Advancements in Animal Breeding: From Mendelian Genetics to Machine Learning
Abstract
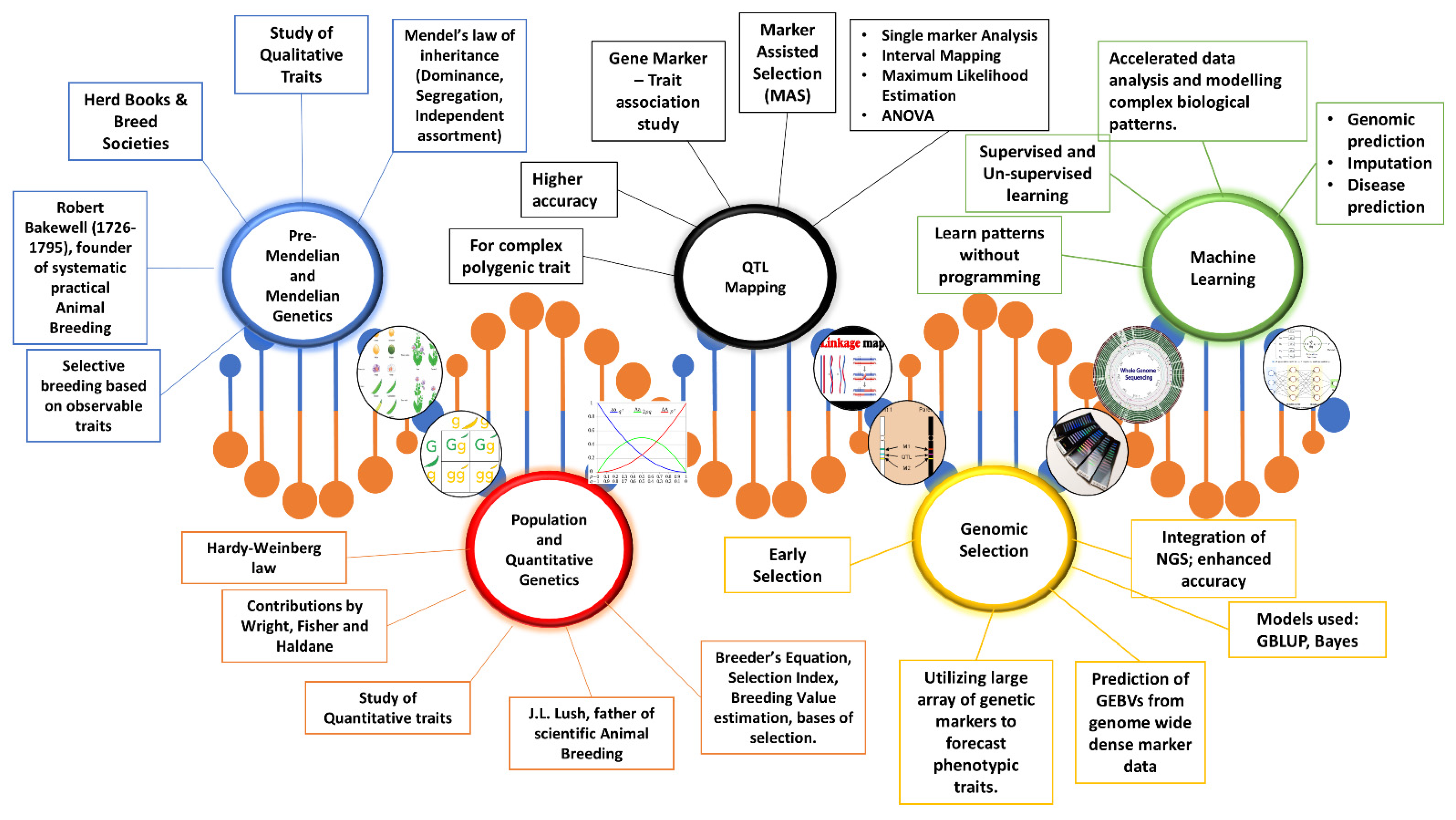
1. Introduction
2. Pre-Mendelian Era in Early Livestock Breeding
3. Quantitative Trait Loci Mapping and Its Applications in Livestock
Applications in Livestock
4. 1000 Genomes Projects in Livestock
4.1. Functional Annotation of Animal Genomes (FAANG) Consortium
4.2. Transcriptome-Wide Association Studies (TWAS) in Livestock
4.3. Mendelian Randomization (MR) Methodology
4.4. Integrative Genomic Analyses Using Phenome-Wide Association Studies (PheWAS)
4.5. FarmGTEx
5. Genomic Selection (GS)
5.1. Overview of the Transition from Phenotype-Based Selection to Genotype-Based Selection
5.2. Genomic Selection
5.3. Implementation in Livestock Breeding
6. Molecular Genetics Advances in Terms of Animal Breeding
6.1. Advances in Sequencing Technologies
6.2. Multi-Omics Approaches
7. Roles of Emerging eRNAs in Animal Breeding
8. Machine Learning and Artificial Intelligence in Genomic Prediction
8.1. Introduction to Machine Learning
8.2. Integrating Machine Learning in Animal Breeding
8.3. Case Studies on Utilizing Machine Learning Approaches in Cattle Breeding
8.3.1. Milk Production
8.3.2. Beef Production
8.3.3. Disease
9. The Concept of Phenomics and Its Advances in Animal Breeding
10. Challenges and Opportunities
11. Future Prospects
12. Conclusions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Pitt, D.; Sevane, N.; Nicolazzi, E.L.; MacHugh, D.E.; Park, S.D.E.; Colli, L.; Martinez, R.; Bruford, M.W.; Orozco-Terwengel, P. Domestication of cattle: Two or three events? Evol. Appl. 2018, 12, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, M.; Kumar, H.; Saravanan, K.A.; Rajawat, D.; Nayak, S.S.; Ghildiyal, K.; Dutt, T. Trajectory of livestock genomics in South Asia: A comprehensive review. Gene 2022, 843, 146808. [Google Scholar]
- Wykes, D.L. Robert Bakewell (1725–1795) of Dishley: Farmer and livestock improver. Agric. Hist. Rev. 2004, 52, 38–55. [Google Scholar]
- Mendel, G. Experiments on Plant Hybridization: Versuche über Pfalnzen-Hybriden; Masarykova Univerzita: Brno, Czech Republic, 1866. [Google Scholar]
- Watson, J.D.; Crick, F.H.C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Whalen, I.; Banzhaf, W.; Al Mamun, H.A.; Gondro, C. Evolution in Action: Past, Present and Future: A Festschrift in Honor of Erik D. Goodman; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Fernando, R.L.; Grossman, M. Marker assisted selection using best linear unbiased prediction. Genet. Sel. Evol. 1989, 21, 467–477. [Google Scholar]
- de Koning, D.J. Meuwissen et al. on Genomic Selection. Genetics 2016, 203, 5–7. [Google Scholar]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of Next-Generation Sequencing Systems. J. Biomed. Biotechnol. 2012, 2012, 251364. [Google Scholar] [CrossRef]
- Bermingham, M.L.; Pong-Wong, R.; Spiliopoulou, A.; Hayward, C.; Rudan, I.; Campbell, H.; Wright, A.F.; Wilson, J.F.; Agakov, F.; Navarro, P.; et al. Application of high-dimensional feature selection: Evaluation for genomic prediction in man. Sci. Rep. 2015, 5, 10312. [Google Scholar] [CrossRef]
- Hayes, B.J.; Lewin, H.A.; Goddard, M.E. The future of livestock breeding: Genomic selection for efficiency, reduced emissions intensity, and adaptation. Trends Genet. 2013, 29, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. The causes of human variability. Eugen. Rev. 1919, 10, 213. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Ma, Q.; Tanasa, B.; Nunez, E.; Chen, A.Y.; Merkurjev, D.; Zhang, J.; Ohgi, K.; Song, X.; et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013, 498, 516–520. [Google Scholar]
- Ehret, A.; Hochstuhl, D.; Krattenmacher, N.; Tetens, J.; Klein, M.; Gronwald, W.; Thaller, G. Short communication: Use of genomic and metabolic information as well as milk performance records for prediction of subclinical ketosis risk via artificial neural networks. J. Dairy Sci. 2015, 98, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Beskorovajni, R.; Jovanovic, R.; Pezo, L.; Popovic, N.; Tolimir, N.; Mihajlovic, L.; Surlan-Momirovic, G. Mathematical modeling for genomic selection in Serbian dairy cattle. Genetika 2021, 53, 1105–1115. [Google Scholar] [CrossRef]
- Yao, C.; Zhu, X.; Weigel, K.A. Semi-supervised learning for genomic prediction of novel traits with small reference populations: An application to residual feed intake in dairy cattle. Genet. Sel. Evol. 2016, 48, 84. [Google Scholar] [CrossRef]
- Abdollahi-Arpanahi, R.; Gianola, D.; Peñagaricano, F. Deep learning versus parametric and ensemble methods for genomic prediction of complex phenotypes. Genet. Sel. Evol. 2020, 52, 12. [Google Scholar] [CrossRef]
- Marques, T.C.; Marques, L.R.; Fernandes, P.B.; de Lima, F.S.; Paim, T.D.P.; Leão, K.M. Machine Learning to Predict Pregnancy in Dairy Cows: An Approach Integrating Automated Activity Monitoring and On-Farm Data. Animals 2024, 14, 1567. [Google Scholar] [CrossRef]
- Sun, C.; Wu, X.-L.; Weigel, K.A.; Rosa, G.J.M.; Bauck, S.; Woodward, B.W.; Schnabel, R.D.; Taylor, J.F.; Gianola, D. An ensemble-based approach to imputation of moderate-density genotypes for genomic selection with application to Angus cattle. Genet. Res. 2012, 94, 133–150. [Google Scholar] [CrossRef]
- Srivastava, S.; Lopez, B.I.; Kumar, H.; Jang, M.; Chai, H.-H.; Park, W.; Park, J.-E.; Lim, D. Prediction of Hanwoo Cattle Phenotypes from Genotypes Using Machine Learning Methods. Animals 2021, 11, 2066. [Google Scholar] [CrossRef]
- Liang, M.; Miao, J.; Wang, X.; Chang, T.; An, B.; Duan, X.; Xu, L.; Gao, X.; Zhang, L.; Li, J.; et al. Application of ensemble learning to genomic selection in chinese simmental beef cattle. J. Anim. Breed. Genet. 2020, 138, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Alexandre, P.A.; Ribeiro, G.; Fukumasu, H.; Sun, W.; Reverter, A.; Li, Y. Identification of Predictor Genes for Feed Efficiency in Beef Cattle by Applying Machine Learning Methods to Multi-Tissue Transcriptome Data. Front. Genet. 2021, 12, 619857. [Google Scholar] [CrossRef]
- An, B.; An, B.; Liang, M.; Liang, M.; Chang, T.; Chang, T.; Duan, X.; Duan, X.; Du, L.; Du, L.; et al. KCRR: A nonlinear machine learning with a modified genomic similarity matrix improved the genomic prediction efficiency. Brief. Bioinform. 2021, 22, bbab132. [Google Scholar] [CrossRef]
- Naderi, S.; Yin, T.; König, S. Random forest estimation of genomic breeding values for disease susceptibility over different disease incidences and genomic architectures in simulated cow calibration groups. J. Dairy Sci. 2016, 99, 7261–7273. [Google Scholar] [CrossRef]
- Swain, S.; Pattnayak, B.K.; Mohanty, M.N.; Jayasingh, S.K.; Patra, K.J.; Panda, C. Smart livestock management: Integrating IoT for cattle health diagnosis and disease prediction through machine learning. Indones. J. Electr. Eng. Comput. Sci. 2024, 34, 1192–1203. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Remington, D.L.; Purugganan, M.D. Candidate Genes, Quantitative Trait Loci, and Functional Trait Evolution in Plants. Int. J. Plant Sci. 2003, 164, S7–S20. [Google Scholar] [CrossRef]
- Mackay, T.F. The genetic architecture of quantitative traits: Lessons from Drosophila. Curr. Opin. Genet. Dev. 2004, 14, 253–257. [Google Scholar] [CrossRef]
- Roff, D.A. A centennial celebration for quantitative genetics. Evolution 2007, 61, 1017–1032. [Google Scholar] [CrossRef]
- Burt, D.W. A comprehensive review on the analysis of QTL in animals. Trends Genet. 2002, 18, 488. [Google Scholar] [CrossRef]
- Panigrahi, M.; Rajawat, D.; Nayak, S.S.; Jain, K.; Vaidhya, A.; Prakash, R.; Sharma, A.; Parida, S.; Bhushan, B.; Dutt, T. Genomic insights into key genes and QTLs involved in cattle reproduction. Gene 2024, 917, 148465. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Banerjee, P.; Joshi, J.; Kapoor, P.; Vijh, R.K. Identification of quantitative trait loci for milk yield in Murrah buffaloes. Indian J. Anim. Sci. 2018, 88, 550–557. [Google Scholar] [CrossRef]
- Goto, T.; Tsudzuki, M. Genetic Mapping of Quantitative Trait Loci for Egg Production and Egg Quality Traits in Chickens: A Review. J. Poult. Sci. 2017, 54, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, M.F.; Hu, Z.-L.; Jiang, Z. Advances in QTL Mapping in Pigs. Int. J. Biol. Sci. 2007, 3, 192–197. [Google Scholar] [CrossRef]
- Bertolini, F.; Servin, B.; Talenti, A.; Rochat, E.; Kim, E.; Oget, C.; Palhiere, I.; Crisa, A.; Catillo, G.; Steri, R. Signatures of selection and environmental adaptation across the goat genome post-domestication. Genet. Sel. Evol. 2018, 50, 57. [Google Scholar] [CrossRef]
- Bidinost, F.; Roldan, D.; Dodero, A.; Cano, E.; Taddeo, H.; Mueller, J.; Poli, M. Wool quantitative trait loci in Merino sheep. Small Rumin. Res. 2007, 74, 113–118. [Google Scholar] [CrossRef]
- Raadsma, H.W.; Fullard, K.J. QTL mapping and gene markers for resistance to infectious diseases in sheep and cattle. In Proceedings of the 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, MG, Brazil, 13–18 August 2006. [Google Scholar]
- Ghildiyal, K.; Nayak, S.S.; Rajawat, D.; Sharma, A.; Chhotaray, S.; Bhushan, B.; Dutt, T.; Panigrahi, M. Genomic insights into the conservation of wild and domestic animal diversity: A review. Gene 2023, 886, 147719. [Google Scholar] [PubMed]
- Hu, Z.-L.; Park, C.A.; Reecy, J.M. Bringing the Animal QTLdb and CorrDB into the future: Meeting new challenges and providing updated services. Nucleic Acids Res. 2021, 50, D956–D961. [Google Scholar] [CrossRef]
- Hayes, B.J.; Daetwyler, H.D. 1000 Bull Genomes Project to Map Simple and Complex Genetic Traits in Cattle: Applications and Outcomes. Annu. Rev. Anim. Biosci. 2019, 7, 89–102. [Google Scholar] [CrossRef]
- Boitard, S.; Boussaha, M.; Capitan, A.; Rocha, D.; Servin, B. Uncovering Adaptation from Sequence Data: Lessons from Genome Resequencing of Four Cattle Breeds. Genetics 2016, 203, 433–450. [Google Scholar] [CrossRef]
- Du, H.; Zhou, L.; Liu, Z.; Zhuo, Y.; Zhang, M.; Huang, Q.; Lu, S.; Xing, K.; Jiang, L.; Liu, J.-F. The 1000 Chinese Indigenous Pig Genomes Project provides insights into the genomic architecture of pigs. Nat. Commun. 2024, 15, 10137. [Google Scholar] [CrossRef]
- Denoyelle, L.; Talouarn, E.; Bardou, P.; Colli, L.; Alberti, A.; Danchin, C.; Del Corvo, M.; Engelen, S.; Orvain, C.; Palhière, I.; et al. VarGoats project: A dataset of 1159 whole-genome sequences to dissect Capra hircus global diversity. Genet. Sel. Evol. 2021, 53, 86. [Google Scholar] [CrossRef]
- Fan, W.; Hou, S.; Zhou, Z. The Duck 1000 Genomes Project: Achievements and perspectives. Anim. Res. One Heal. 2024, 2, 366–376. [Google Scholar] [CrossRef]
- Pineda, P.S.; Flores, E.B.; Villamor, L.P.; Parac, C.J.M.; Khatkar, M.S.; Thu, H.T.; Smith, T.P.L.; Rosen, B.D.; Ajmone-Marsan, P.; Colli, L.; et al. Disentangling river and swamp buffalo genetic diversity: Initial insights from the 1000 Buffalo Genomes Project. GigaScience 2024, 13, giae053. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.C.; Daetwyler, H.D.; Chamberlain, A.J.; Ponce, C.H.; Sargolzaei, M.; Schenkel, F.S.; Sahana, G.; Govignon-Gion, A.; Boitard, S.; Dolezal, M.; et al. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 2018, 50, 362–367. [Google Scholar] [CrossRef] [PubMed]
- The FAANG Consortium; Andersson, L.; Archibald, A.L.; Bottema, C.D.; Brauning, R.; Burgess, S.C.; Burt, D.W.; Casas, E.; Cheng, H.H.; Clarke, L.; et al. Coordinated international action to accelerate genome-to-phenome with FAANG, the Functional Annotation of Animal Genomes project. Genome Biol. 2015, 16, 57. [Google Scholar] [CrossRef]
- Clark, E.L.; Archibald, A.L.; Daetwyler, H.D.; Groenen, M.A.M.; Harrison, P.W.; Houston, R.D.; Kühn, C.; Lien, S.; Macqueen, D.J.; Reecy, J.M.; et al. From FAANG to fork: Application of highly annotated genomes to improve farmed animal production. Genome Biol. 2020, 21, 285. [Google Scholar] [CrossRef]
- Peng, S.; Dahlgren, A.R.; Donnelly, C.G.; Hales, E.N.; Petersen, J.L.; Bellone, R.R.; Kalbfleisch, T.; Finno, C.J. Functional annotation of the animal genomes: An integrated annotation resource for the horse. PLOS Genet. 2023, 19, e1010468. [Google Scholar] [CrossRef]
- Young, R.; Lefevre, L.; Bush, S.J.; Joshi, A.; Singh, S.H.; Jadhav, S.K.; Dhanikachalam, V.; Lisowski, Z.M.; Iamartino, D.; Summers, K.M.; et al. A Gene Expression Atlas of the Domestic Water Buffalo (Bubalus bubalis). Front. Genet. 2019, 10, 668. [Google Scholar] [CrossRef]
- Pan, Z.; Yao, Y.; Yin, H.; Cai, Z.; Wang, Y.; Bai, L.; Kern, C.; Halstead, M.; Chanthavixay, G.; Trakooljul, N.; et al. Pig genome functional annotation enhances the biological interpretation of complex traits and human disease. Nat. Commun. 2021, 12, 5848. [Google Scholar] [CrossRef]
- Kern, C.; Wang, Y.; Xu, X.; Pan, Z.; Halstead, M.; Chanthavixay, G.; Saelao, P.; Waters, S.; Xiang, R.; Chamberlain, A.; et al. Functional annotations of three domestic animal genomes provide vital resources for comparative and agricultural research. Nat. Commun. 2021, 12, 1821. [Google Scholar] [CrossRef] [PubMed]
- Goszczynski, D.E.; Halstead, M.M.; Islas-Trejo, A.D.; Zhou, H.; Ross, P.J. Transcription initiation mapping in 31 bovine tissues reveals complex promoter activity, pervasive transcription, and tissue-specific promoter usage. Genome Res. 2021, 31, 732–744. [Google Scholar]
- Kuang, W.; Zinner, D.; Li, Y.; Yao, X.; Roos, C.; Yu, L. Recent Advances in Genetics and Genomics of Snub-Nosed Monkeys (Rhinopithecus) and Their Implications for Phylogeny, Conservation, and Adaptation. Genes 2023, 14, 985. [Google Scholar] [CrossRef]
- Deng, Y.; Pan, W. Model checking via testing for direct effects in Mendelian Randomization and transcriptome-wide association studies. PLoS Comput. Biol. 2021, 17, e1009266. [Google Scholar] [CrossRef]
- Pathak, G.A.; Singh, K.; Miller-Fleming, T.W.; Wendt, F.R.; Ehsan, N.; Hou, K.; Johnson, R.; Lu, Z.; Gopalan, S.; Yengo, L.; et al. Integrative genomic analyses identify susceptibility genes underlying COVID-19 hospitalization. Nat. Commun. 2021, 12, 4569. [Google Scholar] [CrossRef]
- Li, R.; Li, C.; Chen, H. PigBiobank: A valuable resource for understanding genetic and biological mechanisms of diverse complex traits in pigs. Nucleic Acids Res. 2024, 52, D980–D987. [Google Scholar]
- Stock, J.; Bennewitz, J.; Hinrichs, D.; Wellmann, R. A Review of Genomic Models for the Analysis of Livestock Crossbred Data. Front. Genet. 2020, 11, 568. [Google Scholar] [CrossRef]
- Goddard, M.E.; Hayes, B.J.; Meuwissen, T.H.E. Genomic selection in livestock populations. Genet. Res. 2010, 92, 413–421. [Google Scholar] [CrossRef]
- VanRaden, P. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Berry, D.; Garcia, J.; Garrick, D. Development and implementation of genomic predictions in beef cattle. Anim. Front. 2016, 6, 32–38. [Google Scholar] [CrossRef]
- Wiggans, G.; VanRaden, P.; Cooper, T. The genomic evaluation system in the United States: Past, present, future. J. Dairy Sci. 2011, 94, 3202–3211. [Google Scholar] [CrossRef]
- Lund, M.S.; de Roos, A.P.; de Vries, A.G.; Druet, T.; Ducrocq, V.; Fritz, S.; Guillaume, F.; Guldbrandtsen, B.; Liu, Z.; Reents, R.; et al. A common reference population from four European Holstein populations increases reliability of genomic predictions. Genet. Sel. Evol. 2011, 43, 43. [Google Scholar] [CrossRef]
- Henderson, C.R. Applications of linear models in animal breeding. In Experiments in Plant Hybridization (1865); University of Guelph Press: Guelph, ON, Canada, 1984; pp. 3–47. [Google Scholar]
- Wolc, A.; Dekkers, J.C.M. Application of Bayesian genomic prediction methods to genome-wide association analyses. Genet. Sel. Evol. 2022, 54, 31. [Google Scholar] [CrossRef]
- Nadaf, J.; Riggio, V.; Yu, T.-P.; Pong-Wong, R. Effect of the prior distribution of SNP effects on the estimation of total breeding value. BMC Proc. 2012, 6, S6. [Google Scholar] [CrossRef]
- de Los, G.; Hickey, J.M.; Pong-Wong, R.; Daetwyler, H.D.; Calus, M.P.L. Whole-Genome Regression and Prediction Methods Applied to Plant and Animal Breeding. Genetics 2013, 193, 327–345. [Google Scholar] [CrossRef]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the bayesian alphabet for genomic selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef]
- Whittaker, J.C.; Thompson, R.; Denham, M.C. Marker-assisted selection using ridge regression. Genet. Res. 2000, 75, 249–252. [Google Scholar] [CrossRef]
- Habier, D.; Fernando, R.L.; Garrick, D.J. Genomic BLUP Decoded: A Look into the Black Box of Genomic Prediction. Genetics 2013, 194, 597–607. [Google Scholar] [CrossRef]
- Gianola, D. Priors in Whole-Genome Regression: The Bayesian Alphabet Returns. Genetics 2013, 194, 573–596. [Google Scholar] [CrossRef]
- Rupp, R.; Mucha, S.; Larroque, H.; McEwan, J.; Conington, J. Genomic application in sheep and goat breeding. Anim. Front. 2016, 6, 39–44. [Google Scholar] [CrossRef]
- Gao, N.; Teng, J.; Pan, R.; Li, X.; Ye, S.; Li, J.; Zhang, H.; Zhang, X.; Zhang, Z. Accuracy of whole genome prediction with single-step GBLUP in a Chinese yellow-feathered chicken population. Livest. Sci. 2019, 230, 103817. [Google Scholar] [CrossRef]
- Song, H.; Zhang, J.; Zhang, Q.; Ding, X. Using Different Single-Step Strategies to Improve the Efficiency of Genomic Prediction on Body Measurement Traits in Pig. Front. Genet. 2019, 9, 730. [Google Scholar] [CrossRef]
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef]
- Chen, C.Y.; Misztal, I.; Aguilar, I.; Tsuruta, S.; Meuwissen, T.H.E.; Aggrey, S.E.; Wing, T.; Muir, W.M. Genome-wide marker-assisted selection combining all pedigree phenotypic information with genotypic data in one step: An example using broiler chickens. J. Anim. Sci. 2011, 89, 23–28. [Google Scholar] [CrossRef]
- Wray, N.R.; Goddard, M.E. Increasing long-term response to selection. Genet. Sel. Evol. 1994, 26, 431–451. [Google Scholar]
- Meuwissen, T. Maximizing the response of selection with a predefined rate of inbreeding. J. Anim. Sci. 1997, 75, 934–940. [Google Scholar] [CrossRef]
- Sonesson, A.K.; Meuwissen, T.H.; Goddard, M.E. The use of communal rearing of families and DNA pooling in aquaculture genomic selection schemes. Genet. Sel. Evol. 2010, 42, 41. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Sharma, N.; Singh, A.K.; Gera, M.; Pulicherla, K.K.; Jeong, D.K. Transformation of animal genomics by next-generation sequencing technologies: A decade of challenges and their impact on genetic architecture. Crit. Rev. Biotechnol. 2018, 38, 1157–1175. [Google Scholar] [CrossRef]
- Rautiainen, M.; Nurk, S.; Walenz, B.P.; Logsdon, G.A.; Porubsky, D.; Rhie, A.; Eichler, E.E.; Phillippy, A.M.; Koren, S. Telomere-to-telomere assembly of diploid chromosomes with Verkko. Nat. Biotechnol. 2023, 41, 1474–1482. [Google Scholar] [CrossRef]
- Buermans, H.P.J.; Den Dunnen, J.T. Next generation sequencing technology: Advances and applications. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2014, 1842, 1932–1941. [Google Scholar] [CrossRef]
- Bleidorn, C. Third generation sequencing: Technology and its potential impact on evolutionary biodiversity research. Syst. Biodivers. 2015, 14, 1–8. [Google Scholar] [CrossRef]
- Stevenson, K.R.; Coolon, J.D.; Wittkopp, P.J. Sources of bias in measures of allele-specific expression derived from RNA-seq data aligned to a single reference genome. BMC Genom. 2013, 14, 536. [Google Scholar] [CrossRef]
- Brandt, D.Y.C.; Aguiar, V.R.C.; Bitarello, B.D.; Nunes, K.; Goudet, J.; Meyer, D. Mapping Bias Overestimates Reference Allele Frequencies at the HLA Genes in the 1000 Genomes Project Phase I Data. G3 Genes Genomes Genet. 2015, 5, 931–941. [Google Scholar] [CrossRef]
- Chaisson, M.J.P.; Sanders, A.D.; Zhao, X.; Malhotra, A.; Porubsky, D.; Rausch, T.; Gardner, E.J.; Rodriguez, O.L.; Guo, L.; Collins, R.L.; et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019, 10, 1784. [Google Scholar] [CrossRef]
- Sirén, J.; Eskandar, P.; Ungaro, M.T.; Hickey, G.; Eizenga, J.M.; Novak, A.M.; Chang, X.; Chang, P.-C.; Kolmogorov, M.; Carroll, A.; et al. Personalized pangenome references. Nat. Methods 2024, 21, 2017–2023. [Google Scholar] [CrossRef]
- Leonard, A.S.; Mapel, X.M.; Pausch, H. Pangenome-genotyped structural variation improves molecular phenotype mapping in cattle. Genome Res. 2024, 34, 300–309. [Google Scholar] [CrossRef]
- Li, Z.; Liu, X.; Wang, C.; Li, Z.; Jiang, B.; Zhang, R.; Tong, L.; Qu, Y.; He, S.; Chen, H.; et al. The pig pangenome provides insights into the roles of coding structural variations in genetic diversity and adaptation. Genome Res. 2023, 33, 1833–1847. [Google Scholar] [CrossRef]
- Rice, E.S.; Alberdi, A.; Alfieri, J.; Athrey, G.; Balacco, J.R.; Bardou, P.; Blackmon, H.; Charles, M.; Cheng, H.H.; Fedrigo, O.; et al. A pangenome graph reference of 30 chicken genomes allows genotyping of large and complex structural variants. BMC Biol. 2023, 21, 267. [Google Scholar] [CrossRef]
- Sanyal, A.; Lajoie, B.R.; Jain, G.; Dekker, J. The long-range interaction landscape of gene promoters. Nature 2012, 489, 109–113. [Google Scholar] [CrossRef]
- Bai, L.; Liang, R.; Yang, Y.; Hou, X.; Wang, Z.; Zhu, S.; Wang, C.; Tang, Z.; Li, K. MicroRNA-21 Regulates PI3K/Akt/mTOR Signaling by Targeting TGFβI during Skeletal Muscle Development in Pigs. PLoS ONE 2015, 10, e0119396. [Google Scholar] [CrossRef]
- Zhao, W.; Mu, Y.; Ma, L.; Wang, C.; Tang, Z.; Yang, S.; Zhou, R.; Hu, X.; Li, M.-H.; Li, K. Systematic identification and characterization of long intergenic non-coding RNAs in fetal porcine skeletal muscle development. Sci. Rep. 2015, 5, 8957. [Google Scholar] [CrossRef]
- Marino, R.; Albenzio, M.; della Malva, A.; Caroprese, M.; Santillo, A.; Sevi, A. Changes in meat quality traits and sarcoplasmic proteins during aging in three different cattle breeds. Meat Sci. 2014, 98, 178–186. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yu, Y.; Zhang, Y.; Song, J.; Mitra, A.; Zhang, Y.; Wang, Y.; Sun, D.; Zhang, S. Genome-Wide Bovine H3K27me3 Modifications and the Regulatory Effects on Genes Expressions in Peripheral Blood Lymphocytes. PLoS ONE 2012, 7, e39094. [Google Scholar] [CrossRef] [PubMed]
- Kadarmideen, H.N. Genomics to systems biology in animal and veterinary sciences: Progress, lessons and opportunities. Livest. Sci. 2014, 166, 232–248. [Google Scholar] [CrossRef]
- Woelders, H.; Pas, M.T.; Bannink, A.; Veerkamp, R.; Smits, M. Systems biology in animal sciences. Animal 2011, 5, 1036–1047. [Google Scholar] [CrossRef]
- Saravanan, K.; Panigrahi, M.; Kumar, H.; Nayak, S.S.; Rajawat, D.; Bhushan, B.; Dutt, T. Progress and future perspectives of livestock genomics in India: A mini review. Anim. Biotechnol. 2022, 34, 1979–1987. [Google Scholar] [CrossRef]
- Jin, W.; Jiang, G.; Yang, Y.; Yang, J.; Yang, W.; Wang, D.; Niu, X.; Zhong, R.; Zhang, Z.; Gong, J. Animal-eRNAdb: A comprehensive animal enhancer RNA database. Nucleic Acids Res. 2021, 50, D46–D53. [Google Scholar] [CrossRef]
- Mikhaylichenko, O.; Bondarenko, V.; Harnett, D.; Schor, I.E.; Males, M.; Viales, R.R.; Furlong, E.E.M. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 2018, 32, 42–57. [Google Scholar] [CrossRef]
- Sartorelli, V.; Lauberth, S.M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 2020, 27, 521–528. [Google Scholar] [CrossRef]
- Tsai, P.-F.; Dell’orso, S.; Rodriguez, J.; Vivanco, K.O.; Ko, K.-D.; Jiang, K.; Juan, A.H.; Sarshad, A.A.; Vian, L.; Tran, M.; et al. A Muscle-Specific Enhancer RNA Mediates Cohesin Recruitment and Regulates Transcription In trans. Mol. Cell 2018, 71, 129–141.e8. [Google Scholar] [CrossRef]
- Benhammou, J.N.; Ko, A.; Alvarez, M.; Kaikkonen, M.U.; Rankin, C.; Garske, K.M.; Padua, D.; Bhagat, Y.; Kaminska, D.; Kärjä, V.; et al. Novel lipid long intervening noncoding RNA, oligodendrocyte maturation-associated long intergenic noncoding RNA, regulates the liver steatosis gene stearoyl-coenzyme A desaturase as an enhancer RNA. Hepatol. Commun. 2019, 3, 1356–1372. [Google Scholar] [PubMed]
- Hu, H.; Miao, Y.R.; Jia, L.H.; Yu, Q.Y.; Zhang, Q.; Guo, A.Y. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019, 47, D33–D38. [Google Scholar] [PubMed]
- Hu, Z.-L.; Park, C.A.; Reecy, J.M. Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Res. 2019, 47, D701–D710. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yang, Y.; Zhao, C.; Yang, K.; Wang, D.; Yang, J.; Niu, X.; Gong, J. Animal-ImputeDB: A comprehensive database with multiple animal reference panels for genotype imputation. Nucleic Acids Res. 2019, 48, D659–D667. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, W.; Pan, X.; Liao, W.; Shen, Q.; Cai, J.; Gong, W.; Tian, Y.; Xu, D.; Li, Y.; et al. Pig-eRNAdb: A comprehensive enhancer and eRNA dataset of pigs. Sci. Data 2024, 11, 157. [Google Scholar] [CrossRef]
- Awad, M.; Khanna, R. Machine learning. In Efficient Learning Machines; Apress: Berkeley, CA, USA, 2015; pp. 1–18. [Google Scholar]
- Sarker, I.H. Machine Learning: Algorithms, Real-World Applications and Research Directions. SN Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef]
- Kumar, H.; Panigrahi, M.; Panwar, A.; Rajawat, D.; Nayak, S.S.; Saravanan, K.; Kaisa, K.; Parida, S.; Bhushan, B.; Dutt, T. Machine-Learning Prospects for Detecting Selection Signatures Using Population Genomics Data. J. Comput. Biol. 2022, 29, 943–960. [Google Scholar] [CrossRef]
- Jadhav, P.; Patil, V.; Gore, S. A comparative study of linear regression and regression tree. SSRN Electron. J. 2020. [Google Scholar] [CrossRef]
- Shabtay, L.; Fournier-Viger, P.; Yaari, R.; Dattner, I. A guided FP-Growth algorithm for mining multitude-targeted item-sets and class association rules in imbalanced data. Inf. Sci. 2021, 553, 353–375. [Google Scholar] [CrossRef]
- Xiao, T.; Xia, T.; Yang, Y.; Huang, C.; Wang, X. Learning from massive noisy labeled data for image classification. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Li, Y.; Wu, F.-X.; Ngom, A. A review on machine learning principles for multi-view biological data integration. Brief. Bioinform. 2016, 19, 325–340. [Google Scholar] [CrossRef]
- David, L.; Arús-Pous, J.; Karlsson, J.; Engkvist, O.; Bjerrum, E.J.; Kogej, T.; Kriegl, J.M.; Beck, B.; Chen, H. Applications of Deep-Learning in Exploiting Large-Scale and Heterogeneous Compound Data in Industrial Pharmaceutical Research. Front. Pharmacol. 2019, 10, 1303. [Google Scholar] [CrossRef]
- Tarjan, L.; Senk, I.; Pracner, D.; Rajkovic, D.; Strbac, L. Possibilities for applying machine learning in dairy cattle breeding. In Proceedings of the 2021 20th International Symposium INFOTEH-JAHORINA (INFOTEH), Jahorina, Bosnia, 17–19 March 2021. [Google Scholar]
- Caudai, C.; Galizia, A.; Geraci, F.; Le Pera, L.; Morea, V.; Salerno, E.; Via, A.; Colombo, T. AI applications in functional genomics. Comput. Struct. Biotechnol. J. 2021, 19, 5762–5790. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shi, S.; Wang, G.; Luo, W.; Wei, X.; Qiu, A.; Luo, F.; Ding, X. Using machine learning to improve the accuracy of genomic prediction of reproduction traits in pigs. J. Anim. Sci. Biotechnol. 2022, 13, 60. [Google Scholar] [CrossRef]
- Lee, H.-J.; Lee, J.H.; Gondro, C.; Koh, Y.J.; Lee, S.H. deepGBLUP: Joint deep learning networks and GBLUP framework for accurate genomic prediction of complex traits in Korean native cattle. Genet. Sel. Evol. 2023, 55, 56. [Google Scholar] [CrossRef]
- Chafai, N.; Hayah, I.; Houaga, I.; Badaoui, B. A review of machine learning models applied to genomic prediction in animal breeding. Front. Genet. 2023, 14, 1150596. [Google Scholar] [CrossRef]
- Zeng, S.; Zeng, S.; Mao, Z.; Mao, Z.; Ren, Y.; Ren, Y.; Wang, D.; Wang, D.; Xu, D.; Xu, D.; et al. G2PDeep: A web-based deep-learning framework for quantitative phenotype prediction and discovery of genomic markers. Nucleic Acids Res. 2021, 49, W228–W236. [Google Scholar] [CrossRef] [PubMed]
- Zitnik, M.; Nguyen, F.; Wang, B.; Leskovec, J.; Goldenberg, A.; Hoffman, M.M. Machine learning for integrating data in biology and medicine: Principles, practice, and opportunities. Inf. Fusion 2018, 50, 71–91. [Google Scholar] [CrossRef]
- Tadist, K.; Najah, S.; Nikolov, N.S.; Mrabti, F.; Zahi, A. Feature selection methods and genomic big data: A systematic review. J. Big Data 2019, 6, 79. [Google Scholar] [CrossRef]
- Grisart, B.; Coppieters, W.; Farnir, F.; Karim, L.; Ford, C.; Berzi, P.; Cambisano, N.; Mni, M.; Reid, S.; Simon, P.; et al. Positional Candidate Cloning of a QTL in Dairy Cattle: Identification of a Missense Mutation in the Bovine DGAT1 Gene with Major Effect on Milk Yield and Composition. Genome Res. 2002, 12, 222–231. [Google Scholar] [CrossRef]
- Casas, E.; Shackelford, S.D.; Keele, J.W.; Koohmaraie, M.; Smith, T.P.L.; Stone, R.T. Detection of quantitative trait loci for growth and carcass composition in cattle. J. Anim. Sci. 2003, 81, 2976–2983. [Google Scholar] [CrossRef]
- Pushpa; Magotra, A.; Bangar, Y.C.; Patil, C.S.; Kamaldeep; Sindhu, V.; Malik, D.; Chaudhary, P.; Garg, A.R.; Kumar, S. Association of CXCR1 gene polymorphism with clinical mastitis, reproductive disorders and performance traits in Hardhenu (Bos taurus × Bos indicus) cattle. Reprod. Domest. Anim. 2023, 58, 1234–1243. [Google Scholar]
- Olsen, H.G.; Knutsen, T.M.; Lewandowska-Sabat, A.M.; Grove, H.; Nome, T.; Svendsen, M.; Arnyasi, M.; Sodeland, M.; Sundsaasen, K.K.; Dahl, S.R.; et al. Fine mapping of a QTL on bovine chromosome 6 using imputed full sequence data suggests a key role for the group-specific component (GC) gene in clinical mastitis and milk production. Genet. Sel. Evol. 2016, 48, 79. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; He, H.; Wei, C. Cellular and potential molecular mechanisms underlying transovarial transmission of the obligate symbiont Sulcia in cicadas. Environ. Microbiol. 2023, 25, 836–852. [Google Scholar] [CrossRef]
- Zhao, H.; Hu, R.; Li, F.; Yue, X. Five SNPs Within the FGF5 Gene Significantly Affect Both Wool Traits and Growth Performance in Fine-Wool Sheep (Ovis aries). Front. Genet. 2021, 12, 732097. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, F.; Gao, G.; Yan, X.; Liu, H.; Liu, Z.; Wang, Z.; He, L.; Lv, Q.; Wang, Z.; et al. Genome-Wide Association Study of Body Weight Traits in Inner Mongolia Cashmere Goats. Front. Veter. Sci. 2021, 8, 752746. [Google Scholar] [CrossRef]
- Edwards, D.B.; Ernst, C.W.; Raney, N.E.; Doumit, M.E.; Hoge, M.D.; Bates, R.O. Quantitative trait locus mapping in an F2 Duroc × Pietrain resource population: II. Carcass and meat quality traits. J. Anim. Sci. 2008, 86, 254–266. [Google Scholar]
- Liu, X.; Li, Y.I.; Pritchard, J.K. Trans Effects on Gene Expression Can Drive Omnigenic Inheritance. Cell 2019, 177, 1022–1034.e6. [Google Scholar] [CrossRef]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Danielsen, J.M.R.; Yang, Y.-G.; Qi, Y. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef]
- Nayak, S.S.; Panigrahi, M.; Rajawat, D.; Ghildiyal, K.; Sharma, A.; Jain, K.; Bhushan, B.; Dutt, T. Deciphering climate resilience in Indian cattle breeds by selection signature analyses. Trop. Anim. Heal. Prod. 2024, 56, 46. [Google Scholar] [CrossRef]
- Houle, D.; Govindaraju, D.R.; Omholt, S. Phenomics: The next challenge. Nat. Rev. Genet. 2010, 11, 855–866. [Google Scholar] [CrossRef]
- Pérez-Enciso, M.; Steibel, J.P. Phenomes: The current frontier in animal breeding. Genet. Sel. Evol. 2021, 53, 22. [Google Scholar] [CrossRef] [PubMed]

| Category | Reference | Focus | Models/Algorithms Used | Key Findings |
|---|---|---|---|---|
| Milk Production | [17] | Predicting yield and fertility traits in dairy cattle | ANN (Broyden-Fletcher-Goldfarb-Shanno algorithm) | R2 values: 0.444–0.989; strong predictive accuracy. |
| [18] | Genomic prediction of residual feed intake in HF cattle | Self-trained vs. measured phenotypes | Reduced prediction accuracy as training set size increased. Maximum accuracy improvement: 5.9%. | |
| [19] | Predicting sire conception rate in Holstein cattle | Deep learning (MLP, CNN), Ensemble (RF, Gradient Boosting), Parametric (GBLUP, Bayes B) | Gradient Boosting achieved highest correlation (0.36), followed by Bayes B (0.34). | |
| [20] | Predicting pregnancy likelihood in dairy cows using AAM and on-farm data | Random Forest | Effective in reducing prediction errors by integrating diverse data sources. | |
| Beef Production | [21] | Accuracy of genomic imputation tools in Angus cattle | Beagle, Fimpute, IMPUTE, AlphaImpute, AdaBoost-like | Beagle and Fimpute showed top accuracy (0.8677–0.9858). |
| [22] | Predicting carcass traits in Hanwoo cattle | XGB, GBLUP | XGB excelled in carcass weight and marbling score; GBLUP performed better for backfat thickness and eye muscle area. | |
| [15] | Identifying SNPs for genomic relationship matrices in Brahman cattle | Random Forest, Gradient Boosting, XGBoost | Machine learning refined genomic selection by identifying key genetic markers. | |
| [23] | Forecasting genomic values in Simmental beef cattle | Adaboost.RT with SVR | Achieved 5.4–14.9% accuracy improvement over GBLUP. | |
| [24] | Classifying Nellore cattle into feed-efficiency groups | RF, XGBoost, RX | RX identified 117 significant genes with higher classification accuracy than traditional methods. | |
| [25] | Comparing heritabilities and genetic architectures in Simmental cattle | GBLUP, Bayes B, KcRR, SVR | Machine learning showed improved accuracy for traits like carcass weight and live weight. | |
| Disease | [16] | Predicting subclinical ketosis risk in dairy cows | ANN | Predictive accuracy up to 0.643 for metabolic and milk performance data. |
| [26] | Forecasting genetic risks for binary disease traits | gBLUP, RF | RF highlighted strengths in disease risk prediction; useful for exploring genetic risks. | |
| [27] | Integrating IoT data for cattle disease prediction | RF, NBM, lazy-IBk, PART, SVM | Random Forest showed highest accuracy for diagnosing diseases like milk fever, lameness, and metabolic conditions. |
| Species | Trait | Chromosome (QTL Location) | QTL IDs (Respective to CHR) | Key Findings | Reference |
|---|---|---|---|---|---|
| Cattle | Milk fat percentage | BTA14 | QTL ID: 10581 | DGAT1 on BTA14 influences milk fat content | [126] |
| Cattle | Growth rate, carcass traits | BTA16, BTA20, BTA21 | QTL ID: 1355, 1357, 1358 | QTLs linked to growth, meat quality | [127] |
| Cattle | Disease resistance (Clinical Mastitis) | BTA6, BTA2 | QTL ID: 137487, 283197 | QTLs on BTA6, CXCR1 gene on BTA2 associated with resistance against clinical mastitis | [128,129] |
| Sheep | Litter size, reproduction | OAR5, OARX | QTL ID: 13837, 281743 | GDF9 and BMP15 mutations linked to prolificacy | [130] |
| Sheep | Greasy fleece weight | OAR6 | QTL ID: 238780 | Five FGF5 SNPs affect wool traits and growth in fine-wool sheep. | [131] |
| Goat | Body Weight | CHI25 | QTL ID: 255224 | MAPK3 genes associated with the QTL | [132] |
| Pigs | Growth, leanness, meat quality | SSC6, SSC11, SSC16 | QTL ID: 3650, 3211, 3214 | QTLs for backfat thickness, subcutaneous fat thickness, muscle mass | [133] |
| Poultry | Egg production, body weight | GGA1, GGAZ | QTL ID: 177426, 19583 | QTLs affecting egg number, weight gain | [134,135] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panigrahi, M.; Rajawat, D.; Nayak, S.S.; Bose, A.; Bharia, N.; Singh, S.; Sharma, A.; Dutt, T. Advancements in Animal Breeding: From Mendelian Genetics to Machine Learning. Int. J. Mol. Sci. 2025, 26, 11352. https://doi.org/10.3390/ijms262311352
Panigrahi M, Rajawat D, Nayak SS, Bose A, Bharia N, Singh S, Sharma A, Dutt T. Advancements in Animal Breeding: From Mendelian Genetics to Machine Learning. International Journal of Molecular Sciences. 2025; 26(23):11352. https://doi.org/10.3390/ijms262311352
Chicago/Turabian StylePanigrahi, Manjit, Divya Rajawat, Sonali Sonejita Nayak, Anal Bose, Nishu Bharia, Shreyasi Singh, Anurodh Sharma, and Triveni Dutt. 2025. "Advancements in Animal Breeding: From Mendelian Genetics to Machine Learning" International Journal of Molecular Sciences 26, no. 23: 11352. https://doi.org/10.3390/ijms262311352
APA StylePanigrahi, M., Rajawat, D., Nayak, S. S., Bose, A., Bharia, N., Singh, S., Sharma, A., & Dutt, T. (2025). Advancements in Animal Breeding: From Mendelian Genetics to Machine Learning. International Journal of Molecular Sciences, 26(23), 11352. https://doi.org/10.3390/ijms262311352





