Wounds and the Microbiota: The Healing Interplay Between Host and Microbial Communities
Abstract
1. Introduction
2. Anatomy and Physiology of the Skin
2.1. The Epidermis
2.2. The Dermis
2.3. The Hypodermis
2.4. Functional Integration
3. Phases of Wound Healing: Cellular Dynamics and Microbiome Modulation [50,51]
3.1. Hemostasis: Platelet Activation and Microbial Sensing
3.2. Inflammation: Cytokine Regulation and Biofilm-Mediated Persistence
3.3. Proliferation: Fibroblast Activation and Microbial-Derived Mediators
3.4. Remodeling: ECM Turnover and Microbial Resolution
4. Chronic Wound Healing and the Skin Microbiome
4.1. Chronic Wound Healing: Pathophysiological Context
4.2. The Skin and Wound Microbiome Composition
4.2.1. Commensal Communities and Protective Roles
4.2.2. Dysbiosis and Pathogenic Shifts
4.2.3. Functional Interplay and Spatial Zonation
4.3. Virome and Mycobiome Contributions
5. Molecular Mechanisms of Microbiota-Mediated Wound Modulation
5.1. Pattern-Recognition Receptor Signaling
5.2. Quorum Sensing and Biofilm-Driven Pathogenicity
5.3. Antimicrobial Peptides and Host–Microbe Equilibrium
5.4. Reactive Oxygen Species (ROS) and Redox Signaling
5.5. Immunometabolic and Hypoxic Adaptations
5.6. Integrative View
6. Wound Healing and Skin Microbiota
6.1. The Abundant Bacteria Implicated in Wounds
6.2. Coagulase-Negative Staphylococci (CoNS)
6.2.1. Staphylococcus epidermidis
6.2.2. Staphylococcus hominis
6.2.3. Staphylococcus aureus
6.2.4. Pseudomonas aeruginosa
6.2.5. Streptococcus pyogenes
6.2.6. Lactobacilli
6.2.7. Lactobacillus plantarum
6.2.8. Escherichia coli
6.3. Other Bacteria Implicated in Wounds
Enterococcus faecalis
6.4. Relationship Between Skin Microbiome and Wound Healing
6.5. Interaction Between Perforin-2 and Skin Microbiota in Wound Healing
7. Therapeutic and Diagnostic Applications of the Wound Microbiome
7.1. Probiotics and Postbiotics
7.2. Quorum-Sensing Inhibitors and Anti-Biofilm Strategies
7.3. Bacteriophage-Based Therapeutics
7.4. Engineered Antimicrobial Peptides and Peptidomimetics
7.5. Microbiome-Informed Diagnostics and Omics-Driven Stratification
7.6. Integration into Clinical Practice
8. Biofilms
9. Analytical Advances, Methodological Challenges, and Future Perspectives in Wound–Microbiome Research
9.1. Overview of Microbiome Identification Techniques
9.2. Culture-Dependent Methods and Culturomics
9.3. Metagenomic Sequencing
9.4. Next-Generation Sequencing and Its Applications
9.5. Whole Genome Sequencing
9.6. Whole 16S rRNA Gene Sequencing
9.7. Technical and Methodological Constraints
9.8. Biological Complexity and Host Variability
9.9. Translational and Clinical Barriers
9.10. Future Directions
- Longitudinal multi-omics studies correlating microbial dynamics with host immune and metabolic signatures.
- Functional modeling using 3-D organotypic skin systems and humanized-microbiota animal models to define causality.
- Systems-biology frameworks integrating microbial networks, metabolite fluxes, and host signaling pathways.
- Precision therapeutics, including synthetic microbial consortia, bioengineered peptides, and immunometabolic modulators that restore ecological balance.
- AI-driven diagnostics combining clinical and microbiome data to stratify chronic-wound patients by risk and treatment responsiveness [103].
10. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Canchy, L.; Kerob, D.; Demessant, A.L.; Amici, J.M. Wound healing and microbiome, an unexpected relationship. J. Eur. Acad. Dermatol. Venereol. 2023, 37, 7–15. [Google Scholar] [CrossRef]
- Raziyeva, K.; Kim, Y.; Zharkinbekov, Z.; Kassymbek, K.; Jimi, S.; Saparov, A. Immunology of Acute and Chronic Wound Healing. Biomolecules 2021, 11, 700. [Google Scholar] [CrossRef]
- Coerper, S.; Beckert, S.; Küper, M.A.; Jekov, M.; Königsrainer, A. Fifty percent area reduction after 4 weeks of treatment is a reliable indicator for healing—Analysis of a single-center cohort of 704 diabetic patients. J. Diabetes Its Complicat. 2009, 23, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Leaper, D.J.; Durani, P. Topical antimicrobial therapy of chronic wounds healing by secondary intention using iodine products. Int. Wound J. 2008, 5, 361–368. [Google Scholar] [CrossRef]
- Goh, O.Q.; Ganesan, G.; Graves, N.; Ng, Y.Z.; Harding, K.; Tan, K.B. Incidence of chronic wounds in Singapore, a multiethnic Asian country, between 2000 and 2017: A retrospective cohort study using a nationwide claims database. BMJ Open 2020, 10, e039411. [Google Scholar] [CrossRef] [PubMed]
- Martinengo, L.; Olsson, M.; Bajpai, R.; Soljak, M.; Upton, Z.; Schmidtchen, A.; Car, J.; Järbrink, K. Prevalence of chronic wounds in the general population: Systematic review and meta-analysis of observational studies. Ann. Epidemiol. 2019, 29, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; DiPietro, L.A. Factors Affecting Wound Healing. J. Dent. Res. 2010, 89, 219–229. [Google Scholar] [CrossRef]
- Coleman, S.; Nelson, E.A.; Vowden, P.; Vowden, K.; Adderley, U.; Sunderland, L.; Harker, J.; Conroy, T.; Fiori, S.; Bezer, N.; et al. Development of a generic wound care assessment minimum data set. J. Tissue Viability 2017, 26, 226–240. [Google Scholar] [CrossRef]
- Xu, Z.; Hsia, H.C. The Impact of Microbial Communities on Wound Healing: A Review. Ann. Plast. Surg. 2018, 81, 113–123. [Google Scholar] [CrossRef]
- Borrego-Ruiz, A.; Borrego, J.J. Microbial Dysbiosis in the Skin Microbiome and Its Psychological Consequences. Microorganisms 2024, 12, 1908. [Google Scholar] [CrossRef]
- Zielińska, M.; Pawłowska, A.; Orzeł, A.; Sulej, L.; Muzyka-Placzyńska, K.; Baran, A.; Filipecka-Tyczka, D.; Pawłowska, P.; Nowińska, A.; Bogusławska, J.; et al. Wound Microbiota and Its Impact on Wound Healing. Int. J. Mol. Sci. 2023, 24, 17318. [Google Scholar] [CrossRef] [PubMed]
- Kanitakis, J. Anatomy, histology and immunohistochemistry of normal human skin. Eur. J. Dermatol. 2002, 12, 390–399, quiz 400-391. [Google Scholar]
- Oláh, A.; Szöllősi, A.G.; Bíró, T. The channel physiology of the skin. Rev. Physiol. Biochem. Pharmacol. 2012, 163, 65–131. [Google Scholar] [CrossRef]
- Venus, M.; Waterman, J.; McNab, I. Basic physiology of the skin. Surg. Oxf. Int. Ed. 2010, 28, 469–472. [Google Scholar] [CrossRef]
- Yousef, H.; Alhajj, M.; Fakoya, A.O.; Sharma, S. Anatomy, Skin (Integument), Epidermis. In StatPearls; Copyright© 2025; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2025. [Google Scholar]
- Lotfollahi, Z. The anatomy, physiology and function of all skin layers and the impact of ageing on the skin. Wound Pract. Res. J. Aust. Wound Manag. Assoc. 2024, 32, 6–10. [Google Scholar] [CrossRef]
- Lebre, M.C.; Van Der Aar, A.M.; Van Baarsen, L.; Van Capel, T.M.; Schuitemaker, J.H.; Kapsenberg, M.L.; de Jong, E.C. Human keratinocytes express functional Toll-like receptor 3, 4, 5, and 9. J. Investig. Dermatol. 2007, 127, 331–341. [Google Scholar] [CrossRef]
- Lai, Y.; Cogen, A.L.; Radek, K.A.; Park, H.J.; Macleod, D.T.; Leichtle, A.; Ryan, A.F.; Di Nardo, A.; Gallo, R.L. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J. Investig. Dermatol. 2010, 130, 2211–2221. [Google Scholar] [CrossRef] [PubMed]
- Piipponen, M.; Li, D.; Landén, N.X. The Immune Functions of Keratinocytes in Skin Wound Healing. Int. J. Mol. Sci. 2020, 21, 8790. [Google Scholar] [CrossRef]
- Chessa, C.; Bodet, C.; Jousselin, C.; Wehbe, M.; Lévêque, N.; Garcia, M. Antiviral and immunomodulatory properties of antimicrobial peptides produced by human keratinocytes. Front. Microbiol. 2020, 11, 1155. [Google Scholar] [CrossRef]
- Song, P.I.; Neparidze, N.; Armstrong, C.A.; Ansel, J.C.; Park, Y.-M.; Abraham, T.; Harten, B.; Zivony, A. Human Keratinocytes Express Functional CD14 and Toll-Like Receptor 4. J. Investig. Dermatol. 2002, 119, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Morizane, S.; Mukai, T.; Sunagawa, K.; Tachibana, K.; Kawakami, Y.; Ouchida, M. “Input/output cytokines” in epidermal keratinocytes and the involvement in inflammatory skin diseases. Front. Immunol. 2023, 14, 1239598. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Di Nardo, A.; Nakatsuji, T.; Leichtle, A.; Yang, Y.; Cogen, A.L.; Wu, Z.R.; Hooper, L.V.; Schmidt, R.R.; von Aulock, S.; et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. 2009, 15, 1377–1382. [Google Scholar] [CrossRef]
- Cogen, A.L.; Yamasaki, K.; Sanchez, K.M.; Dorschner, R.A.; Lai, Y.; MacLeod, D.T.; Torpey, J.W.; Otto, M.; Nizet, V.; Kim, J.E.; et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Investig. Dermatol. 2010, 130, 192–200. [Google Scholar] [CrossRef]
- Coates, M.; Blanchard, S.; MacLeod, A.S. Innate antimicrobial immunity in the skin: A protective barrier against bacteria, viruses, and fungi. PLoS Pathog. 2018, 14, e1007353. [Google Scholar] [CrossRef]
- Kan, C.; Abe, M.; Yamanaka, M.; Ishikawa, O. Hypoxia-induced increase of matrix metalloproteinase-1 synthesis is not restored by reoxygenation in a three-dimensional culture of human dermal fibroblasts. J. Dermatol. Sci. 2003, 32, 75–82. [Google Scholar] [CrossRef]
- Zheng, H.; Cheng, X.; Jin, L.; Shan, S.; Yang, J.; Zhou, J. Recent advances in strategies to target the behavior of macrophages in wound healing. Biomed. Pharmacother. 2023, 165, 115199. [Google Scholar] [CrossRef]
- Rodrigues, M.; Kosaric, N.; Bonham, C.A.; Gurtner, G.C. Wound Healing: A Cellular Perspective. Physiol. Rev. 2019, 99, 665–706. [Google Scholar] [CrossRef]
- Hong, H.; Tian, X.Y. The Role of Macrophages in Vascular Repair and Regeneration after Ischemic Injury. Int. J. Mol. Sci. 2020, 21, 6328. [Google Scholar] [CrossRef]
- Jetten, N.; Verbruggen, S.; Gijbels, M.J.; Post, M.J.; De Winther, M.P.; Donners, M.M. Anti-inflammatory M2, but not pro-inflammatory M1 macrophages promote angiogenesis in vivo. Angiogenesis 2014, 17, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Spiller, K.L.; Anfang, R.R.; Spiller, K.J.; Ng, J.; Nakazawa, K.R.; Daulton, J.W.; Vunjak-Novakovic, G. The role of macrophage phenotype in vascularization of tissue engineering scaffolds. Biomaterials 2014, 35, 4477–4488. [Google Scholar] [CrossRef] [PubMed]
- Namour, F.; Galien, R.; Van Kaem, T.; Van der Aa, A.; Vanhoutte, F.; Beetens, J.; Van ‘t Klooster, G. Safety, Pharmacokinetics and Pharmacodynamics of GLPG0974, a potent and selective FFA2 antagonist, in healthy male subjects. Br. J. Clin. Pharmacol. 2016, 82, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Krzyszczyk, P.; Schloss, R.; Palmer, A.; Berthiaume, F. The Role of Macrophages in Acute and Chronic Wound Healing and Interventions to Promote Pro-wound Healing Phenotypes. Front. Physiol. 2018, 9, 419. [Google Scholar] [CrossRef] [PubMed]
- Botusan, I.R.; Sunkari, V.G.; Savu, O.; Catrina, A.I.; Grünler, J.; Lindberg, S.; Pereira, T.; Ylä-Herttuala, S.; Poellinger, L.; Brismar, K.; et al. Stabilization of HIF-1alpha is critical to improve wound healing in diabetic mice. Proc. Natl. Acad. Sci. USA 2008, 105, 19426–19431. [Google Scholar] [CrossRef]
- Tareen, S.H.K.; Kutmon, M.; Adriaens, M.E.; Mariman, E.C.M.; de Kok, T.M.; Arts, I.C.W.; Evelo, C.T. Exploring the cellular network of metabolic flexibility in the adipose tissue. Genes Nutr. 2018, 13, 17. [Google Scholar] [CrossRef]
- Lin, Y.; Lee, H.; Berg, A.H.; Lisanti, M.P.; Shapiro, L.; Scherer, P.E. The Lipopolysaccharide-activated Toll-like Receptor (TLR)-4 Induces Synthesis of the Closely Related Receptor TLR-2 in Adipocytes *. J. Biol. Chem. 2000, 275, 24255–24263. [Google Scholar] [CrossRef]
- Loffreda, S.; Yang, S.Q.; Lin, H.Z.; Karp, C.L.; Brengman, M.L.; Wang, D.J.; Klein, A.S.; Bulkley, G.B.; Bao, C.; Noble, P.W.; et al. Leptin regulates proinflammatory immune responses. Faseb J. 1998, 12, 57–65. [Google Scholar] [CrossRef]
- Liu, L.; Yu, J.; Liu, Y.; Xie, L.; Hu, F.; Liu, H. Hypoxia-driven angiogenesis and metabolic reprogramming in vascular tumors. Front. Cell Dev. Biol. 2025, 13, 1572909. [Google Scholar] [CrossRef]
- Nie, J.; Fu, X.; Han, W. Microenvironment-dependent homeostasis and differentiation of epidermal basal undifferentiated keratinocytes and their clinical applications in skin repair. J. Eur. Acad. Dermatol. Venereol. 2013, 27, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Konieczny, P.; Xing, Y.; Sidhu, I.; Subudhi, I.; Mansfield, K.P.; Hsieh, B.; Biancur, D.E.; Larsen, S.B.; Cammer, M.; Li, D.; et al. Interleukin-17 governs hypoxic adaptation of injured epithelium. Science 2022, 377, eabg9302. [Google Scholar] [CrossRef]
- Wang, G.; Sweren, E.; Andrews, W.; Li, Y.; Chen, J.; Xue, Y.; Wier, E.; Alphonse, M.P.; Luo, L.; Miao, Y.; et al. Commensal microbiome promotes hair follicle regeneration by inducing keratinocyte HIF-1α signaling and glutamine metabolism. Sci. Adv. 2023, 9, eabo7555. [Google Scholar] [CrossRef] [PubMed]
- Sanford, J.A.; Gallo, R.L. Functions of the skin microbiota in health and disease. Semin Immunol. 2013, 25, 370–3777. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chiang, H.-I.; Jiang, S.B.; Nagarajan, H.; Zengler, K.; Gallo, R.L. The microbiome extends to subepidermal compartments of normal skin. Nat Commun. 2013, 4, 1431. [Google Scholar] [CrossRef]
- Tokudome, Y. Influence of oral administration of lactic acid bacteria metabolites on skin barrier function and water content in a murine model of atopic dermatitis. Nutrients 2018, 10, 1858. [Google Scholar] [CrossRef]
- Fitz-Gibbon, S.; Tomida, S.; Chiu, B.-H.; Nguyen, L.; Du, C.; Liu, M.; Elashoff, D.; Erfe, M.C.; Loncaric, A.; Kim, J.; et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Investig. Dermatol. 2013, 133, 2152–2160. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253, Erratum in Nat. Rev. Microbiol. 2011, 9, 626. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Cosseau, C.; Romano-Bertrand, S.; Duplan, H.; Lucas, O.; Ingrassia, I.; Pigasse, C.; Roques, C.; Jumas-Bilak, E. Proteobacteria from the human skin microbiota: Species-level diversity and hypotheses. One Health 2016, 2, 33–41. [Google Scholar] [CrossRef]
- Torrens, C.; Marí, R.; Alier, A.; Puig, L.; Santana, F.; Corvec, S. Cutibacterium acnes in primary reverse shoulder arthroplasty: From skin to deep layers. J. Shoulder Elb. Surg. 2019, 28, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Fazli, M.; Bjarnsholt, T.; Kirketerp-Møller, K.; Jørgensen, B.; Andersen, A.S.; Krogfelt, K.A.; Givskov, M.; Tolker-Nielsen, T. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 2009, 47, 4084–4089. [Google Scholar] [CrossRef]
- Barnes, C.J.; Asplund, M.; Clausen, M.-L.; Rasmussen, L.; Olesen, C.M.; Yüksel, Y.T.; Andersen, P.S.; Litman, T.; Holmstrøm, K.; Bay, L.; et al. A simplified bacterial community found within the epidermis than at the epidermal surface of atopic dermatitis patients and healthy controls. BMC Microbiol. 2023, 23, 273. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.X.; Zhang, L.-J.; Gallo, R.L. Dermal White Adipose Tissue: A Newly Recognized Layer of Skin Innate Defense. J. Investig. Dermatol. 2019, 139, 1002–1009. [Google Scholar] [CrossRef]
- Zhang, L.-J.; Guerrero-Juarez, C.F.; Hata, T.; Bapat, S.P.; Ramos, R.; Plikus, M.V.; Gallo, R.L. Innate immunity. Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science 2015, 347, 67–71. [Google Scholar] [CrossRef]
- Pang, M.; Yao, Z.; Chen, C.; Lei, X.; Cheng, B. Human-microorganism mutualism theory: Possible mechanisms for the delayed chronic wound healing process. Med. Hypotheses 2020, 141, 109720. [Google Scholar] [CrossRef]
- Meddahi-Pellé, A.; Legrand, A.; Marcellan, A.; Louedec, L.; Letourneur, D.; Leibler, L. Organ repair, hemostasis, and in vivo bonding of medical devices by aqueous solutions of nanoparticles. Angew. Chem. Int. Ed. Engl. 2014, 53, 6369–6373. [Google Scholar] [CrossRef]
- Mamun, A.A.; Shao, C.; Geng, P.; Wang, S.; Xiao, J. Recent advances in molecular mechanisms of skin wound healing and its treatments. Front. Immunol. 2024, 15, 1395479. [Google Scholar] [CrossRef]
- Gillitzer, R.; Goebeler, M. Chemokines in cutaneous wound healing. J. Leukoc. Biol. 2001, 69, 513–521. [Google Scholar] [CrossRef]
- Pasparakis, M.; Haase, I.; Nestle, F.O. Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 2014, 14, 289–301. [Google Scholar] [CrossRef]
- Ellis, S.; Lin, E.J.; Tartar, D. Immunology of Wound Healing. Curr. Dermatol. Rep. 2018, 7, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Rademacher, F.; Simanski, M.; Hesse, B.; Dombrowsky, G.; Vent, N.; Gläser, R.; Harder, J. Staphylococcus epidermidis Activates Aryl Hydrocarbon Receptor Signaling in Human Keratinocytes: Implications for Cutaneous Defense. J. Innate Immun. 2019, 11, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef]
- Kaisho, T.; Akira, S. Toll-like receptor function and signaling. J. Allergy Clin. Immunol. 2006, 117, 979–987. [Google Scholar] [CrossRef]
- Wilgus, T.A.; Roy, S.; McDaniel, J.C. Neutrophils and Wound Repair: Positive Actions and Negative Reactions. Adv. Wound Care 2013, 2, 379–388. [Google Scholar] [CrossRef]
- Alonso, J.E.; Lee, J.; Burgess, A.R.; Browner, B.D. The management of complex orthopedic injuries. Surg. Clin. N. Am. 1996, 76, 879–903. [Google Scholar] [CrossRef]
- Martins, S.G.; Zilhão, R.; Thorsteinsdóttir, S.; Carlos, A.R. Linking Oxidative Stress and DNA Damage to Changes in the Expression of Extracellular Matrix Components. Front. Genet. 2021, 12, 673002. [Google Scholar] [CrossRef]
- Wang, Z.; Qi, F.; Luo, H.; Xu, G.; Wang, D. Inflammatory Microenvironment of Skin Wounds. Front. Immunol. 2022, 13, 789274. [Google Scholar] [CrossRef]
- Ganesh, G.V.; Ramkumar, K.M. Macrophage mediation in normal and diabetic wound healing responses. Inflamm. Res. 2020, 69, 347–363. [Google Scholar] [CrossRef]
- Smigiel, K.S.; Parks, W.C. Macrophages, wound healing, and fibrosis: Recent insights. Curr. Rheumatol. Rep. 2018, 20, 17. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yang, P.; Gao, M.; Yu, T.; Shi, Y.; Zhang, M.; Yao, M.; Liu, Y.; Zhang, X. NLRP3 activation induced by neutrophil extracellular traps sustains inflammatory response in the diabetic wound. Clin. Sci. 2019, 133, 565–582. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Huang, J.; Li, Y.; Dong, L. Symbiotic bacteria-mediated imbalance and repair of immune homeostasis: Exploring novel mechanisms of microbiome-host interactions in atopic dermatitis. Front. Immunol. 2025, 16, 1649857. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Gambello, M.J.; Iglewski, B.H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 1991, 173, 3000–3009. [Google Scholar] [CrossRef] [PubMed]
- van der Plas, M.J.; Bhongir, R.K.; Kjellström, S.; Siller, H.; Kasetty, G.; Mörgelin, M.; Schmidtchen, A. Pseudomonas aeruginosa elastase cleaves a C-terminal peptide from human thrombin that inhibits host inflammatory responses. Nat. Commun. 2016, 7, 11567. [Google Scholar] [CrossRef] [PubMed]
- Qiu, B.; Yuan, P.; Du, X.; Jin, H.; Du, J.; Huang, Y. Hypoxia inducible factor-1α; is an important regulator of macrophage biology. Heliyon 2023, 9, e17167. [Google Scholar] [CrossRef] [PubMed]
- Trøstrup, H.; Lerche, C.J.; Christophersen, L.J.; Thomsen, K.; Jensen, P.; Hougen, H.P.; Høiby, N.; Moser, C. Pseudomonas aeruginosa biofilm hampers murine central wound healing by suppression of vascular epithelial growth factor. Int. Wound J. 2018, 15, 123–132. [Google Scholar] [CrossRef]
- Lau, K.; Paus, R.; Tiede, S.; Day, P.; Bayat, A. Exploring the role of stem cells in cutaneous wound healing. Exp. Dermatol. 2009, 18, 921–933. [Google Scholar] [CrossRef]
- Mirastschijski, U.; Schnabel, R.; Claes, J.; Schneider, W.; Agren, M.S.; Haaksma, C.; Tomasek, J.J. Matrix metalloproteinase inhibition delays wound healing and blocks the latent transforming growth factor-beta1-promoted myofibroblast formation and function. Wound Repair Regen. 2010, 18, 223–234. [Google Scholar] [CrossRef]
- Mirastschijski, U.; Jokuszies, A.; Vogt, P. Skin Wound Healing: Repair Biology, Wound and Scar Treatment; Elsevier Saunders: Philadelphia, PA, USA, 2012; pp. 267–296. [Google Scholar]
- Sinno, H.; Prakash, S. Complements and the Wound Healing Cascade: An Updated Review. Plast. Surg. Int. 2013, 2013, 146764. [Google Scholar] [CrossRef]
- Wynn, T.A.; Ramalingam, T.R. Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat. Med. 2012, 18, 1028–1040. [Google Scholar] [CrossRef]
- Rockey, D.C.; Bell, P.D.; Hill, J.A. Fibrosis—A common pathway to organ injury and failure. N. Engl. J. Med. 2015, 372, 1138–1149. [Google Scholar] [CrossRef]
- D’Urso, M.; Kurniawan, N.A. Mechanical and Physical Regulation of Fibroblast–Myofibroblast Transition: From Cellular Mechanoresponse to Tissue Pathology. Front. Bioeng. Biotechnol. 2020, 8, 609653. [Google Scholar] [CrossRef]
- Li, Y.; Reddy, M.A.; Miao, F.; Shanmugam, N.; Yee, J.K.; Hawkins, D.; Ren, B.; Natarajan, R. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J. Biol. Chem. 2008, 283, 26771–26781. [Google Scholar] [CrossRef]
- Landén, N.X.; Li, D.; Ståhle, M. Transition from inflammation to proliferation: A critical step during wound healing. Cell. Mol. Life Sci. 2016, 73, 3861–3885. [Google Scholar] [CrossRef]
- Tai, Y.; Woods, E.; Dally, J.; Kong, D.; Steadman, R.; Moseley, R.; Midgley, A. Myofibroblasts: Function, Formation, and Scope of Molecular Therapies for Skin Fibrosis. Biomolecules 2021, 11, 1095. [Google Scholar] [CrossRef]
- Darby, I.A.; Laverdet, B.; Bonté, F.; Desmoulière, A. Fibroblasts and myofibroblasts in wound healing. Clin. Cosmet. Investig. Dermatol. 2014, 7, 301–311. [Google Scholar] [CrossRef]
- Barrientos, S.; Stojadinovic, O.; Golinko, M.S.; Brem, H.; Tomic-Canic, M. Growth factors and cytokines in wound healing. Wound Repair. Regen. 2008, 16, 585–601. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.-j.; Fan, M.; Gao, W. Identification of potential hub genes associated with skin wound healing based on time course bioinformatic analyses. BMC Surg. 2021, 21, 303. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Guarino, M.; Hernández-Bule, M.L.; Bacci, S. Cellular and Molecular Processes in Wound Healing. Biomedicines 2023, 11, 2526. [Google Scholar] [CrossRef]
- Tyavambiza, C.; Meyer, M.; Meyer, S. Cellular and molecular events of wound healing and the potential of silver based nanoformulations as wound healing agents. Bioengineering 2022, 9, 712. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Kong, H.H.; Oh, J. State of residency: Microbial strain diversity in the skin. J. Investig. Dermatol. 2022, 142, 1260–1264. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, Y.; Li, Y.; Lu, Y.; Xiong, K.; Zhong, Q.; Wang, J. Bacteriophage-Resistant Mutant of Enterococcus faecalis Is Impaired in Biofilm Formation. Front. Microbiol. 2022, 13, 913023. [Google Scholar] [CrossRef]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-E.; Zheng, P.; Ye, S.-Z.; Ma, X.; Liu, E.; Pang, Y.-B.; He, Q.-Y.; Zhang, Y.-X.; Li, W.-Q.; Zeng, J.-H. Microbiome: Role in inflammatory skin diseases. J. Inflamm. Res. 2024, 17, 1057–1082. [Google Scholar] [CrossRef]
- Prasad, A.S.B.; Shruptha, P.; Prabhu, V.; Srujan, C.; Nayak, U.Y.; Anuradha, C.K.R.; Ramachandra, L.; Keerthana, P.; Joshi, M.B.; Murali, T.S.; et al. Pseudomonas aeruginosa virulence proteins pseudolysin and protease IV impede cutaneous wound healing. Lab. Investig. 2020, 100, 1532–1550. [Google Scholar] [CrossRef]
- Singh, S.; Young, A.; McNaught, C.-E. The physiology of wound healing. Surgery 2017, 35, 473–477. [Google Scholar] [CrossRef]
- Hurlow, J.; Bowler, P.G. Acute and chronic wound infections: Microbiological, immunological, clinical and therapeutic distinctions. J. Wound Care 2022, 31, 436–445. [Google Scholar] [CrossRef]
- Wolcott, R.D.; Rhoads, D.D.; Dowd, S.E. Biofilms and chronic wound inflammation. J. Wound Care 2008, 17, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Woo, K.; Ayello, E.A.; Sibbald, R.G. The edge effect: Current therapeutic options to advance the wound edge. Adv. Ski. Wound Care 2007, 20, 99–117. [Google Scholar] [CrossRef]
- Stojadinovic, A.; Carlson, J.W.; Schultz, G.S.; Davis, T.A.; Elster, E.A. Topical advances in wound care. Gynecol. Oncol. 2008, 111, S70–S80. [Google Scholar] [CrossRef] [PubMed]
- Attinger, C.E.; Janis, J.E.; Steinberg, J.; Schwartz, J.; Al-Attar, A.; Couch, K. Clinical approach to wounds: Débridement and wound bed preparation including the use of dressings and wound-healing adjuvants. Plast. Reconstr. Surg. 2006, 117, 72s–109s. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, M. Body of knowledge around the diabetic foot and limb salvage. J. Cardiovasc. Surg. 2012, 53, 605–616. [Google Scholar]
- Wysocki, A.B.; Staiano-Coico, L.; Grinnell, F. Wound fluid from chronic leg ulcers contains elevated levels of metalloproteinases MMP-2 and MMP-9. J. Investig. Dermatol. 1993, 101, 64–68. [Google Scholar] [CrossRef]
- Stanley, A.; Osler, T. Senescence and the healing rates of venous ulcers. J. Vasc. Surg. 2001, 33, 1206–1211. [Google Scholar] [CrossRef]
- Mendez, M.V.; Raffetto, J.D.; Phillips, T.; Menzoian, J.O.; Park, H.Y. The proliferative capacity of neonatal skin fibroblasts is reduced after exposure to venous ulcer wound fluid: A potential mechanism for senescence in venous ulcers. J. Vasc. Surg. 1999, 30, 734–743. [Google Scholar] [CrossRef]
- Fivenson, D.P.; Faria, D.T.; Nickoloff, B.J.; Poverini, P.J.; Kunkel, S.; Burdick, M.; Strieter, R.M. Chemokine and inflammatory cytokine changes during chronic wound healing. Wound Repair Regen. 1997, 5, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Harris, I.R.; Yee, K.C.; Walters, C.E.; Cunliffe, W.J.; Kearney, J.N.; Wood, E.J.; Ingham, E. Cytokine and protease levels in healing and non-healing chronic venous leg ulcers. Exp. Dermatol. 1995, 4, 342–349. [Google Scholar] [CrossRef]
- Yager, D.R.; Nwomeh, B.C. The proteolytic environment of chronic wounds. Wound Repair Regen. 1999, 7, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.A.F. Oxidative Stress and “Senescent” Fibroblasts in Non-Healing Wounds as Potential Therapeutic Targets. J. Investig. Dermatol. 2008, 128, 2361–2364. [Google Scholar] [CrossRef]
- Stanley, A.C.; Park, H.Y.; Phillips, T.J.; Russakovsky, V.; Menzoian, J.O. Reduced growth of dermal fibroblasts from chronic venous ulcers can be stimulated with growth factors. J. Vasc. Surg. 1997, 26, 994–999; discussion 999–1001. [Google Scholar] [CrossRef]
- Loots, M.A.; Lamme, E.N.; Zeegelaar, J.; Mekkes, J.R.; Bos, J.D.; Middelkoop, E. Differences in cellular infiltrate and extracellular matrix of chronic diabetic and venous ulcers versus acute wounds. J. Investig. Dermatol. 1998, 111, 850–857. [Google Scholar] [CrossRef]
- Vande Berg, J.S.; Rudolph, R.; Hollan, C.; Haywood-Reid, P.L. Fibroblast senescence in pressure ulcers. Wound Repair Regen. 1998, 6, 38–49. [Google Scholar] [CrossRef]
- Vande Berg, J.S.; Rose, M.A.; Haywood-Reid, P.L.; Rudolph, R.; Payne, W.G.; Robson, M.C. Cultured pressure ulcer fibroblasts show replicative senescence with elevated production of plasmin, plasminogen activator inhibitor-1, and transforming growth factor-beta1. Wound Repair Regen. 2005, 13, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Bishop, A. Role of oxygen in wound healing. J. Wound Care 2008, 17, 399–402. [Google Scholar] [CrossRef]
- Rodriguez, P.G.; Felix, F.N.; Woodley, D.T.; Shim, E.K. The role of oxygen in wound healing: A review of the literature. Dermatol. Surg. 2008, 34, 1159–1169. [Google Scholar] [CrossRef]
- Harris-Tryon, T.A.; Grice, E.A. Microbiota and maintenance of skin barrier function. Science 2022, 376, 940–945. [Google Scholar] [CrossRef]
- Townsend, E.C.; Kalan, L.R. The dynamic balance of the skin microbiome across the lifespan. Biochem. Soc. Trans. 2023, 51, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A. Fibrotic disease and the TH1/TH2 paradigm. Nat. Rev. Immunol. 2004, 4, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.C.; Brutscher, A.; Ehrström, M.; Melican, K. Tissue resident cells differentiate S. aureus from S. epidermidis via IL-1β following barrier disruption in healthy human skin. PLoS Pathog. 2024, 20, e1012056. [Google Scholar] [CrossRef]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010, 465, 346–349. [Google Scholar] [CrossRef]
- Ommori, R.; Shinkuma, S.; Asada, H. Staphylococcus epidermidis augments human β-defensin-3 synthesis through the transforming growth factor alpha-epidermal growth factor receptor cascade. J. Dermatol. Sci. 2024, 116, 34–40. [Google Scholar] [CrossRef]
- Ioannou, P.; Katsoulieris, E.; Afratis, N.A. Matrix Dynamics and Microbiome Crosstalk: Matrix Metalloproteinases as Key Players in Disease and Therapy. Int. J. Mol. Sci. 2025, 26, 3621. [Google Scholar] [CrossRef]
- Coluccio, A.; Lopez Palomera, F.; Spero, M.A. Anaerobic bacteria in chronic wounds: Roles in disease, infection and treatment failure. Wound Repair Regen. 2024, 32, 840–857. [Google Scholar] [CrossRef]
- Xu, C.; Akakuru, O.U.; Ma, X.; Zheng, J.; Zheng, J.; Wu, A. Nanoparticle-Based Wound Dressing: Recent Progress in the Detection and Therapy of Bacterial Infections. Bioconjugate Chem. 2020, 31, 1708–1723. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, C.; Satyamoorthy, K.; Murali, T.S. Microbial Interplay in Skin and Chronic Wounds. Curr. Clin. Microbiol. Rep. 2022, 9, 21–31. [Google Scholar] [CrossRef]
- Hammerschmidt, S.; Rohde, M.; Preissner, K.T. Extracellular Matrix Interactions with Gram-Positive Pathogens. Microbiol. Spectr. 2019, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.K.; Cheng, N.C.; Cheng, C.M. Biofilms in Chronic Wounds: Pathogenesis and Diagnosis. Trends Biotechnol. 2019, 37, 505–517. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Ito, T.; Tamai, M.; Nakagawa, S.; Nakamura, Y. The role of Staphylococcus aureus quorum sensing in cutaneous and systemic infections. Inflamm. Regen. 2024, 44, 9. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Plésiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef]
- Gurjala, A.N.; Schierle, C.F.; Galiano, R.D.; Leung, K.P.; Mustoe, T.A. Animal Models of Biofilm-Infected Wound Healing. In Advances in Wound Care: Volume 1; Mary Ann Liebert, Inc.: New Rochelle, NY, USA, 2010; pp. 305–310. [Google Scholar]
- Hoffman, L.R.; Richardson, A.R.; Houston, L.S.; Kulasekara, H.D.; Martens-Habbena, W.; Klausen, M.; Burns, J.L.; Stahl, D.A.; Hassett, D.J.; Fang, F.C.; et al. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog. 2010, 6, e1000712. [Google Scholar] [CrossRef] [PubMed]
- Khatib-Massalha, E.; Bhattacharya, S.; Massalha, H.; Biram, A.; Golan, K.; Kollet, O.; Kumari, A.; Avemaria, F.; Petrovich-Kopitman, E.; Gur-Cohen, S. Lactate released by inflammatory bone marrow neutrophils induces their mobilization via endothelial GPR81 signaling. Nat. Commun. 2020, 11, 3547. [Google Scholar] [CrossRef]
- Mu, X.; Shi, W.; Xu, Y.; Xu, C.; Zhao, T.; Geng, B.; Yang, J.; Pan, J.; Hu, S.; Zhang, C. Tumor-derived lactate induces M2 macrophage polarization via the activation of the ERK/STAT3 signaling pathway in breast cancer. Cell Cycle 2018, 17, 428–438. [Google Scholar] [CrossRef]
- Paolini, L.; Clément, C.; Beauvillain, C.; Preisser, L.; Blanchard, S.; Pignon, P.; Seegers, V.; Chevalier, L.M.; Campone, M.; Wernert, R.; et al. Lactic acidosis together with GM-CSF and M-CSF induces human macrophages toward an inflammatory protumor phenotype. Cancer Immunol. Res. 2020, 8, 383–395. [Google Scholar] [CrossRef]
- Penny, H.L.; Sieow, J.L.; Adriani, G.; Yeap, W.H.; See Chi Ee, P.; San Luis, B.; Lee, B.; Lee, T.; Mak, S.Y.; Ho, Y.S. Warburg metabolism in tumor-conditioned macrophages promotes metastasis in human pancreatic ductal adenocarcinoma. Oncoimmunology 2016, 5, e1191731. [Google Scholar] [CrossRef] [PubMed]
- Helm, O.; Held-Feindt, J.; Grage-Griebenow, E.; Reiling, N.; Ungefroren, H.; Vogel, I.; Krüger, U.; Becker, T.; Ebsen, M.; Röcken, C. Tumor-associated macrophages exhibit pro-and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int. J. Cancer 2014, 135, 843–861. [Google Scholar] [CrossRef] [PubMed]
- de-Brito, N.M.; Duncan-Moretti, J.; da-Costa, H.C.; Saldanha-Gama, R.; Paula-Neto, H.A.; Dorighello, G.G.; Simoes, R.L.; Barja-Fidalgo, C. Aerobic glycolysis is a metabolic requirement to maintain the M2-like polarization of tumor-associated macrophages. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2020, 1867, 118604. [Google Scholar] [CrossRef]
- Llibre, A.; Kucuk, S.; Gope, A.; Certo, M.; Mauro, C. Lactate: A key regulator of the immune response. Immunity 2025, 58, 535–554. [Google Scholar] [CrossRef]
- Lee, D.C.; Sohn, H.A.; Park, Z.-Y.; Oh, S.; Kang, Y.K.; Lee, K.-m.; Kang, M.; Jang, Y.J.; Yang, S.-J.; Hong, Y.K. A lactate-induced response to hypoxia. Cell 2015, 161, 595–609. [Google Scholar] [CrossRef]
- Moser, C.; Pedersen, H.T.; Lerche, C.J.; Kolpen, M.; Line, L.; Thomsen, K.; Høiby, N.; Jensen, P.Ø. Biofilms and host response—helpful or harmful. APMIS 2017, 125, 320–338. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, X.; Sun, L.; He, T.; Wei, R.; Pang, M.; Wang, R. Staphylococcus aureus Bacteriophage Suppresses LPS-Induced Inflammation in MAC-T Bovine Mammary Epithelial Cells. Front. Microbiol. 2018, 9, 1614. [Google Scholar] [CrossRef] [PubMed]
- Souza, E.B.; Pinto, A.R.; Fongaro, G. Bacteriophages as Potential Clinical Immune Modulators. Microorganisms 2023, 11, 2222. [Google Scholar] [CrossRef] [PubMed]
- Popescu, M.C.; Pennetzdorfer, N.; Hargil, A.; Kaber, G.; Bollyky, P.L. Pf bacteriophage inhibits neutrophil migration in the lung. bioRxiv 2022. bioRxiv: 2022.2010.2012.511980. [Google Scholar]
- Goodridge, H.S.; Underhill, D.M. Fungal Recognition by TLR2 and Dectin-1. Handb. Exp. Pharmacol. 2008, 183, 87–109. [Google Scholar] [CrossRef]
- Yang, F.; Wang, L.; Song, D.; Zhang, L.; Wang, X.; Du, D.; Jiang, X. Signaling pathways and targeted therapy for rosacea. Front. Immunol. 2024, 15, 1367994. [Google Scholar] [CrossRef]
- Kroeze, K.L.; Boink, M.A.; Sampat-Sardjoepersad, S.C.; Waaijman, T.; Scheper, R.J.; Gibbs, S. Autocrine regulation of re-epithelialization after wounding by chemokine receptors CCR1, CCR10, CXCR1, CXCR2, and CXCR3. J. Investig. Dermatol. 2012, 132, 216–225. [Google Scholar] [CrossRef]
- Hammad, A.S.; Zahedy, S.H.; Elqasass, S.S.; Said, S.S.; Elgamal, A.M.; Mahmoud, N.N.; Al-Asmakh, M. In Vitro Analysis of the Dynamic Role of the Bacterial Virulence Factors in Skin Wound Healing. Int. J. Mol. Sci. 2025, 26, 10472. [Google Scholar] [CrossRef]
- Wu, H.M.; Zhao, C.C.; Xie, Q.M.; Xu, J.; Fei, G.H. TLR2-Melatonin Feedback Loop Regulates the Activation of NLRP3 Inflammasome in Murine Allergic Airway Inflammation. Front. Immunol. 2020, 11, 172. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Vuong, C.; Saenz, H.L.; Götz, F.; Otto, M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 2000, 182, 1688–1693. [Google Scholar] [CrossRef]
- Boles, B.R.; Horswill, A.R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008, 4, e1000052. [Google Scholar] [CrossRef] [PubMed]
- Gambello, M.J.; Kaye, S.; Iglewski, B.H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 1993, 61, 1180–1184. [Google Scholar] [CrossRef]
- Schuster, M.; Lostroh, C.P.; Ogi, T.; Greenberg, E.P. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J. Bacteriol. 2003, 185, 2066–2079. [Google Scholar] [CrossRef]
- Schuster, M.; Urbanowski, M.L.; Greenberg, E.P. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 2004, 101, 15833–15839. [Google Scholar] [CrossRef] [PubMed]
- Naga, N.G.; Shaaban, M.I.; El-Metwally, M.M. An insight on the powerful of bacterial quorum sensing inhibition. Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 2071–2081. [Google Scholar] [CrossRef]
- MacLeod, A.S.; Mansbridge, J.N. The innate immune system in acute and chronic wounds. Adv. Wound Care 2016, 5, 65–78. [Google Scholar] [CrossRef]
- Su, Y.; Huang, J.; Zhao, X.; Lu, H.; Wang, W.; Yang, X.O.; Shi, Y.; Wang, X.; Lai, Y.; Dong, C. Interleukin-17 receptor D constitutes an alternative receptor for interleukin-17A important in psoriasis-like skin inflammation. Sci. Immunol. 2019, 4, eaau9657. [Google Scholar] [CrossRef]
- Heilborn, J.D.; Nilsson, M.F.; Sørensen, O.; Ståhle-Bäckdahl, M.; Kratz, G.; Weber, G.; Borregaard, N. The cathelicidin anti-microbial peptide LL-37 is involved in re-epithelialization of human skin wounds and is lacking in chronic ulcer epithelium. J. Investig. Dermatol. 2003, 120, 379–389. [Google Scholar] [CrossRef]
- Carretero, M.; Escámez, M.J.; García, M.; Duarte, B.; Holguín, A.; Retamosa, L.; Jorcano, J.L.; Del Río, M.; Larcher, F. In vitro and in vivo wound healing-promoting activities of human cathelicidin LL-37. J. Investig. Dermatol. 2008, 128, 223–236. [Google Scholar] [CrossRef]
- Lindsay, S.; Oates, A.; Bourdillon, K. The detrimental impact of extracellular bacterial proteases on wound healing. Int. Wound J. 2017, 14, 1237–1247. [Google Scholar] [CrossRef] [PubMed]
- Schilrreff, P.; Alexiev, U. Chronic Inflammation in Non-Healing Skin Wounds and Promising Natural Bioactive Compounds Treatment. Int. J. Mol. Sci. 2022, 23, 4928. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Chen, Z.; Tian, P.; Han, Q.; Zhang, J.; Zhang, A.-M.; Song, Y. Wound healing mechanism of antimicrobial peptide cathelicidin-DM. Front. Bioeng. Biotechnol. 2022, 10, 977159. [Google Scholar] [CrossRef]
- Cano Sanchez, M.; Lancel, S.; Boulanger, E.; Neviere, R. Targeting Oxidative Stress and Mitochondrial Dysfunction in the Treatment of Impaired Wound Healing: A Systematic Review. Antioxidants 2018, 7, 98. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.; Werner, S. Oxidative stress in normal and impaired wound repair. Pharmacol. Res. 2008, 58, 165–171. [Google Scholar] [CrossRef]
- Dunnill, C.; Patton, T.; Brennan, J.; Barrett, J.; Dryden, M.; Cooke, J.; Leaper, D.; Georgopoulos, N.T. Reactive oxygen species (ROS) and wound healing: The functional role of ROS and emerging ROS-modulating technologies for augmentation of the healing process. Int. Wound J. 2017, 14, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Sen, C.K.; Roy, S. Redox signals in wound healing. Biochim. Biophys. Acta 2008, 1780, 1348–1361. [Google Scholar] [CrossRef]
- David, J.A.; Rifkin, W.J.; Rabbani, P.S.; Ceradini, D.J. The Nrf2/Keap1/ARE pathway and oxidative stress as a therapeutic target in type II diabetes mellitus. J. Diabetes Res. 2017, 2017, 4826724. [Google Scholar] [CrossRef]
- Bitar, M.S.; Al-Mulla, F. A defect in Nrf2 signaling constitutes a mechanism for cellular stress hypersensitivity in a genetic rat model of type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E1119–E1129. [Google Scholar] [CrossRef]
- Sherrill, C.; Fahey, R.C. Import and metabolism of glutathione by Streptococcus mutans. J. Bacteriol. 1998, 180, 1454–1459. [Google Scholar] [CrossRef]
- Wiederholt, K.M.; Steele, J.L. Glutathione accumulation in lactococci. J. Dairy Sci. 1994, 77, 1183–1188. [Google Scholar] [CrossRef]
- Han, F.; Wang, W.; Shen, K.; Wang, P.; Wang, Y.; Han, S.; Hu, D.; Guan, H.; Wu, G. A novel Lactobacillus delbrueckii-based topical spray promotes wound healing and inhibits dermal fibrosis: A probiotic biomaterial for integrated scar management. Chem. Eng. J. 2025, 525, 169786. [Google Scholar] [CrossRef]
- Zhang, S.P.; Tong, M.; Mo, J.; Dong, Z.Y.; Huang, Y.F. M2 macrophages activate the IL-10/JAK2/STAT3 pathway to induce pathological microangiogenesis in the nucleus pulposus exacerbating intervertebral disc degeneration. J. Orthop. Surg. Res. 2025, 20, 532. [Google Scholar] [CrossRef]
- Zhang, S.-M.; Wei, C.-Y.; Wang, Q.; Wang, L.; Lu, L.; Qi, F.-Z. M2-polarized macrophages mediate wound healing by regulating connective tissue growth factor via AKT, ERK1/2, and STAT3 signaling pathways. Mol. Biol. Rep. 2021, 48, 6443–6456. [Google Scholar] [CrossRef]
- Palmieri, E.M.; Gonzalez-Cotto, M.; Baseler, W.A.; Davies, L.C.; Ghesquière, B.; Maio, N.; Rice, C.M.; Rouault, T.A.; Cassel, T.; Higashi, R.M.; et al. Nitric oxide orchestrates metabolic rewiring in M1 macrophages by targeting aconitase 2 and pyruvate dehydrogenase. Nat. Commun. 2020, 11, 698. [Google Scholar] [CrossRef]
- MacMicking, J.; Xie, Q.W.; Nathan, C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997, 15, 323–350. [Google Scholar] [CrossRef]
- Hu, T.; Liu, C.-H.; Lei, M.; Zeng, Q.; Li, L.; Tang, H.; Zhang, N. Metabolic regulation of the immune system in health and diseases: Mechanisms and interventions. Signal Transduct. Target. Ther. 2024, 9, 268. [Google Scholar] [CrossRef]
- Maslowski, K.M. Metabolism at the centre of the host-microbe relationship. Clin. Exp. Immunol. 2019, 197, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Franchi, L.; Kamada, N.; Nakamura, Y.; Burberry, A.; Kuffa, P.; Suzuki, S.; Shaw, M.H.; Kim, Y.G.; Núñez, G. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat. Immunol. 2012, 13, 449–456. [Google Scholar] [CrossRef]
- Lopes, F.B.; Sarandy, M.M.; Novaes, R.D.; Valacchi, G.; Gonçalves, R.V. OxInflammatory Responses in the Wound Healing Process: A Systematic Review. Antioxidants 2024, 13, 823. [Google Scholar] [CrossRef] [PubMed]
- Adnan, S.B.; Maarof, M.; Fauzi, M.B.; Md Fadilah, N.I. Antimicrobial Peptides in Wound Healing and Skin Regeneration: Dual Roles in Immunity and Microbial Defense. Int. J. Mol. Sci. 2025, 26, 5920. [Google Scholar] [CrossRef]
- Han, S.; Zhuang, H.; Lee, P.Y.; Li, M.; Yang, L.; Nigrovic, P.A.; Reeves, W.H. NF-E2-Related Factor 2 Regulates Interferon Receptor Expression and Alters Macrophage Polarization in Lupus. Arthritis Rheumatol. 2020, 72, 1707–1720. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Huang, J.; Zeng, A.; Long, X.; Yu, N.; Wang, X. The role of the skin microbiome in wound healing. Burn. Trauma 2024, 12, tkad059. [Google Scholar] [CrossRef]
- Roth, R.R.; James, W.D. Microbial Ecology of the Skin. Annu. Rev. Microbiol. 1988, 42, 441–464. [Google Scholar] [CrossRef]
- Fluhr, J.W.; Kao, J.; Jain, M.; Ahn, S.K.; Feingold, K.R.; Elias, P.M. Generation of free fatty acids from phospholipids regulates stratum corneum acidification and integrity. J. Investig. Dermatol. 2001, 117, 44–51. [Google Scholar] [CrossRef]
- Hülpüsch, C.; Tremmel, K.; Hammel, G.; Bhattacharyya, M.; de Tomassi, A.; Nussbaumer, T.; Neumann, A.U.; Reiger, M.; Traidl-Hoffmann, C. Skin pH-dependent Staphylococcus aureus abundance as predictor for increasing atopic dermatitis severity. Allergy 2020, 75, 2888–2898. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Hunt, R.L.; Villaruz, A.E.; Fisher, E.L.; Liu, R.; Liu, Q.; Cheung, G.Y.C.; Li, M.; Otto, M. Commensal Staphylococcus epidermidis contributes to skin barrier homeostasis by generating protective ceramides. Cell Host Microbe 2022, 30, 301–313.e309. [Google Scholar] [CrossRef]
- Maheswary, T.; Nurul, A.A.; Fauzi, M.B. The Insights of Microbes’ Roles in Wound Healing: A Comprehensive Review. Pharmaceutics 2021, 13, 981. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, W.; Wu, Y.; Ma, X.; Zhou, W.; Lai, Y. Lipopeptide 78 from Staphylococcus epidermidis Activates β-Catenin To Inhibit Skin Inflammation. J. Immunol. 2019, 202, 1219–1228. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Paulino, L.C.; Tseng, C.-H.; Strober, B.E.; Blaser, M.J. Molecular Analysis of Fungal Microbiota in Samples from Healthy Human Skin and Psoriatic Lesions. J. Clin. Microbiol. 2006, 44, 2933–2941. [Google Scholar] [CrossRef]
- Lacey, N.; Delaney, S.; Kavanagh, K.; Powell, F.C. Mite-related bacterial antigens stimulate inflammatory cells in rosacea. Br. J. Dermatol. 2007, 157, 474–481. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Zhou, J.; Yao, D.; Qian, Z.; Hou, S.; Li, L.; Jenkins, A.T.A.; Fan, Y. Bacteria-responsive intelligent wound dressing: Simultaneous In situ detection and inhibition of bacterial infection for accelerated wound healing. Biomaterials 2018, 161, 11–23. [Google Scholar] [CrossRef] [PubMed]
- DeLeon, S.; Clinton, A.; Fowler, H.; Everett, J.; Horswill, A.R.; Rumbaugh, K.P. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 2014, 82, 4718–4728. [Google Scholar] [CrossRef] [PubMed]
- Alves, P.M.; Al-Badi, E.; Withycombe, C.; Jones, P.M.; Purdy, K.J.; Maddocks, S.E. Interaction between Staphylococcus aureus and Pseudomonas aeruginosa is beneficial for colonisation and pathogenicity in a mixed biofilm. Pathog. Dis. 2018, 76, fty003. [Google Scholar] [CrossRef] [PubMed]
- Korgaonkar, A.; Trivedi, U.; Rumbaugh, K.P.; Whiteley, M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. USA 2013, 110, 1059–1064. [Google Scholar] [CrossRef]
- Orazi, G.; Ruoff, K.L.; O’Toole, G.A. Pseudomonas aeruginosa increases the sensitivity of biofilm-grown Staphylococcus aureus to membrane-targeting antiseptics and antibiotics. MBio 2019, 10, e01501-19. [Google Scholar] [CrossRef]
- Sadeghpour Heravi, F.; Zakrzewski, M.; Vickery, K.; Armstrong, D.G.; Hu, H. Bacterial diversity of diabetic foot ulcers: Current status and future prospectives. J. Clin. Med. 2019, 8, 1935. [Google Scholar] [CrossRef]
- Naik, S.; Bouladoux, N.; Linehan, J.L.; Han, S.J.; Harrison, O.J.; Wilhelm, C.; Conlan, S.; Himmelfarb, S.; Byrd, A.L.; Deming, C.; et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 2015, 520, 104–108. [Google Scholar] [CrossRef]
- Linehan, J.L.; Harrison, O.J.; Han, S.J.; Byrd, A.L.; Vujkovic-Cvijin, I.; Villarino, A.V.; Sen, S.K.; Shaik, J.; Smelkinson, M.; Tamoutounour, S.; et al. Non-classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell 2018, 172, 784–796.e718. [Google Scholar] [CrossRef]
- Mochtar, C.F.; Bakhtiar, M.; Hamzah, H.; Pratama, A.; Rasyidah, J.; Rakhmatia, J.; Alfiannur, M.; Amalia, N.; Putri, N.; Damis, N. Topical gel preparation from Staphylococcus epidermidis isolate extract accelerated healing on incision wound model in rats. Indones. J. Pharmacol. Ther. 2024, 5, 1. [Google Scholar] [CrossRef]
- Mohammedsaeed, W.; Samyah, B.; Nikhat, M.; and Almaramhy, H.H. An ex-vivo study to investigate the potential of Staphylococcus epidermidis lysate to improve wound healing in diabetic patients. J. Taibah Univ. Sci. 2022, 16, 895–902. [Google Scholar] [CrossRef]
- Ohshima, H.; Kurosumi, M.; Kanto, H. New solution of beauty problem by Staphylococcus hominis: Relevance between skin microbiome and skin condition in healthy subject. Ski. Res. Technol. 2021, 27, 692–700. [Google Scholar] [CrossRef]
- Severn, M.M.; Williams, M.R.; Shahbandi, A.; Bunch, Z.L.; Lyon, L.M.; Nguyen, A.; Zaramela, L.S.; Todd, D.A.; Zengler, K.; Cech, N.B.; et al. The Ubiquitous Human Skin Commensal Staphylococcus hominis Protects against Opportunistic Pathogens. mBio 2022, 13, e0093022. [Google Scholar] [CrossRef]
- Xu, W.; Dielubanza, E.; Maisel, A.; Leung, K.; Mustoe, T.; Hong, S.; Galiano, R. Staphylococcus aureus impairs cutaneous wound healing by activating the expression of a gap junction protein, connexin-43 in keratinocytes. Cell. Mol. Life Sci. 2021, 78, 935–947. [Google Scholar] [CrossRef]
- Gurjala, A.N.; Geringer, M.R.; Seth, A.K.; Hong, S.J.; Smeltzer, M.S.; Galiano, R.D.; Leung, K.P.; Mustoe, T.A. Development of a novel, highly quantitative in vivo model for the study of biofilm-impaired cutaneous wound healing. Wound Repair Regen. 2011, 19, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Kirker, K.R.; Secor, P.R.; James, G.A.; Fleckman, P.; Olerud, J.E.; Stewart, P.S. Loss of viability and induction of apoptosis in human keratinocytes exposed to Staphylococcus aureus biofilms in vitro. Wound Repair Regen. 2009, 17, 690–699. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.T.; Daut, J.; Schalinski, E.; Fischer-Medert, T.; Hellmeyer, L. Severe Lactational Mastitis With Complicated Wound Infection Caused by Streptococcus pyogenes. J. Hum. Lact. 2021, 37, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Hübner, G.; Brauchle, M.; Smola, H.; Madlener, M.; Fässler, R.; Werner, S. Differential regulation of pro-inflammatory cytokines during wound healing in normal and glucocorticoid-treated mice. Cytokine 1996, 8, 548–556. [Google Scholar] [CrossRef]
- Celik, C.; Lee, S.T.T.; Tanoto, F.R.; Veleba, M.; Kline, K.; Thibault, G. Decoding the complexity of delayed wound healing following Enterococcus faecalis infection. eLife 2024, 13, RP95113. [Google Scholar] [CrossRef]
- Chong, K.K.L.; Tay, W.H.; Janela, B.; Yong, A.M.H.; Liew, T.H.; Madden, L.; Keogh, D.; Barkham, T.M.S.; Ginhoux, F.; Becker, D.L.; et al. Enterococcus faecalis Modulates Immune Activation and Slows Healing During Wound Infection. J. Infect. Dis. 2017, 216, 1644–1654. [Google Scholar] [CrossRef]
- Petkovšek, Ž.; Eleršič, K.; Gubina, M.; Žgur-Bertok, D.; Starčič Erjavec, M. Virulence Potential of Escherichia coli Isolates from Skin and Soft Tissue Infections. J. Clin. Microbiol. 2009, 47, 1811–1817. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, X.; Liu, J.; Tao, Y.; Wang, T.; Li, L. Lactobacillus Plantarum Promotes Wound Healing by Inhibiting the NLRP3 Inflammasome and Pyroptosis Activation in Diabetic Foot Wounds. J. Inflamm. Res. 2024, 17, 1707–1720. [Google Scholar] [CrossRef]
- Mohtashami, M.; Mohamadi, M.; Azimi-Nezhad, M.; Saeidi, J.; Nia, F.F.; Ghasemi, A. Lactobacillus bulgaricus and Lactobacillus plantarum improve diabetic wound healing through modulating inflammatory factors. Biotechnol. Appl. Biochem. 2021, 68, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.-H.; Chou, C.-H.; Huang, T.-Y.; Wang, H.-L.; Chien, P.-J.; Chang, W.-W.; Lee, H.-T. Heat-Killed Lactobacilli Preparations Promote Healing in the Experimental Cutaneous Wounds. Cells 2021, 10, 3264. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-Negative Staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef]
- Parlet, C.P.; Brown, M.M.; Horswill, A.R. Commensal Staphylococci Influence Staphylococcus aureus Skin Colonization and Disease. Trends Microbiol. 2019, 27, 497–507. [Google Scholar] [CrossRef]
- Ersanli, C.; Tzora, A.; Voidarou, C.C.; Skoufos, S.; Zeugolis, D.I.; Skoufos, I. Biodiversity of Skin Microbiota as an Important Biomarker for Wound Healing. Biology 2023, 12, 1187. [Google Scholar] [CrossRef]
- Dinić, M.; Verpile, R.; Burgess, J.L.; Ming, J.; Marjanovic, J.; Beliz, C.N.; Plano, L.; Hower, S.; Thaller, S.R.; Banerjee, S.; et al. Multi-drug resistant Staphylococcus epidermidis from chronic wounds impair healing in human wound model. Wound Repair Regen. 2024, 32, 799–810. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Hata, T.R.; Tong, Y.; Cheng, J.Y.; Shafiq, F.; Butcher, A.M.; Salem, S.S.; Brinton, S.L.; Rudman Spergel, A.K.; Johnson, K.; et al. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat. Med. 2021, 27, 700–709. [Google Scholar] [CrossRef]
- Herman, R.; Kinniment-Williams, B.; Rudden, M.; James, A.G.; Wilkinson, A.J.; Murphy, B.; Thomas, G.H. Identification of a staphylococcal dipeptidase involved in the production of human body odor. J. Biol. Chem. 2024, 300, 107928. [Google Scholar] [CrossRef]
- Doichinova, T.; Gancheva, G.; Totsev, N. Staphylococcal Meningitis: Review of Five Cases. JMED Res. 2014, 2014, 5. [Google Scholar]
- Jung, Y.J.; Chang, M.C. Bacterial meningitis and cauda equina syndrome after trans-sacral epiduroscopic laser decompression: A case report. Medicine 2019, 98, e14874. [Google Scholar] [CrossRef]
- Shan, W.; Wang, Y.; Zhang, Z.; Xing, J.; Xu, J.; Xiao, W.; Shen, Y.; Guo, S.; Que, H. Qingre Baidu mixture-induced effect of AI-2 on Staphylococcus aureus and Pseudomonas aeruginosa biofilms in chronic and refractory wounds. Exp. Ther. Med. 2019, 17, 3343–3350. [Google Scholar] [CrossRef]
- Simonetti, O.; Rizzetto, G.; Radi, G.; Molinelli, E.; Cirioni, O.; Giacometti, A.; Offidani, A. New Perspectives on Old and New Therapies of Staphylococcal Skin Infections: The Role of Biofilm Targeting in Wound Healing. Antibiotics 2021, 10, 1377. [Google Scholar] [CrossRef]
- Hachem, R.; Parikh, U.M.; Reitzel, R.; Rosenblatt, J.; Kaul, A.; Vargas-Cruz, N.; Hill, L.; Moore, L.; Meyer, J.; Chaftari, A.M.; et al. Novel antimicrobial ointment for infected wound healing in an in vitro and in vivo porcine model. Wound Repair Regen. 2021, 29, 830–842. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.M.C.; Dean, S.N.; Propst, C.N.; Bishop, B.M.; van Hoek, M.L. Komodo dragon-inspired synthetic peptide DRGN-1 promotes wound-healing of a mixed-biofilm infected wound. NPJ Biofilms Microbiomes 2017, 3, 9. [Google Scholar] [CrossRef]
- Pastar, I.; Nusbaum, A.G.; Gil, J.; Patel, S.B.; Chen, J.; Valdes, J.; Stojadinovic, O.; Plano, L.R.; Tomic-Canic, M.; Davis, S.C. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS ONE 2013, 8, e56846. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Namikawa, H.; Fujimoto, H.; Nakaie, K.; Takizawa, E.; Okada, Y.; Fujita, A.; Kawaguchi, H.; Nakamura, Y.; Abe, J.; et al. Clinical Characteristics of Methicillin-resistant Coagulase-negative Staphylococcal Bacteremia in a Tertiary Hospital. Intern. Med. 2017, 56, 781–785. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Daley, A.J.; Istivan, T.S.; Garland, S.M.; Deighton, M.A. Antibiotic susceptibility of coagulase-negative staphylococci isolated from very low birth weight babies: Comprehensive comparisons of bacteria at different stages of biofilm formation. Ann. Clin. Microbiol. Antimicrob. 2010, 9, 16. [Google Scholar] [CrossRef]
- Severn, M.M.; Horswill, A.R. Staphylococcus epidermidis and its dual lifestyle in skin health and infection. Nat. Rev. Microbiol. 2023, 21, 97–111. [Google Scholar] [CrossRef]
- Brown, M.M.; Horswill, A.R. Staphylococcus epidermidis-Skin friend or foe? PLoS Pathog. 2020, 16, e1009026. [Google Scholar] [CrossRef]
- Constantinides, M.G.; Link, V.M.; Tamoutounour, S.; Wong, A.C.; Perez-Chaparro, P.J.; Han, S.J.; Chen, Y.E.; Li, K.; Farhat, S.; Weckel, A.; et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 2019, 366, aax6624. [Google Scholar] [CrossRef]
- Petreaca, M.L.; Do, D.; Dhall, S.; McLelland, D.; Serafino, A.; Lyubovitsky, J.; Schiller, N.; Martins-Green, M.M. Deletion of a tumor necrosis superfamily gene in mice leads to impaired healing that mimics chronic wounds in humans. Wound Repair Regen. 2012, 20, 353–366. [Google Scholar] [CrossRef]
- Lee, J.Y.H.; Monk, I.R.; Gonçalves da Silva, A.; Seemann, T.; Chua, K.Y.L.; Kearns, A.; Hill, R.; Woodford, N.; Bartels, M.D.; Strommenger, B.; et al. Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat. Microbiol. 2018, 3, 1175–1185. [Google Scholar] [CrossRef]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; Murray, P.R.; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef]
- Fredheim, E.G.A.; Flægstad, T.; Askarian, F.; Klingenberg, C. Colonisation and interaction between S. epidermidis and S. aureus in the nose and throat of healthy adolescents. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 123–129. [Google Scholar] [CrossRef]
- Boldock, E.; Surewaard, B.G.J.; Shamarina, D.; Na, M.; Fei, Y.; Ali, A.; Williams, A.; Pollitt, E.J.G.; Szkuta, P.; Morris, P.; et al. Human skin commensals augment Staphylococcus aureus pathogenesis. Nat. Microbiol. 2018, 3, 881–890. [Google Scholar] [CrossRef]
- Mulcahy, L.R.; Isabella, V.M.; Lewis, K. Pseudomonas aeruginosa Biofilms in Disease. Microb. Ecol. 2014, 68, 1–12. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Kirketerp-Møller, K.; Jensen, P.; Madsen, K.G.; Phipps, R.; Krogfelt, K.; Høiby, N.; Givskov, M. Why chronic wounds will not heal: A novel hypothesis. Wound Repair Regen. 2008, 16, 2–10. [Google Scholar] [CrossRef]
- Bliss, S.K.; Butcher, B.A.; Denkers, E.Y. Rapid recruitment of neutrophils containing prestored IL-12 during microbial infection. J. Immunol. 2000, 165, 4515–4521. [Google Scholar] [CrossRef]
- Römling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012, 272, 541–561. [Google Scholar] [CrossRef]
- Mielko, K.A.; Jabłoński, S.J.; Milczewska, J.; Sands, D.; Łukaszewicz, M.; Młynarz, P. Metabolomic studies of Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 2019, 35, 178. [Google Scholar] [CrossRef]
- Kanno, E.; Kawakami, K.; Miyairi, S.; Tanno, H.; Suzuki, A.; Kamimatsuno, R.; Takagi, N.; Miyasaka, T.; Ishii, K.; Gotoh, N.; et al. Promotion of acute-phase skin wound healing by Pseudomonas aeruginosa C-HSL. Int. Wound J. 2016, 13, 1325–1335. [Google Scholar] [CrossRef]
- Kanno, E.; Kawakami, K.; Ritsu, M.; Ishii, K.; Tanno, H.; Toriyabe, S.; Imai, Y.; Maruyama, R.; Tachi, M. Wound healing in skin promoted by inoculation with seudomonas aeruginosa PAO1: The critical role of tumor necrosis factor-α secreted from infiltrating neutrophils. Wound Repair Regen. 2011, 19, 608–621. [Google Scholar] [CrossRef]
- Kanwal, S.; Vaitla, P. Streptococcus Pyogenes. In StatPearls; Copyright© 2025; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2025. [Google Scholar]
- Langley, G.; Hao, Y.; Pondo, T.; Miller, L.; Petit, S.; Thomas, A.; Lindegren, M.L.; Farley, M.M.; Dumyati, G.; Como-Sabetti, K.; et al. The Impact of Obesity and Diabetes on the Risk of Disease and Death due to Invasive Group A Streptococcus Infections in Adults. Clin. Infect. Dis. 2016, 62, 845–852. [Google Scholar] [CrossRef]
- Avire, N.J.; Whiley, H.; Ross, K. A Review of Streptococcus pyogenes: Public Health Risk Factors, Prevention and Control. Pathogens 2021, 10, 248. [Google Scholar] [CrossRef]
- Casabonne, C.; González, A.; Aquili, V.; Balagué, C. Antibacterial activity of silver sulfadiazine against Streptococcus pyogenes. Wounds Int. 2022, 13, 10–14. [Google Scholar]
- Usanga, V.; Elom, M.; Okon, U.; Nworie, A.; Ukwah, B.; Kalu, M.; Simon, A. Antibacterial Activity of Honey on Staphylococcus aureus, Escherichia coli and Streptococcus pyogenes Isolated from Wounds. J. Adv. Microbiol. 2020, 20, 51–57. [Google Scholar] [CrossRef]
- Bae, S.; Gu, H.; Gwon, M.-G.; An, H.-J.; Han, S.-M.; Lee, S.-J.; Leem, J.; Park, K.-K. Therapeutic Effect of Bee Venom and Melittin on Skin Infection Caused by Streptococcus pyogenes. Toxins 2022, 14, 663. [Google Scholar] [CrossRef]
- El-Kased, R.F.; Amer, R.I.; Attia, D.; Elmazar, M.M. Honey-based hydrogel: In vitro and comparative In vivo evaluation for burn wound healing. Sci. Rep. 2017, 7, 9692. [Google Scholar] [CrossRef]
- Reczyńska-Kolman, K.; Hartman, K.; Kwiecień, K.; Brzychczy-Włoch, M.; Pamuła, E. Composites Based on Gellan Gum, Alginate and Nisin-Enriched Lipid Nanoparticles for the Treatment of Infected Wounds. Int. J. Mol. Sci. 2021, 23, 321. [Google Scholar] [CrossRef]
- Pot, B.; Ludwig, W.; Kersters, K.; Schleifer, K.-H. Taxonomy of Lactic Acid Bacteria. In Bacteriocins of Lactic Acid Bacteria: Microbiology, Genetics and Applications; De Vuyst, L., Vandamme, E.J., Eds.; Springer: Boston, MA, USA, 1994; pp. 13–90. [Google Scholar]
- Kleerebezem, M.; Hols, P.; Bernard, E.; Rolain, T.; Zhou, M.; Siezen, R.J.; Bron, P.A. The extracellular biology of the lactobacilli. FEMS Microbiol. Rev. 2010, 34, 199–230. [Google Scholar] [CrossRef]
- Tsai, Y.-L.; Lin, T.-L.; Chang, C.-J.; Wu, T.-R.; Lai, W.-F.; Lu, C.-C.; Lai, H.-C. Probiotics, prebiotics and amelioration of diseases. J. Biomed. Sci. 2019, 26, 3. [Google Scholar] [CrossRef]
- Hammes, W.P.; Vogel, R.F. The genus Lactobacillus. In The Genera of Lactic Acid Bacteria; Wood, B.J.B., Holzapfel, W.H., Eds.; Springer: Boston, MA, USA, 1995; pp. 19–54. [Google Scholar]
- Li, H.; Zhou, X.; Lui, T.; Wu, R.; Huang, Z.-f.; Sun, C.-w.; Liu, Z.-a.; Zheng, S.-y.; Lai, W.; Lou, H.; et al. The clinical value of metagenomic next-generation sequencing for rapid microbial identification of chronic granulation wound infections. Arch. Med. Sci. 2023, 19, 1162–1167. [Google Scholar] [CrossRef]
- Knackstedt, R.; Knackstedt, T.; Gatherwright, J. The role of topical probiotics on wound healing: A review of animal and human studies. Int. Wound J. 2020, 17, 1687–1694. [Google Scholar] [CrossRef]
- Fijan, S.; Frauwallner, A.; Langerholc, T.; Krebs, B.; ter Haar, J.A.; Heschl, A.; Mičetić Turk, D.; Rogelj, I. Efficacy of Using Probiotics with Antagonistic Activity against Pathogens of Wound Infections: An Integrative Review of Literature. BioMed Res. Int. 2019, 2019, 7585486. [Google Scholar] [CrossRef]
- Lopes, E.G.; Moreira, D.A.; Gullón, P.; Gullón, B.; Cardelle-Cobas, A.; Tavaria, F.K. Topical application of probiotics in skin: Adhesion, antimicrobial and antibiofilm in vitro assays. J. Appl. Microbiol. 2017, 122, 450–461. [Google Scholar] [CrossRef]
- Klarin, B.; Johansson, M.L.; Molin, G.; Larsson, A.; Jeppsson, B. Adhesion of the probiotic bacterium Lactobacillus plantarum 299v onto the gut mucosa in critically ill patients: A randomised open trial. Crit. Care 2005, 9, R285–R293. [Google Scholar] [CrossRef]
- Petrof, E.O.; Claud, E.C.; Sun, J.; Abramova, T.; Guo, Y.; Waypa, T.S.; He, S.M.; Nakagawa, Y.; Chang, E.B. Bacteria-free solution derived from Lactobacillus plantarum inhibits multiple NF-kappaB pathways and inhibits proteasome function. Inflamm. Bowel Dis. 2009, 15, 1537–1547. [Google Scholar] [CrossRef]
- Seddik, H.A.; Bendali, F.; Gancel, F.; Fliss, I.; Spano, G.; Drider, D. Lactobacillus plantarum and Its Probiotic and Food Potentialities. Probiotics Antimicrob. Proteins 2017, 9, 111–122. [Google Scholar] [CrossRef]
- Von Mollendorff, J.; Vaz-Velho, M.; Todorov, S. Boza, a Traditional Cereal-Based Fermented Beverage: A Rich Source of Probiotics and Bacteriocin-Producing Lactic Acid Bacteria. In Functional Properties of Traditional Foods; Springer: Boston, MA, USA, 2016; pp. 157–188. [Google Scholar]
- da Silva Sabo, S.; Vitolo, M.; González, J.M.D.; Oliveira, R.P.S. Overview of Lactobacillus plantarum as a promising bacteriocin producer among lactic acid bacteria. Food Res. Int. 2014, 64, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.L.; Nobaek, S.; Berggren, A.; Nyman, M.; Björck, I.; Ahrné, S.; Jeppsson, B.; Molin, G. Survival of Lactobacillus plantarum DSM 9843 (299v), and effect on the short-chain fatty acid content of faeces after ingestion of a rose-hip drink with fermented oats. Int. J. Food Microbiol. 1998, 42, 29–38. [Google Scholar] [CrossRef]
- Valdéz, J.C.; Peral, M.C.; Rachid, M.; Santana, M.; Perdigón, G. Interference of Lactobacillus plantarum with Pseudomonas aeruginosa in vitro and in infected burns: The potential use of probiotics in wound treatment. Clin. Microbiol. Infect. 2005, 11, 472–479. [Google Scholar] [CrossRef]
- Soleymanzadeh Moghadam, S.; Mohammad, N.; Ghooshchian, M.; FathiZadeh, S.; Khodaii, Z.; Faramarzi, M.; Fagheei Aghmiyuni, Z.; Roudbari, M.; Pazouki, A.; Mousavi Shabestari, T. Comparison of the effects of Lactobacillus plantarum versus imipenem on infected burn wound healing. Med. J. Islam. Repub. Iran 2020, 34, 94. [Google Scholar] [CrossRef]
- Hao, R.; Liu, Q.; Wang, L.; Jian, W.; Cheng, Y.; Zhang, Q.; Hayer, K.; Kamarudin Raja Idris, R.; Zhang, Y.; Lu, H.; et al. Anti-inflammatory effect of Lactiplantibacillus plantarum T1 cell-free supernatants through suppression of oxidative stress and NF-κB- and MAPK-signaling pathways. Appl. Environ. Microbiol. 2023, 89, e0060823. [Google Scholar] [CrossRef]
- Salazar, J.J.; Ennis, W.J.; Koh, T.J. Diabetes medications: Impact on inflammation and wound healing. J. Diabetes Complicat. 2016, 30, 746–752. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Allocati, N.; Masulli, M.; Alexeyev, M.F.; Di Ilio, C. Escherichia coli in Europe: An Overview. Int. J. Environ. Res. Public Health 2013, 10, 6235–6254. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, W.F.; Silva, P.M.S.; Silva, R.C.S.; Silva, G.M.M.; Machado, G.; Coelho, L.; Correia, M.T.S. Staphylococcus aureus and Staphylococcus epidermidis infections on implants. J. Hosp. Infect. 2018, 98, 111–117. [Google Scholar] [CrossRef]
- Ding, X.; Tang, Q.; Xu, Z.; Xu, Y.; Zhang, H.; Zheng, D.; Wang, S.; Tan, Q.; Maitz, J.; Maitz, P.K.; et al. Challenges and innovations in treating chronic and acute wound infections: From basic science to clinical practice. Burn. Trauma 2022, 10, tkac014. [Google Scholar] [CrossRef]
- Oliveira, A.; Sousa, J.C.; Silva, A.C.; Melo, L.D.R.; Sillankorva, S. Chestnut Honey and Bacteriophage Application to Control Pseudomonas aeruginosa and Escherichia coli Biofilms: Evaluation in an ex vivo Wound Model. Front. Microbiol. 2018, 9, 1725. [Google Scholar] [CrossRef]
- Sharma, G.; Sharma, S.; Sharma, P.; Chandola, D.; Dang, S.; Gupta, S.; Gabrani, R. Escherichia coli biofilm: Development and therapeutic strategies. J. Appl. Microbiol. 2016, 121, 309–319. [Google Scholar] [CrossRef]
- Choi, H.J.; Ahn, J.H.; Park, S.-H.; Do, K.H.; Kim, J.; Moon, Y. Enhanced Wound Healing by Recombinant Escherichia coli Nissle 1917 via Human Epidermal Growth Factor Receptor in Human Intestinal Epithelial Cells: Therapeutic Implication Using Recombinant Probiotics. Infect. Immun. 2012, 80, 1079–1087. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.P.; Kirsner, R.S. Angiogenesis in wound repair: Angiogenic growth factors and the extracellular matrix. Microsc. Res. Tech. 2003, 60, 107–114. [Google Scholar] [CrossRef]
- Ashrafi, M.; Xu, Y.; Muhamadali, H.; White, I.; Wilkinson, M.; Hollywood, K.; Baguneid, M.; Goodacre, R.; Bayat, A. A microbiome and metabolomic signature of phases of cutaneous healing identified by profiling sequential acute wounds of human skin: An exploratory study. PLoS ONE 2020, 15, e0229545. [Google Scholar] [CrossRef]
- Tomic-Canic, M.; Burgess, J.L.; O’Neill, K.E.; Strbo, N.; Pastar, I. Skin Microbiota and its Interplay with Wound Healing. Am. J. Clin. Dermatol. 2020, 21, 36–43. [Google Scholar] [CrossRef]
- Di Domizio, J.; Belkhodja, C.; Chenuet, P.; Fries, A.; Murray, T.; Mondéjar, P.M.; Demaria, O.; Conrad, C.; Homey, B.; Werner, S.; et al. The commensal skin microbiota triggers type I IFN–dependent innate repair responses in injured skin. Nat. Immunol. 2020, 21, 1034–1045. [Google Scholar] [CrossRef]
- Ghoul, M.; Mitri, S. The Ecology and Evolution of Microbial Competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef]
- Penhallow, K. A review of studies that examine the impact of infection on the normal wound-healing process. J. Wound Care 2005, 14, 123–126. [Google Scholar] [CrossRef]
- Johnson, T.R.; Gómez, B.I.; McIntyre, M.K.; Dubick, M.A.; Christy, R.J.; Nicholson, S.E.; Burmeister, D.M. The Cutaneous Microbiome and Wounds: New Molecular Targets to Promote Wound Healing. Int. J. Mol. Sci. 2018, 19, 2699. [Google Scholar] [CrossRef]
- Versey, Z.; da Cruz Nizer, W.S.; Russell, E.; Zigic, S.; DeZeeuw, K.G.; Marek, J.E.; Overhage, J.; Cassol, E. Biofilm-Innate Immune Interface: Contribution to Chronic Wound Formation. Front. Immunol. 2021, 12, 648554. [Google Scholar] [CrossRef]
- Jefferson, K.K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar] [CrossRef]
- Pastar, I.; O’Neill, K.; Padula, L.; Head, C.R.; Burgess, J.L.; Chen, V.; Garcia, D.; Stojadinovic, O.; Hower, S.; Plano, G.V.; et al. Staphylococcus epidermidis Boosts Innate Immune Response by Activation of Gamma Delta T Cells and Induction of Perforin-2 in Human Skin. Front. Immunol. 2020, 11, 550946, Erratum in Front. Immunol. 2021, 12, 741437. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.K.; Patel, K.H.; Huang, R.Y.; Lee, C.N.; Moochhala, S.M. The Gut-Skin Microbiota Axis and Its Role in Diabetic Wound Healing-A Review Based on Current Literature. Int. J. Mol. Sci. 2022, 23, 2375. [Google Scholar] [CrossRef]
- Pastar, I.; Sawaya, A.P.; Marjanovic, J.; Burgess, J.L.; Strbo, N.; Rivas, K.E.; Wikramanayake, T.C.; Head, C.R.; Stone, R.C.; Jozic, I.; et al. Intracellular Staphylococcus aureus triggers pyroptosis and contributes to inhibition of healing due to perforin-2 suppression. J. Clin. Investig. 2021, 131, e133727. [Google Scholar] [CrossRef]
- Strbo, N.; Pastar, I.; Romero, L.; Chen, V.; Vujanac, M.; Sawaya, A.P.; Jozic, I.; Ferreira, A.D.F.; Wong, L.L.; Head, C.; et al. Single cell analyses reveal specific distribution of anti-bacterial molecule Perforin-2 in human skin and its modulation by wounding and Staphylococcus aureus infection. Exp. Dermatol. 2019, 28, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Togo, C.; Zidorio, A.P.; Gonçalves, V.; Botelho, P.; de Carvalho, K.; Dutra, E. Does Probiotic Consumption Enhance Wound Healing? A Systematic Review. Nutrients 2022, 14, 111. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Dunaway, S.; Champer, J.; Kim, J.; Alikhan, A. Changing our microbiome: Probiotics in dermatology. Br. J. Dermatol. 2020, 182, 39–46. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.C.; Isolauri, E.; Gueimonde, M. Microbial–Host Interactions: Selecting the Right Probiotics and Prebiotics for Infants. Nestle Nutr. Workshop Ser. Pediatr. Program 2009, 64, 201–213. [Google Scholar] [CrossRef]
- Kuhn, T.; Aljohmani, A.; Frank, N.; Zielke, L.; Mehanny, M.; Laschke, M.W.; Koch, M.; Hoppstädter, J.; Kiemer, A.K.; Yildiz, D.; et al. A cell-free, biomimetic hydrogel based on probiotic membrane vesicles ameliorates wound healing. J. Control. Release 2024, 365, 969–980. [Google Scholar] [CrossRef]
- Ishi, S.; Kanno, E.; Tanno, H.; Kurosaka, S.; Shoji, M.; Imai, T.; Yamaguchi, K.; Kotsugai, K.; Niiyama, M.; Kurachi, H.; et al. Cutaneous wound healing promoted by topical administration of heat-killed Lactobacillus plantarum KB131 and possible contribution of CARD9-mediated signaling. Sci. Rep. 2023, 13, 15917. [Google Scholar] [CrossRef]
- Bădăluță, V.A.; Curuțiu, C.; Dițu, L.M.; Holban, A.M.; Lazăr, V. Probiotics in Wound Healing. Int. J. Mol. Sci. 2024, 25, 5723. [Google Scholar] [CrossRef]
- Ong, J.S.; Taylor, T.D.; Yong, C.C.; Khoo, B.Y.; Sasidharan, S.; Choi, S.B.; Ohno, H.; Liong, M.T. Lactobacillus plantarum USM8613 Aids in Wound Healing and Suppresses Staphylococcus aureus Infection at Wound Sites. Probiotics Antimicrob. Proteins 2020, 12, 125–137. [Google Scholar] [CrossRef]
- El-Ghazely, M.H.; Mahmoud, W.H.; Atia, M.A.; Eldip, E.M. Effect of probiotic administration in the therapy of pediatric thermal burn. Ann. Burn. Fire Disasters 2016, 29, 268–272. [Google Scholar]
- Hessle, C.; Andersson, B.; Wold, A.E. Gram-Positive Bacteria Are Potent Inducers of Monocytic Interleukin-12 (IL-12) while Gram-Negative Bacteria Preferentially Stimulate IL-10 Production. Infect. Immun. 2000, 68, 3581–3586. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef]
- De Almeida, C.V.; Antiga, E.; Lulli, M. Oral and Topical Probiotics and Postbiotics in Skincare and Dermatological Therapy: A Concise Review. Microorganisms 2023, 11, 1420. [Google Scholar] [CrossRef]
- Pattapulavar, V.; Ramanujam, S.; Kini, B.; Christopher, J.G. Probiotic-derived postbiotics: A perspective on next-generation therapeutics. Front. Nutr. 2025, 12, 1624539. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Dernst, A.; Martin, B.; Lorenzi, L.; Cadefau-Fabregat, M.; Phulphagar, K.; Wagener, A.; Budden, C.; Stair, N.; Wagner, T.; et al. Butyrate and propionate are microbial danger signals that activate the NLRP3 inflammasome in human macrophages upon TLR stimulation. Cell Rep. 2024, 43, 114736. [Google Scholar] [CrossRef]
- Castro, P.R.; Bittencourt, L.F.F.; Larochelle, S.; Andrade, S.P.; Mackay, C.R.; Slevin, M.; Moulin, V.J.; Barcelos, L.S. GPR43 regulates sodium butyrate-induced angiogenesis and matrix remodeling. Am. J. Physiol. Heart Circ. Physiol. 2021, 320, H1066–H1079. [Google Scholar] [CrossRef]
- Golkar, N.; Ashoori, Y.; Heidari, R.; Omidifar, N.; Abootalebi, S.N.; Mohkam, M.; Gholami, A. A novel effective formulation of bioactive compounds for wound healing: Preparation, in vivo characterization, and comparison of various postbiotics cold creams in a rat model. Evid. -Based Complement. Altern. Med. 2021, 2021, 8577116. [Google Scholar] [CrossRef]
- Erkihun, M.; Asmare, Z.; Endalamew, K.; Getie, B.; Kiros, T.; Berhan, A. Medical Scope of Biofilm and Quorum Sensing During Biofilm Formation: Systematic Review. Bacteria 2024, 3, 118–135. [Google Scholar] [CrossRef]
- Luo, J.; Dong, B.; Wang, K.; Cai, S.; Liu, T.; Cheng, X.; Lei, D.; Chen, Y.; Li, Y.; Kong, J.; et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. PLoS ONE 2017, 12, e0176883. [Google Scholar] [CrossRef]
- Ren, Y.; Zhu, R.; You, X.; Li, D.; Guo, M.; Fei, B.; Liu, Y.; Yang, X.; Liu, X.; Li, Y. Quercetin: A promising virulence inhibitor of Pseudomonas aeruginosa LasB in vitro. Appl. Microbiol. Biotechnol. 2024, 108, 57. [Google Scholar] [CrossRef]
- Tan, L.; Li, S.R.; Jiang, B.; Hu, X.M.; Li, S. Therapeutic Targeting of the Staphylococcus aureus Accessory Gene Regulator (agr) System. Front. Microbiol. 2018, 9, 55. [Google Scholar] [CrossRef]
- Kaplan, J.B.; LoVetri, K.; Cardona, S.T.; Madhyastha, S.; Sadovskaya, I.; Jabbouri, S.; Izano, E.A. Recombinant human DNase I decreases biofilm and increases antimicrobial susceptibility in staphylococci. J. Antibiot. 2012, 65, 73–77. [Google Scholar] [CrossRef]
- Ahsan, A.; Thomas, N.; Barnes, T.J.; Subramaniam, S.; Loh, T.C.; Joyce, P.; Prestidge, C.A. Lipid Nanocarriers-Enabled Delivery of Antibiotics and Antimicrobial Adjuvants to Overcome Bacterial Biofilms. Pharmaceutics 2024, 16, 396. [Google Scholar] [CrossRef]
- Goswami, A.G.; Basu, S.; Banerjee, T.; Shukla, V.K. Biofilm and wound healing: From bench to bedside. Eur. J. Med. Res. 2023, 28, 157. [Google Scholar] [CrossRef]
- Ghaznavi-Rad, E.; Komijani, M.; Moradabadi, A.; Rezaei, M.; Shaykh-Baygloo, N. Isolation of a lytic bacteriophage against extensively drug-resistant Acinetobacter baumannii infections and its dramatic effect in rat model of burn infection. J. Clin. Lab. Anal. 2022, 36, e24497. [Google Scholar] [CrossRef]
- Rahimian, M.; Jafari-Gharabaghlou, D.; Mohammadi, E.; Zarghami, N. A New Insight into Phage Combination Therapeutic Approaches Against Drug-Resistant Mixed Bacterial Infections. PHAGE 2024, 5, 203–222. [Google Scholar] [CrossRef]
- Soontarach, R.; Srimanote, P.; Voravuthikunchai, S.P.; Chusri, S. Antibacterial and Anti-Biofilm Efficacy of Endolysin LysAB1245 against a Panel of Important Pathogens. Pharmaceuticals 2024, 17, 155. [Google Scholar] [CrossRef]
- Mi, B.; Liu, J.; Liu, Y.; Hu, L.; Liu, Y.; Panayi, A.C.; Zhou, W.; Liu, G. The Designer Antimicrobial Peptide A-hBD-2 Facilitates Skin Wound Healing by Stimulating Keratinocyte Migration and Proliferation. Cell Physiol. Biochem. 2018, 51, 647–663. [Google Scholar] [CrossRef]
- Xiao, Q.; Luo, Y.; Shi, W.; Lu, Y.; Xiong, R.; Wu, X.; Huang, H.; Zhao, C.; Zeng, J.; Chen, C. The effects of LL-37 on virulence factors related to the quorum sensing system of Pseudomonas aeruginosa. Ann. Transl. Med. 2022, 10, 284. [Google Scholar] [CrossRef]
- Xhindoli, D.; Pacor, S.; Benincasa, M.; Scocchi, M.; Gennaro, R.; Tossi, A. The human cathelicidin LL-37—A pore-forming antibacterial peptide and host-cell modulator. Biochim. Biophys. Acta (BBA)—Biomembr. 2016, 1858, 546–566. [Google Scholar] [CrossRef]
- Dean, S.N.; Bishop, B.M.; Van Hoek, M.L. Susceptibility of Pseudomonas aeruginosa Biofilm to Alpha-Helical Peptides: D-enantiomer of LL-37. Front. Microbiol. 2011, 2, 128. [Google Scholar] [CrossRef]
- Mihai, M.M.; Bălăceanu-Gurău, B.; Ion, A.; Holban, A.M.; Gurău, C.D.; Popescu, M.N.; Beiu, C.; Popa, L.G.; Popa, M.I.; Dragomirescu, C.C.; et al. Host-Microbiome Crosstalk in Chronic Wound Healing. Int. J. Mol. Sci. 2024, 25, 4629. [Google Scholar] [CrossRef]
- Wolcott, R.D.; Hanson, J.D.; Rees, E.J.; Koenig, L.D.; Phillips, C.D.; Wolcott, R.A.; Cox, S.B.; White, J.S. Analysis of the chronic wound microbiota of 2963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2016, 24, 163–174. [Google Scholar] [CrossRef]
- Omeir, K.; Ancira, J.; Gabrilska, R.; Tipton, C.; Miller, C.; Noe, A.; Subasinghe, K.; Rowe, M.; Phillips, N.; Wolcott, J.; et al. Heritable Tissue-Specific Gene Expression Associates with Chronic Wound Microbial Species. Wound Repair Regen. 2025, 33, e70055. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, L.; Toma, M.A.; Li, D.; Bian, X.; Pastar, I.; Tomic-Canic, M.; Sommar, P.; Xu Landén, N. Integrative small and long RNA omics analysis of human healing and nonhealing wounds discovers cooperating microRNAs as therapeutic targets. eLife 2022, 11, e80322. [Google Scholar] [CrossRef]
- Schmidt, B.M. Emerging Diabetic Foot Ulcer Microbiome Analysis Using Cutting Edge Technologies. J. Diabetes Sci. Technol. 2022, 16, 353–363. [Google Scholar] [CrossRef]
- Rozas, M.; Brillet, F.; Callewaert, C.; Paetzold, B. MinION™ Nanopore Sequencing of Skin Microbiome 16S and 16S-23S rRNA Gene Amplicons. Front. Cell. Infect. Microbiol. 2022, 11, 806476. [Google Scholar] [CrossRef]
- Stec, A.; Sikora, M.; Maciejewska, M.; Paralusz-Stec, K.; Michalska, M.; Sikorska, E.; Rudnicka, L. Bacterial Metabolites: A Link between Gut Microbiota and Dermatological Diseases. Int. J. Mol. Sci. 2023, 24, 3494. [Google Scholar] [CrossRef]
- Xiong, L.; Huang, Y.X.; Mao, L.; Xu, Y.; Deng, Y.Q. Targeting gut microbiota and its associated metabolites as a potential strategy for promoting would healing in diabetes. World J. Diabetes 2025, 16, 98788. [Google Scholar] [CrossRef]
- Rembe, J.-D.; Garabet, W.; Lackmann, J.-W.; Alizadehrahrouei, S.; Augustin, M.; Dissemond, J.; Ibing, W.; Köhrer, K.; Pfeffer, K.; Rommerskirchen, A.; et al. Assessment and Monitoring of the Wound Micro-Environment in Chronic Wounds Using Standardized Wound Swabbing for Individualized Diagnostics and Targeted Interventions. Biomedicines 2024, 12, 2187. [Google Scholar] [CrossRef]
- Cordaillat-Simmons, M.; Rouanet, A.; Pot, B. Live biotherapeutic products: The importance of a defined regulatory framework. Exp. Mol. Med. 2020, 52, 1397–1406. [Google Scholar] [CrossRef]
- Rodovalho, V.d.R.; Luz, B.S.R.d.; Rabah, H.; Do Carmo, F.L.R.; Folador, E.L.; Nicolas, A.; Jardin, J.; Briard-Bion, V.; Blottière, H.; Lapaque, N. Extracellular vesicles produced by the probiotic Propionibacterium freudenreichii CIRM-BIA 129 mitigate inflammation by modulating the NF-κB pathway. Front. Microbiol. 2020, 11, 1544. [Google Scholar] [CrossRef]
- Lee, S.; Jung, S.Y.; Yoo, D.; Go, D.; Park, J.Y.; Lee, J.M.; Um, W. Alternatives of mesenchymal stem cell-derived exosomes as potential therapeutic platforms. Front. Bioeng. Biotechnol. 2024, 12, 1478517. [Google Scholar] [CrossRef]
- Piazzesi, A.; Scanu, M.; Ciprandi, G.; Putignani, L. Modulations of the skin microbiome in skin disorders: A narrative review from a wound care perspective. Int. Wound J. 2024, 21, e70087. [Google Scholar] [CrossRef]
- Omar, A.; Wright, J.B.; Schultz, G.; Burrell, R.; Nadworny, P. Microbial biofilms and chronic wounds. Microorganisms 2017, 5, 9. [Google Scholar] [CrossRef]
- Sen, C.K.; Roy, S.; Mathew-Steiner, S.S.; Gordillo, G.M. Biofilm Management in Wound Care. Plast. Reconstr. Surg. 2021, 148, 275e–288e. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Sweren, E.; Liu, H.; Wier, E.; Alphonse, M.P.; Chen, R.; Islam, N.; Li, A.; Xue, Y.; Chen, J. Bacteria induce skin regeneration via IL-1β signaling. Cell Host Microbe 2021, 29, 777–791.e776. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, J.; Zhu, W.; Kuang, Y.; Liu, T.; Zhang, W.; Chen, X.; Peng, C. Skin and Gut Microbiome in Psoriasis: Gaining Insight Into the Pathophysiology of It and Finding Novel Therapeutic Strategies. Front. Microbiol. 2020, 11, 589726. [Google Scholar] [CrossRef]
- Grogan, M.D.; Bartow-McKenney, C.; Flowers, L.; Knight, S.A.B.; Uberoi, A.; Grice, E.A. Research Techniques Made Simple: Profiling the Skin Microbiota. J. Investig. Dermatol. 2019, 139, 747–752.e741. [Google Scholar] [CrossRef]
- Antibiotic susceptibility diagnostics for the future. Nat. Microbiol. 2019, 4, 1603. [CrossRef] [PubMed]
- Wensel, C.R.; Pluznick, J.L.; Salzberg, S.L.; Sears, C.L. Next-generation sequencing: Insights to advance clinical investigations of the microbiome. J. Clin. Investig. 2022, 132, e154944. [Google Scholar] [CrossRef]
- Pérez-Cobas, A.E.; Gomez-Valero, L.; Buchrieser, C. Metagenomic approaches in microbial ecology: An update on whole-genome and marker gene sequencing analyses. Microb. Genom. 2020, 6, mgen000409. [Google Scholar] [CrossRef]
- Azli, B.; Razak, M.N.; Omar, A.R.; Mohd Zain, N.A.; Abdul Razak, F.; Nurulfiza, I. Metagenomics Insights Into the Microbial Diversity and Microbiome Network Analysis on the Heterogeneity of Influent to Effluent Water. Front. Microbiol. 2022, 13, 779196. [Google Scholar] [CrossRef]
- Be, N.A.; Allen, J.E.; Brown, T.S.; Gardner, S.N.; McLoughlin, K.S.; Forsberg, J.A.; Kirkup, B.C.; Chromy, B.A.; Luciw, P.A.; Elster, E.A.; et al. Microbial profiling of combat wound infection through detection microarray and next-generation sequencing. J. Clin. Microbiol. 2014, 52, 2583–2594. [Google Scholar] [CrossRef]
- Santiago-Rodriguez, T.M.; Le François, B.; Macklaim, J.M.; Doukhanine, E.; Hollister, E.B. The Skin Microbiome: Current Techniques, Challenges, and Future Directions. Microorganisms 2023, 11, 1222. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, H.; Zhang, X.; Wang, X.; Sun, W.; Zhang, X.; Chi, B.; Go, Y.; Chan, X.H.F.; Wu, J.; et al. Microecology in vitro model replicates the human skin microbiome interactions. Nat. Commun. 2025, 16, 3085. [Google Scholar] [CrossRef]
- Diakite, A.; Dubourg, G.; Dione, N.; Afouda, P.; Bellali, S.; Ngom, I.I.; Valles, C.; Tall, M.l.; Lagier, J.-C.; Raoult, D. Optimization and standardization of the culturomics technique for human microbiome exploration. Sci. Rep. 2020, 10, 9674. [Google Scholar] [CrossRef]
- Ndiaye, C.; Bassene, H.; Fonkou, M.D.M.; Fenollar, F.; Lagier, J.C.; Raoult, D.; Sokhna, C. The Application of Culturomics to Explore African Skin Microbiota. Am. J. Trop. Med. Hyg. 2024, 111, 1331–1337. [Google Scholar] [CrossRef]
- Brandwein, M.; Steinberg, D.; Meshner, S. Microbial biofilms and the human skin microbiome. npj Biofilms Microbiomes 2016, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Li, Z.; Gu, Z.; Liu, X.; Krutmann, J.; Wang, J.; Xia, J. Shotgun metagenomic sequencing reveals skin microbial variability from different facial sites. Front. Microbiol. 2022, 13, 933189. [Google Scholar] [CrossRef]
- Barnard, E.; Shi, B.; Kang, D.; Craft, N.; Li, H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci. Rep. 2016, 6, 39491. [Google Scholar] [CrossRef]
- Mathieu, A.; Vogel, T.M.; Simonet, P. The future of skin metagenomics. Res. Microbiol. 2014, 165, 69–76. [Google Scholar] [CrossRef]
- Jost, G.; Schwendener, S.; Liassine, N.; Perreten, V. Methicillin-resistant Macrococcus canis in a human wound. Infect. Genet. Evol. 2021, 96, 105125. [Google Scholar] [CrossRef]
- Whittle, E.; Yonkus, J.A.; Jeraldo, P.; Alva-Ruiz, R.; Nelson, H.; Kendrick, M.L.; Grys, T.E.; Patel, R.; Truty, M.J.; Chia, N. Optimizing Nanopore Sequencing for Rapid Detection of Microbial Species and Antimicrobial Resistance in Patients at Risk of Surgical Site Infections. mSphere 2022, 7, e0096421. [Google Scholar] [CrossRef]
- Morsli, M.; Salipante, F.; Magnan, C.; Dunyach-Remy, C.; Sotto, A.; Lavigne, J.P. Direct metagenomics investigation of non-surgical hard-to-heal wounds: A review. Ann. Clin. Microbiol. Antimicrob. 2024, 23, 39. [Google Scholar] [CrossRef]
- Sanjar, F.; Weaver, A.J.; Peacock, T.J.; Nguyen, J.Q.; Brandenburg, K.S.; Leung, K.P. Identification of Metagenomics Structure and Function Associated With Temporal Changes in Rat (Rattus norvegicus) Skin Microbiome During Health and Cutaneous Burn. J. Burn. Care Res. 2020, 41, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Minasyan, H. Sepsis: Mechanisms of bacterial injury to the patient. Scand. J. Trauma Resusc. Emerg. Med. 2019, 27, 19. [Google Scholar] [CrossRef] [PubMed]
- Horiba, K.; Kawada, J.I.; Okuno, Y.; Tetsuka, N.; Suzuki, T.; Ando, S.; Kamiya, Y.; Torii, Y.; Yagi, T.; Takahashi, Y.; et al. Comprehensive detection of pathogens in immunocompromised children with bloodstream infections by next-generation sequencing. Sci. Rep. 2018, 8, 3784. [Google Scholar] [CrossRef] [PubMed]
- 16S and ITS rRNA Sequencing. Available online: https://sapac.illumina.com/areas-of-interest/microbiology/microbial-sequencing-methods/16s-rrna-sequencing.html (accessed on 16 October 2025).
- Bhaskaran, S.; Saikumar, C. A Review of Next Generation Sequencing Methods and its Applications in Laboratory Diagnosis. J. Pure Appl. Microbiol. 2022, 16, 825–833. [Google Scholar] [CrossRef]
- Kong, H.H. Details Matter: Designing Skin Microbiome Studies. J. Investig. Dermatol. 2016, 136, 900–902. [Google Scholar] [CrossRef]
- Han, A.; Zenilman, J.M.; Melendez, J.H.; Shirtliff, M.E.; Agostinho, A.; James, G.; Stewart, P.S.; Mongodin, E.F.; Rao, D.; Rickard, A.H.; et al. The importance of a multifaceted approach to characterizing the microbial flora of chronic wounds. Wound Repair Regen. 2011, 19, 532–541. [Google Scholar] [CrossRef]
- Kalan, L.; Grice, E.A. Fungi in the Wound Microbiome. Adv. Wound Care 2017, 7, 247–255. [Google Scholar] [CrossRef]
- Lipof, J.S.; Jones, C.M.C.; Daiss, J.; Oh, I. Comparative study of culture, next-generation sequencing, and immunoassay for identification of pathogen in diabetic foot ulcer. J. Orthop. Res. 2021, 39, 2638–2645. [Google Scholar] [CrossRef]
- Nong, Y.; Walsh, D.M.; Maloh, J.; Dadlani, M.; Sivamani, R. Whole-Genome Shotgun Metagenomic Sequencing Reveals Shifts in the Skin Microbiome and Bacteriophages of Psoriasis: An Extended Analysis of Published Data. J. Psoriasis Psoriatic Arthritis 2024, 9, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, S.; Meola, M.; Egli, A. Combination of Whole Genome Sequencing and Metagenomics for Microbiological Diagnostics. Int. J. Mol. Sci. 2022, 23, 9834. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, A.S. Whole Genome Sequencing: Applications in Clinical Bacteriology. Med. Princ. Pract. 2024, 33, 185–197. [Google Scholar] [CrossRef]
- Chen, Y.; Knight, R.; Gallo, R.L. Evolving approaches to profiling the microbiome in skin disease. Front. Immunol. 2023, 14, 1151527. [Google Scholar] [CrossRef]
- Schürch, A.C.; Arredondo-Alonso, S.; Willems, R.J.L.; Goering, R.V. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene–based approaches. Clin. Microbiol. Infect. 2018, 24, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Jiang, G.; Li, Y.; Zhao, J.; Li, J.; Li, S.; Zhou, Z.; Xie, E.; Li, H. Multi-Omics Analysis of Wound Microbiome and Staphylococcus aureus in Pressure Ulcer. bioRxiv 2025. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.-Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
- Fida, M.; Khalil, S.; Abu Saleh, O.; Challener, D.W.; Sohail, M.R.; Yang, J.N.; Pritt, B.S.; Schuetz, A.N.; Patel, R. Diagnostic Value of 16S Ribosomal RNA Gene Polymerase Chain Reaction/Sanger Sequencing in Clinical Practice. Clin. Infect. Dis. 2021, 73, 961–968. [Google Scholar] [CrossRef]
- Meisel, J.S.; Hannigan, G.D.; Tyldsley, A.S.; SanMiguel, A.J.; Hodkinson, B.P.; Zheng, Q.; Grice, E.A. Skin Microbiome Surveys Are Strongly Influenced by Experimental Design. J. Investig. Dermatol. 2016, 136, 947–956. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, X.; Chen, A.; Li, S.; Tao, R.; Chen, K.; Huang, P.; Li, L.; Huang, J.; Li, C.; et al. Comparison of the full-length sequence and sub-regions of 16S rRNA gene for skin microbiome profiling. mSystems 2024, 9, e0039924. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, J.L.; Willner, D.; Buttini, M.; Huygens, F.; Pelzer, E.S. Limitations of 16S rRNA Gene Sequencing to Characterize Lactobacillus Species in the Upper Genital Tract. Front. Cell Dev. Biol. 2021, 9, 641921. [Google Scholar] [CrossRef]
- Abellan-Schneyder, I.; Matchado, M.S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K. Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing. mSphere 2021, 6, e01202–e01220. [Google Scholar] [CrossRef]
- Marizzoni, M.; Gurry, T.; Provasi, S.; Greub, G.; Lopizzo, N.; Ribaldi, F.; Festari, C.; Mazzelli, M.; Mombelli, E.; Salvatore, M.; et al. Comparison of Bioinformatics Pipelines and Operating Systems for the Analyses of 16S rRNA Gene Amplicon Sequences in Human Fecal Samples. Front. Microbiol. 2020, 11, 1262. [Google Scholar] [CrossRef] [PubMed]
- Butler, I.; Turner, O.; Mohammed, K.; Akhtar, M.; Evans, D.; Lambourne, J.; Harris, K.; O’Sullivan, D.M.; Sergaki, C. Standardization of 16S rRNA gene sequencing using nanopore long read sequencing technology for clinical diagnosis of culture negative infections. Front. Cell. Infect. Microbiol. 2025, 15, 1517208. [Google Scholar] [CrossRef]
- Carlisle, D.; Cunningham-Oakes, E.; Booth, J.; Frankland, A.; McDowell, M.; Pilgrim, J.; Rzeszutek, A.; Evans, C.; Larkin, S.; Li, A.; et al. Implementing portable, real-time 16S rRNA sequencing in the healthcare sector enhances antimicrobial stewardship. medRxiv 2024. [Google Scholar] [CrossRef]
- Genomics, C. 16S rRNA Sequencing vs. Metagenome Sequencing: A Guide for Selection. Available online: https://www.cd-genomics.com/resource-16s-rRNA-sequencing-vs-metagenome-sequencing.html (accessed on 15 October 2025).
- Suga, H.; Sugaya, M.; Fujita, H.; Asano, Y.; Tada, Y.; Kadono, T.; Sato, S. TLR4, rather than TLR2, regulates wound healing through TGF-β and CCL5 expression. J. Dermatol. Sci. 2014, 73, 117–124. [Google Scholar] [CrossRef]
- Huebener, P.; Schwabe, R.F. Regulation of wound healing and organ fibrosis by toll-like receptors. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2013, 1832, 1005–1017. [Google Scholar] [CrossRef]
- Machado, P.; Ribeiro, F.N.; Giublin, F.C.W.; Mieres, N.G.; Tonin, F.S.; Pontarolo, R.; Sari, M.H.M.; Lazo, R.E.L.; Ferreira, L.M. Next-Generation Wound Care: A Scoping Review on Probiotic, Prebiotic, Synbiotic, and Postbiotic Cutaneous Formulations. Pharmaceuticals 2025, 18, 704. [Google Scholar] [CrossRef]
- Smith, A.; Ghori, N.-U.-H.; Foster, R.; Nicol, M.P.; Barnett, T.; Pickering, J.; Whelan, A.; Strunk, T.; Wood, F.; Raby, E.; et al. Optimisation of the sampling method for skin microbiome studies in healthy children: A pilot cohort study. Front. Microbiomes 2024, 3, 1446394. [Google Scholar] [CrossRef]

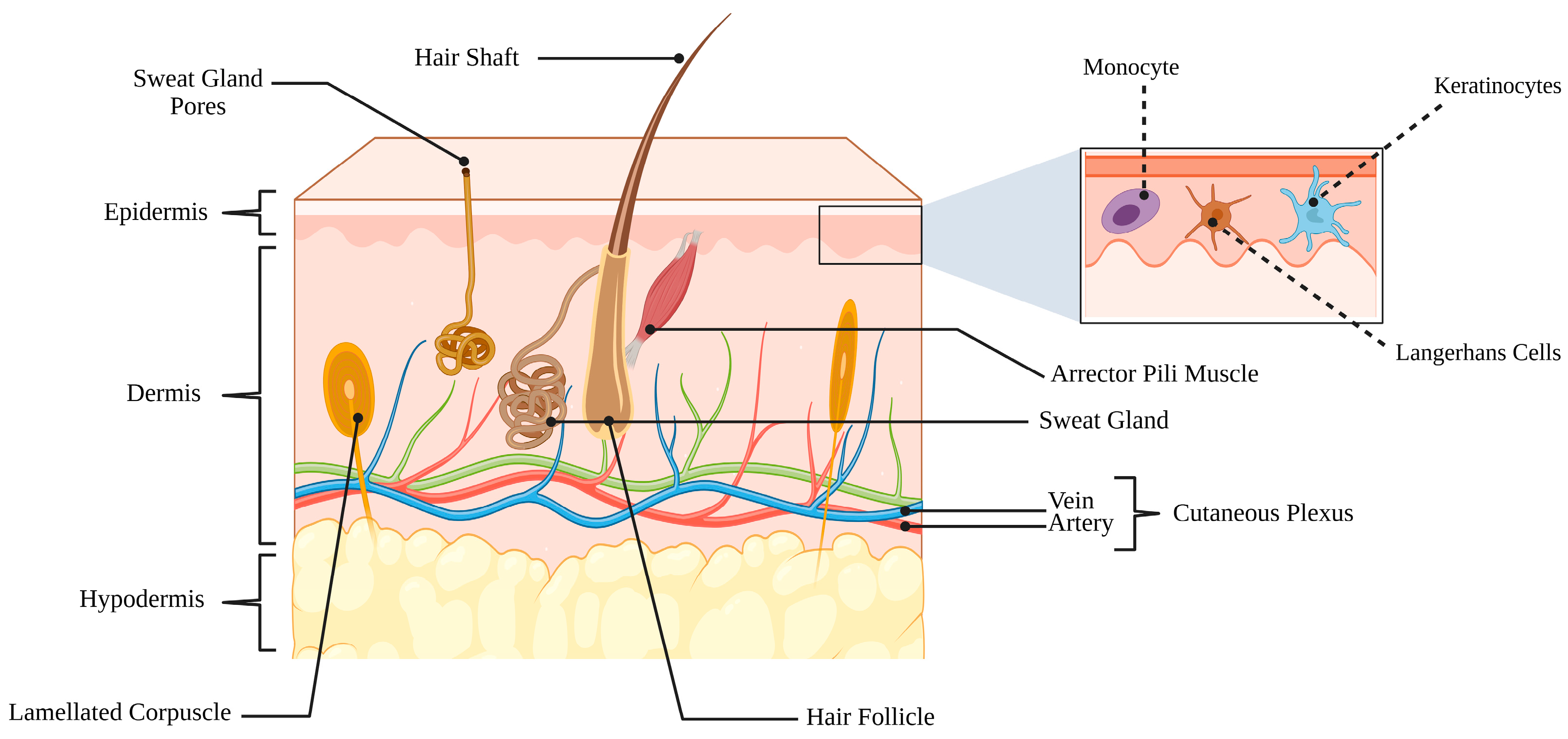
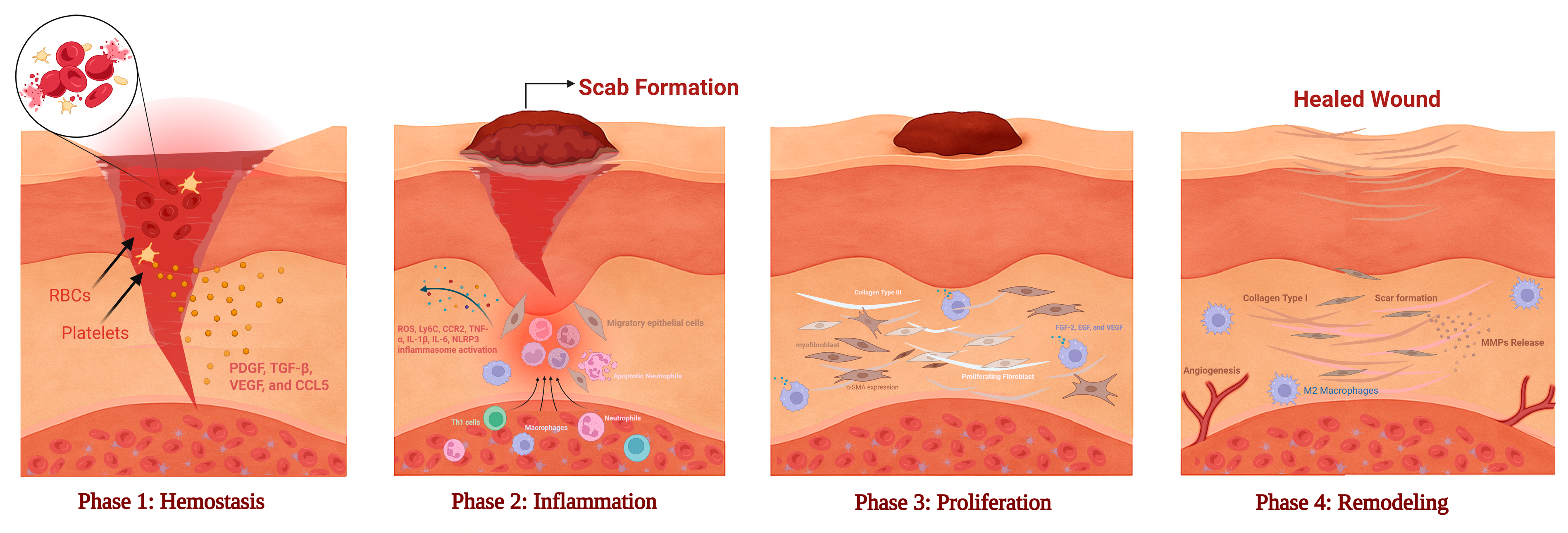
| Bacteria | Type of Study | Influence on Wound Healing | Mechanism/Host Pathway | Reference |
|---|---|---|---|---|
| Staphylococcus epidermidis | In vivo | Promotes wound healing | Dendritic cell–mediated CD8⁺ T cell IL-17A induction → enhanced barrier defence | [202,203] |
| In vivo (rat incision wound model) | Promote wound healing | Topical gel application → enhanced epithelial migration and reduced inflammation | [204] | |
| Ex vivo human skin explant model (diabetic & non-diabetic) | Promote wound healing | Keratinocyte proliferation & migration ↑ → accelerated epithelial tongue length in wounded skin | [205] | |
| Staphylococcus hominis | Clinical trial (human topical application) | Promote wound healing | Topical application → increased abundance on skin → enhanced barrier parameters and skin physiology | [206] |
| In vitro and in vivo (mouse dermonecrosis model) | Secretion of AIPs→ agr quorum-sensing inhibition in S. aureus → reduced α-toxin and protease expression → attenuated tissue injury and enhanced barrier protection | [207] | ||
| Staphylococcus aureus | In vivo and in vitro | Delays wound healing | Activates NF-κB/MAPK signaling → upregulates connexin-43 in keratinocytes → impairs keratinocyte migration and re-epithelialization | [208] |
| In vivo (rabbit ear wound model) | Mature biofilm formation → sustained low-grade inflammation → impaired tissue repair | [209] | ||
| In vitro | Biofilm-conditioned medium → keratinocyte dendrite-like morphology & increased apoptosis → delayed scratch-closure | [210] | ||
| Streptococcus pyogenes | Clinical case report | Delay wound healing | Invasive S. pyogenes infection → skin erosion, abscess formation, and milk fistula → impaired epithelial repair and inflammation | [211] |
| Pseudomonas aeruginosa | In vivo (murine excisional wound model) | Delays wound healing | Secreted proteases pseudolysin (LasB) and protease IV degrade ECM proteins (fibronectin, elastin, collagen) and disrupt keratinocyte migration → suppression of TGF-β and VEGF signaling | [97] |
| Reduced TNF-α, IL-1β, and IL-6 expression → attenuated macrophage recruitment and delayed fibroblast activation | [212] | |||
| Enterococcus faecalis | In vivo | Delays wound healing | E. faecalis formed biofilm-like microcolonies within wounds → activated TLR2–NF-κB pathway → sustained neutrophil and macrophage infiltration → chronic inflammatory microenvironment | [213] |
| E. faecalis persisted within wound tissue → modulated innate immune activation → suppressed macrophage phagocytic function and cytokine signaling (IL-1β, TNF-α) → prolonged inflammatory phase | [214] | |||
| Escherichia coli | Clinical isolates from skin & soft-tissue infections (human) | Delay wound healing | Virulence genes (cnf1, hlyA, ompT) → tissue invasion and immune evasion | [215] |
| Lactobacillus plantarum | In vivo and in vitro | Promotes wound healing | AGEs → NLRP3 inflammasome/Caspase-1/GSDMD activation → IL-1β & IL-18 ↑ → pyroptosis; L. plantarum ↓ NLRP3 → ↓ pyroptosis → improved repair | [216] |
| Lactobacillus bulgaricus & Lactobacillus plantarum | In vivo | Promote wound healing | ↓ pro-inflammatory cytokines (IL-1β, TNF-α) → reduced inflammatory cell infiltration → enhanced fibroblast migration & matrix deposition | [217] |
| Lactobacillus plantarum (GMNL-6) & L. paracasei (GMNL-653) | In vitro and In vivo | Promote wound healing | ↑ MMP-1 early phase → ECM remodelling; ↓ α-SMA/fibrosis in later phase; lipoteichoic acid mimics effect → TGF-β/Smad2 inhibition → reduced fibrosis, faster repair | [218] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Taweel, R.; Hammad, A.S.; Tajammul, A.; Crovella, S.; Al-Asmakh, M. Wounds and the Microbiota: The Healing Interplay Between Host and Microbial Communities. Int. J. Mol. Sci. 2025, 26, 11365. https://doi.org/10.3390/ijms262311365
Al-Taweel R, Hammad AS, Tajammul A, Crovella S, Al-Asmakh M. Wounds and the Microbiota: The Healing Interplay Between Host and Microbial Communities. International Journal of Molecular Sciences. 2025; 26(23):11365. https://doi.org/10.3390/ijms262311365
Chicago/Turabian StyleAl-Taweel, Raghad, Ayat S Hammad, Ali Tajammul, Sergio Crovella, and Maha Al-Asmakh. 2025. "Wounds and the Microbiota: The Healing Interplay Between Host and Microbial Communities" International Journal of Molecular Sciences 26, no. 23: 11365. https://doi.org/10.3390/ijms262311365
APA StyleAl-Taweel, R., Hammad, A. S., Tajammul, A., Crovella, S., & Al-Asmakh, M. (2025). Wounds and the Microbiota: The Healing Interplay Between Host and Microbial Communities. International Journal of Molecular Sciences, 26(23), 11365. https://doi.org/10.3390/ijms262311365







