J. Cell. Mol. Med. 2021, 25, 6289–6303. [Google Scholar] [CrossRef]
- Davegårdh, C.; Hall Wedin, E.; Broholm, C.; Henriksen, T.I.; Pedersen, M.; Pedersen, B.K.; Scheele, C.; Ling, C. Sex Influences DNA Methylation and Gene Expression in Human Skeletal Muscle Myoblasts and Myotubes. Stem Cell Res. Ther. 2019, 10, 26. [Google Scholar] [CrossRef]
- Chukrallah, L.G.; Potgieter, S.; Chueh, L.; Snyder, E.M. Two RNA Binding Proteins, ADAD2 and RNF17, Interact to Form a Heterogeneous Population of Novel Meiotic Germ Cell Granules with Developmentally Dependent Organelle Association. PLoS Genet. 2023, 19, e1010519. [Google Scholar] [CrossRef]
- Anwer, D.; Kerkhoven, E.; Polster, A. Transcriptional Network Dysregulation in Alzheimer’s Disease Revealed by Individual-Specific Gene Regulatory Models. medRxiv 2025, 2025.03.26.25324703. [Google Scholar] [CrossRef]
- Zaitoun, I.; Khatib, H. Assessment of Genomic Imprinting of SLC38A4, NNAT, NAP1L5, and H19 in Cattle. BMC Genet. 2006, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shao, S.; Wang, Y.; Du, Z.; Yu, H.; Li, F.; Qin, Y. Ahdc1 Is a Potent Regulator of Obesity and Energy Metabolism. Am. J. Physiol.-Endocrinol. Metab. 2023, 325, E638–E648. [Google Scholar] [CrossRef]
- Robichaux, M.A.; Cowan, C.W. Signaling Mechanisms of Axon Guidance and Early Synaptogenesis. Curr. Top. Behav. Neurosci. 2013, 16, 19–48. [Google Scholar] [CrossRef]
- O’Donnell, M.; Chance, R.K.; Bashaw, G.J. Axon Growth and Guidance: Receptor Regulation and Signal Transduction. Annu. Rev. Neurosci. 2009, 32, 383–412. [Google Scholar] [CrossRef]
- Russell, S.A.; Bashaw, G.J. Axon Guidance Pathways and the Control of Gene Expression. Dev. Dyn. 2018, 247, 571–580. [Google Scholar] [CrossRef]
- Liu, B.; Liu, Y.; Li, S.; Chen, P.; Zhang, J.; Feng, L. BDNF Promotes Mouse Follicular Development and Reverses Ovarian Aging by Promoting Cell Proliferation. J. Ovarian Res. 2023, 16, 83. [Google Scholar] [CrossRef]
- Hawkey-Noble, A.; Umali, J.; Fowler, G.; French, C.R. Expression of Three P4-Phospholipid Flippases—Atp11a, Atp11b, and Atp11c in Zebrafish (Danio rerio). Gene Expr. Patterns 2020, 36, 119115. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, Y.; Suzuki, C.; Segawa, K.; Uchiyama, Y.; Nagata, S.; Birge, R.; Hirota, Y. Inefficient Development of Syncytiotrophoblasts in the Atp11a-Deficient Mouse Placenta. Proc. Natl. Acad. Sci. USA 2022, 119, e2200582119. [Google Scholar] [CrossRef]
- Thornburg, K.L.; O’Tierney, P.F.; Louey, S. Review: The Placenta Is a Programming Agent for Cardiovascular Disease. Placenta 2010, 31, S54–S59. [Google Scholar] [CrossRef] [PubMed]
- McCarty, K.J.; Owen, M.P.T.; Hart, C.G.; Thompson, R.C.; Burnett, D.D.; King, E.H.; Hopper, R.M.; Lemley, C.O. Effect of Chronic Melatonin Supplementation during Mid to Late Gestation on Maternal Uterine Artery Blood Flow and Subsequent Development of Male Offspring in Beef Cattle. J. Anim. Sci. 2018, 96, 5100–5111. [Google Scholar] [CrossRef]
- Maganhin, C.C.; Simões, R.S.; Fuchs, L.F.P.; Sasso, G.R.S.; Simões, M.J.; Baracat, E.C.; Soares, J.M. Melatonin Influences on Steroidogenic Gene Expression in the Ovary of Pinealectomized Rats. Fertil. Steril. 2014, 102, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.K.; Kelly, S.L.; Kaderbhai, M.A. Cytochrome B5 Modulation of 17α Hydroxylase and 17–20 Lyase (CYP17) Activities in Steroidogenesis. J. Endocrinol. 2005, 187, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Novakovic, B.; Sibson, M.; Ng, H.K.; Manuelpillai, U.; Rakyan, V.; Down, T.; Beck, S.; Fournier, T.; Evain-Brion, D.; Dimitriadis, E.; et al. Placenta-Specific Methylation of the Vitamin D 24-Hydroxylase Gene: Implications for Feedback Autoregulation of Active Vitamin D Levels at the Fetomaternal Interface. J. Biol. Chem. 2009, 284, 14838. [Google Scholar] [CrossRef]
- Fantone, S.; Tossetta, G.; Di Simone, N.; Tersigni, C.; Scambia, G.; Marcheggiani, F.; Giannubilo, S.R.; Marzioni, D. CD93 a Potential Player in Cytotrophoblast and Endothelial Cell Migration. Cell Tissue Res. 2022, 387, 123–130. [Google Scholar] [CrossRef]
- Zhang, H.; Bao, S.; Zhao, X.; Bai, Y.; Lv, Y.; Gao, P.; Li, F.; Zhang, W. Genome-Wide Association Study and Phenotype Prediction of Reproductive Traits in Large White Pigs. Animals 2024, 14, 3348. [Google Scholar] [CrossRef]
- Khan, M.Z.; Chen, W.; Naz, S.; Liu, X.; Liang, H.; Chen, Y.; Kou, X.; Liu, Y.; Ashraf, I.; Han, Y.; et al. Determinant Genetic Markers of Semen Quality in Livestock. Front. Endocrinol. 2024, 15, 1456305. [Google Scholar] [CrossRef]
- Ren, P.; Lu, L.; Cai, S.; Chen, J.; Lin, W.; Han, F. Alternative Splicing: A New Cause and Potential Therapeutic Target in Autoimmune Disease. Front. Immunol. 2021, 12, 713540, Correction in Front. Immunol. 2024, 15, 1513491. [Google Scholar] [CrossRef]
- Zhang, D.; Yue, Y.; Yuan, C.; An, X.; Guo, T.; Chen, B.; Liu, J.; Lu, Z. DIA-Based Proteomic Analysis Reveals MYOZ2 as a Key Protein Affecting Muscle Growth and Development in Hybrid Sheep. Int. J. Mol. Sci. 2024, 25, 2975. [Google Scholar] [CrossRef]
- Huang, Y.; Yuan, C.; Zhao, Y.; Li, C.; Cao, M.; Li, H.; Zhao, Z.; Sun, A.; Basang, W.; Zhu, Y.; et al. Identification and Regulatory Network Analysis of Genes Related to Reproductive Performance in the Hypothalamus and Pituitary of Angus Cattle. Genes 2022, 13, 965. [Google Scholar] [CrossRef]
- National Research Council. Nutrient Requirements of Beef Cattle: Seventh Revised Edition: Update 2000; The National Academies Press: Washington, DC, USA, 1996. [Google Scholar] [CrossRef]
- Lemley, C.O.; Meyer, A.M.; Camacho, L.E.; Neville, T.L.; Newman, D.J.; Caton, J.S.; Vonnahme, K.A. Melatonin Supplementation Alters Uteroplacental Hemodynamics and Fetal Development in an Ovine Model of Intrauterine Growth Restriction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Correa, Z.E.; Cochran, T.; Metcalfe, A.; Burnett, D.D.; Lemley, C.O. Seasonal and Temporal Variation in the Placenta during Melatonin Supplementation in a Bovine Compromised Pregnancy Model. J. Anim. Sci. 2022, 100, skac372. [Google Scholar] [CrossRef]
- Patel, H.; Ewels, P.; Peltzer, A.; Hammarén, R.; Botvinnik, O.; Sturm, G.; Moreno, D.; Vemuri, P.; silviamorins; Pantano, L.; et al. Nf-Core/Rnaseq: Nf-Core/Rnaseq v3.0—Silver Shark; Zenodo: Geneva, Switzerland, 2020. [Google Scholar] [CrossRef]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow Enables Reproducible Computational Workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality Control of RNA-Seq Experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [PubMed]
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L.M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T.F.; Conesa, A. Qualimap: Evaluating next-Generation Sequencing Alignment Data. Bioinformatics 2012, 28, 2678–2679. [Google Scholar] [CrossRef]
- Sayols, S.; Scherzinger, D.; Klein, H. DupRadar: A Bioconductor Package for the Assessment of PCR Artifacts in RNA-Seq Data. BMC Bioinform. 2016, 17, 428. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Prabhakar, L.; Davis G, D.J. Meta-Analysis of Lean and Obese RNA-Seq Datasets to Identify Genes Targeting Obesity. Bioinformation 2023, 19, 331. [Google Scholar] [CrossRef] [PubMed]
- Venny, O.J.C. An Interactive Tool for Comparing Lists with Venn Diagrams. 2007. Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 18 November 2025).
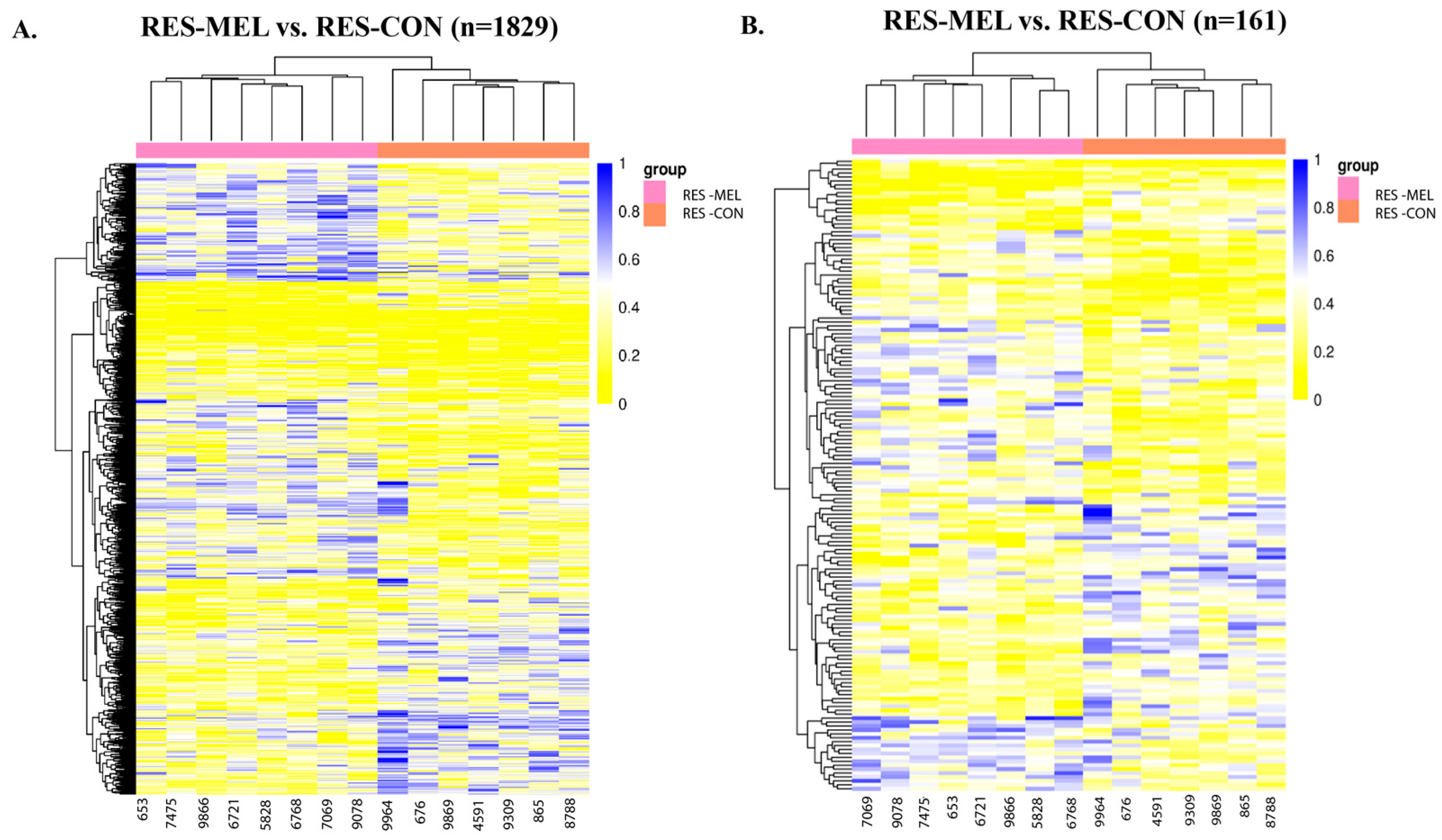
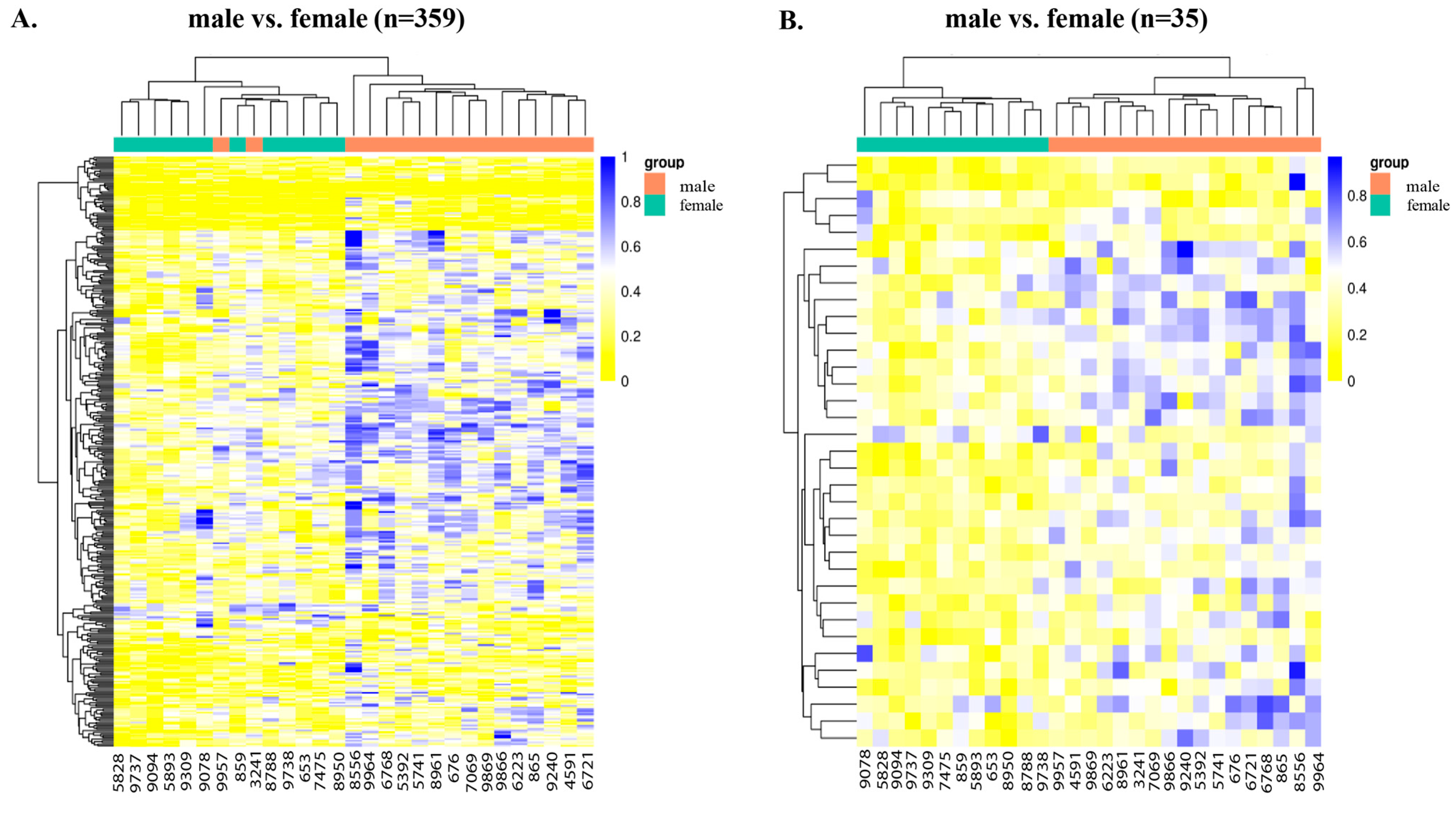
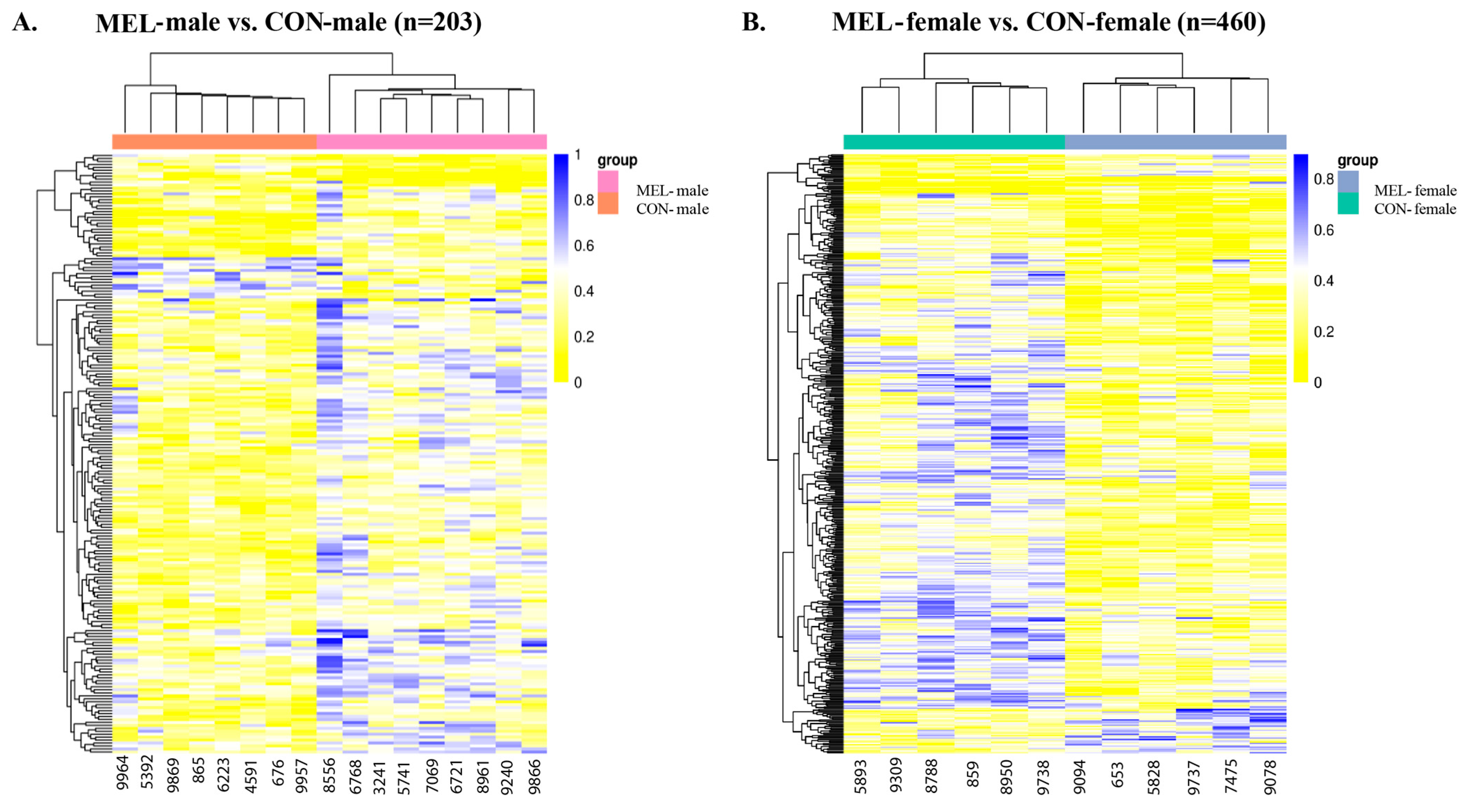


| Group | Methylation Change | Adjusted p-Value | Top Differentially Methylated Genes |
|---|---|---|---|
| RES-CON vs. ADQ-CON | Hypomethylated | padj < 0.05 | PLEKHM2, KLF15, MCM2, SLC22A23, ZNF691, ITFG2, TNPO2, HCFC1, FLNA |
| Hypermethylated | LEFTY2, COX19, COL27A, MIR455, CHAF1B, U2AF2, KDM8 | ||
| ADQ-MEL vs. ADQ-CON | Hypomethylated | padj < 0.05 | CUL4A, INPP4A, TSNARE1, LEFTY2, DAG1, KDM1B, LAMA5, APMAP, PLEKHM1, MSANTD1 |
| Hypermethylated | DNMT3A, CHD7, CHCD5, MYBL2, HDAC4, LAMB1, FLNA, ZCCHC13 | ||
| RES-MEL vs. RES-CON | Hypomethylated | padj < 0.05 | WNT7B, NRDE2, CD93, MIR411A, MIR299, BCL2, ADARB1, AHDC1, ZNF275 |
| Hypermethylated | EZH1, PIK3R1, PLCL2, CUL4A, SPATA13, TPRA1, HCFC1, LAMP1 | ||
| RES-MEL vs. ADQ-MEL | Hypomethylated | padj < 0.05 | ADAP1, PLXND1, HDAC7, SL12A7, PXDN, LAMP1, MAML3, MAD1L1, KCTD7, MYO10 |
| Hypermethylated | OVOL1, ATG7, CYP26C1, LAMP1, IRF8, CEPI12, ATP9, ITBCID5, KIF26A | ||
| Male vs. female | Hypomethylated | padj < 0.05 | ACAD8, ZNF532 |
| Hypermethylated | CAMSAP1, MAP2K4, TBCID13, EZH1, PLEKHA6, ZNF687, PAWR, ASTN2, LMNB2 | ||
| MEL-male vs. CON-male | Hypomethylated | padj < 0.05 | BOP1, TPRA1, PKD1, USP12, EIPR1, FGF14, AP1B1, FGF18, AHDC1, MAD1L1 |
| Hypermethylated | CUL1, LRP5, MED15, HDAC4, PI4KA, NNAT, ADAD2, TAFIC, MYO18A, CAMK2A, MYOM3 | ||
| MEL-female vs. CON-female | Hypomethylated | padj < 0.05 | GREB1L, EEFSEC, ZBED4, ADARB1, HSF5, SLC22A23, PYGM, TMEM185B, TRIM10 |
| Hypermethylated | RREB1, KIF25, MIR2309, LETM1, OSGIN1, CNST |
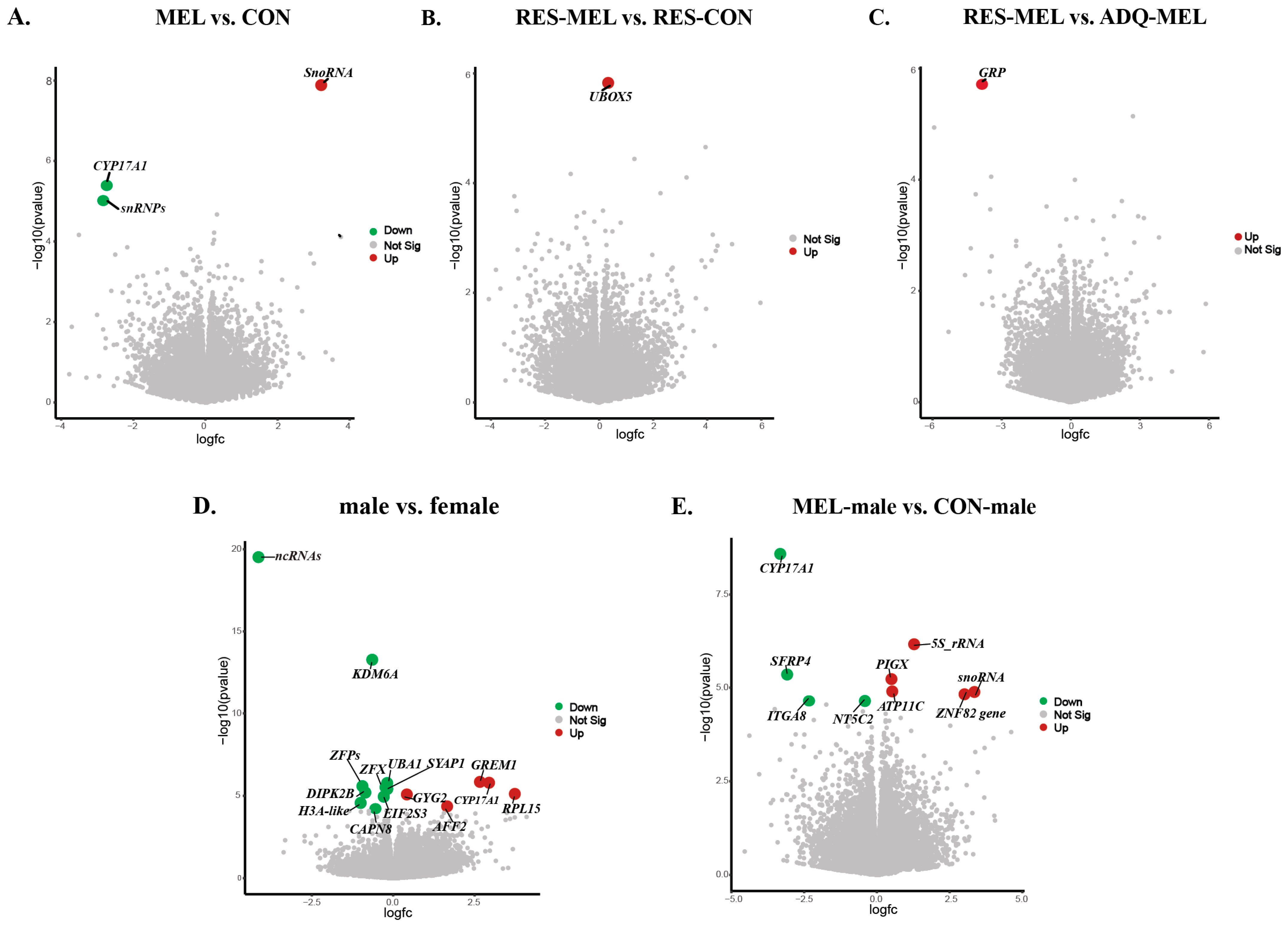
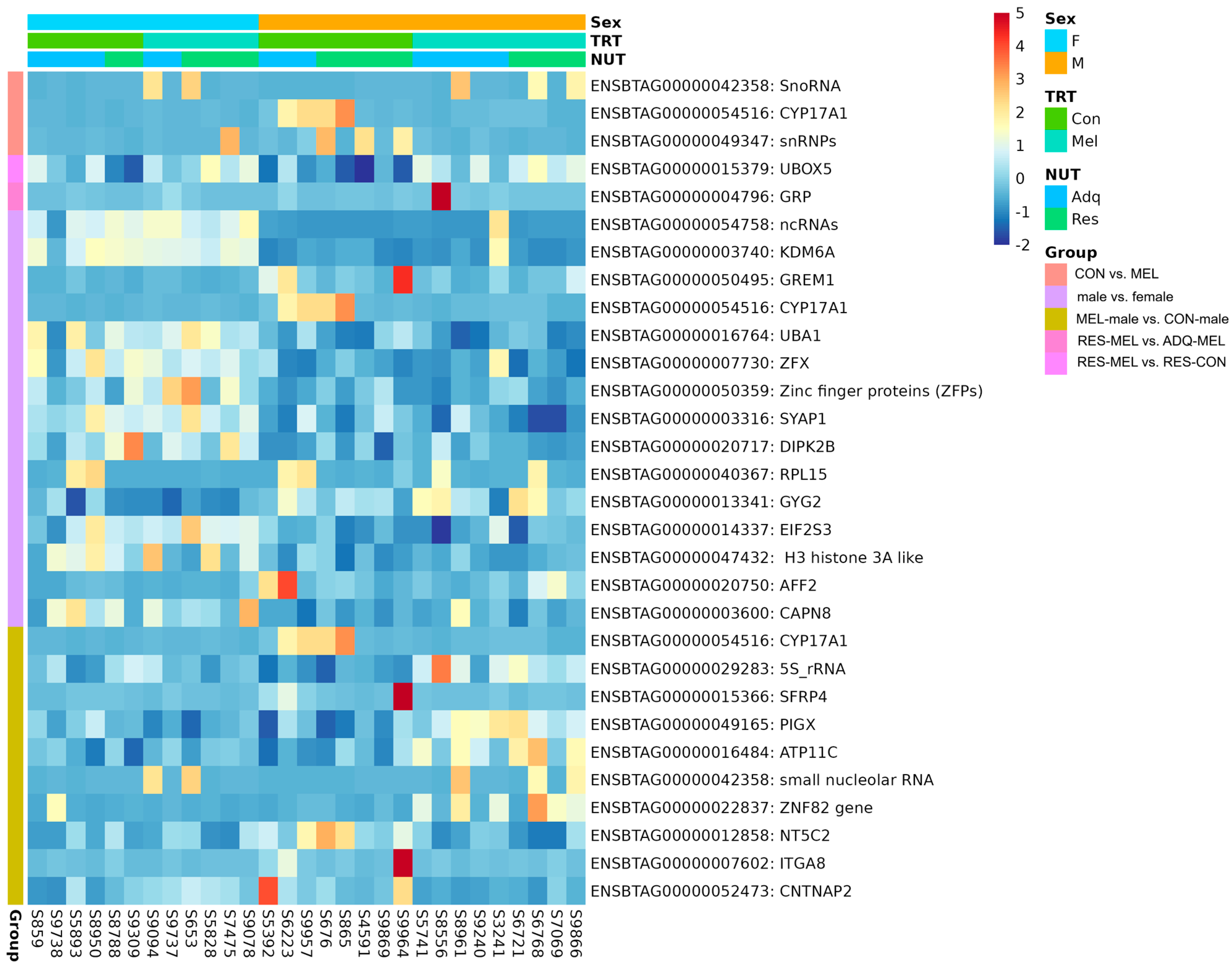
| Group | Gene ID | Gene Name | log2FoldChange | padj | Gene Expression |
|---|---|---|---|---|---|
| MEL vs. CON | ENSBTAG00000042358 | SnoRNA | 3.270511 | 0.000273 | Upregulated |
| ENSBTAG00000054516 | CYP17A1 | −2.73751 | 0.049874 | Downregulated | |
| RES-MEL vs. RES-CON | ENSBTAG00000015379 | UBOX5 | 0.334699 | 0.035745 | Upregulated |
| RES-MEL vs. ADQ-MEL | ENSBTAG00000004796 | GRP | −3.85346 | 0.042907 | Downregulated |
| Male vs. female | ENSBTAG00000054758 | ncRNAs | −4.13781 | 4.77 × 10−16 | Downregulated |
| ENSBTAG00000003740 | KDM6A | −0.64488 | 4.2 × 10−10 | Downregulated | |
| ENSBTAG00000050495 | GREM1 | 2.662567 | 0.005307 | Upregulated | |
| ENSBTAG00000054516 | CYP17A1 | 2.942639 | 0.005307 | Upregulated | |
| ENSBTAG00000016764 | UBA1 | −0.18675 | 0.005307 | Downregulated | |
| ENSBTAG00000007730 | ZFX | −0.23691 | 0.007499 | Downregulated | |
| ENSBTAG00000050359 | ZFPs | −0.87648 | 0.007499 | Downregulated | |
| ENSBTAG00000003316 | SYAP1 | −0.18178 | 0.007499 | Downregulated | |
| ENSBTAG00000020717 | DIPK2B | −0.85649 | 0.011048 | Downregulated | |
| ENSBTAG00000040367 | RPL15 | 3.733833 | 0.011802 | Upregulated | |
| ENSBTAG00000013341 | GYG2 | 0.41884 | 0.011802 | Upregulated | |
| ENSBTAG00000014337 | EIF2S3 | −0.29056 | 0.014704 | Downregulated | |
| ENSBTAG00000047432 | H3A-like | −1.00082 | 0.032569 | Downregulated | |
| ENSBTAG00000020750 | AFF2 | 1.655051 | 0.049011 | Upregulated | |
| ENSBTAG00000003600 | CAPN8 | −0.54618 | 0.050001 | Downregulated | |
| MEL-male vs. CON-male | ENSBTAG00000054516 | CYP17A1 | −3.32335 | 5.04 × 10−5 | Downregulated |
| ENSBTAG00000029283 | 5S_rRNA | 1.278478 | 0.006595 | Upregulated | |
| ENSBTAG00000015366 | SFRP4 | −3.08592 | 0.028455 | Downregulated | |
| ENSBTAG00000049165 | PIGX | 0.499777 | 0.028455 | Upregulated | |
| ENSBTAG00000016484 | ATP11C | 0.527126 | 0.041514 | Upregulated | |
| ENSBTAG00000042358 | snoRNA | 3.351421 | 0.041514 | Upregulated | |
| ENSBTAG00000022837 | ZNF82 | 3.011589 | 0.041514 | Upregulated | |
| ENSBTAG00000012858 | NT5C2 | −0.41284 | 0.049942 | Downregulated | |
| ENSBTAG00000007602 | ITGA8 | −2.33427 | 0.049942 | Downregulated | |
| ENSBTAG00000052473 | CNTNAP2 | −1.74072 | 0.054788 | Downregulated |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajput, S.; Littlejohn, B.; Contreras-Correa, Z.E.; El Daous, H.; Sidelinger, D.; King, H.; Arick, M.; Lemley, C. Maternal Melatonin Supplementation Modulates Placental DNA Methylation and Gene Expression in Nutrient-Restricted Cattle. Int. J. Mol. Sci. 2025, 26, 11387. https://doi.org/10.3390/ijms262311387
Rajput S, Littlejohn B, Contreras-Correa ZE, El Daous H, Sidelinger D, King H, Arick M, Lemley C. Maternal Melatonin Supplementation Modulates Placental DNA Methylation and Gene Expression in Nutrient-Restricted Cattle. International Journal of Molecular Sciences. 2025; 26(23):11387. https://doi.org/10.3390/ijms262311387
Chicago/Turabian StyleRajput, Shiveeli, Brittni Littlejohn, Zully E. Contreras-Correa, Hala El Daous, Darcie Sidelinger, Heath King, Mark Arick, and Caleb Lemley. 2025. "Maternal Melatonin Supplementation Modulates Placental DNA Methylation and Gene Expression in Nutrient-Restricted Cattle" International Journal of Molecular Sciences 26, no. 23: 11387. https://doi.org/10.3390/ijms262311387
APA StyleRajput, S., Littlejohn, B., Contreras-Correa, Z. E., El Daous, H., Sidelinger, D., King, H., Arick, M., & Lemley, C. (2025). Maternal Melatonin Supplementation Modulates Placental DNA Methylation and Gene Expression in Nutrient-Restricted Cattle. International Journal of Molecular Sciences, 26(23), 11387. https://doi.org/10.3390/ijms262311387







