A Duplex qPCR Assay Targeting the fadA Gene Enables Robust Detection of Fusobacterium in Clinical Samples
Abstract
1. Introduction
2. Results
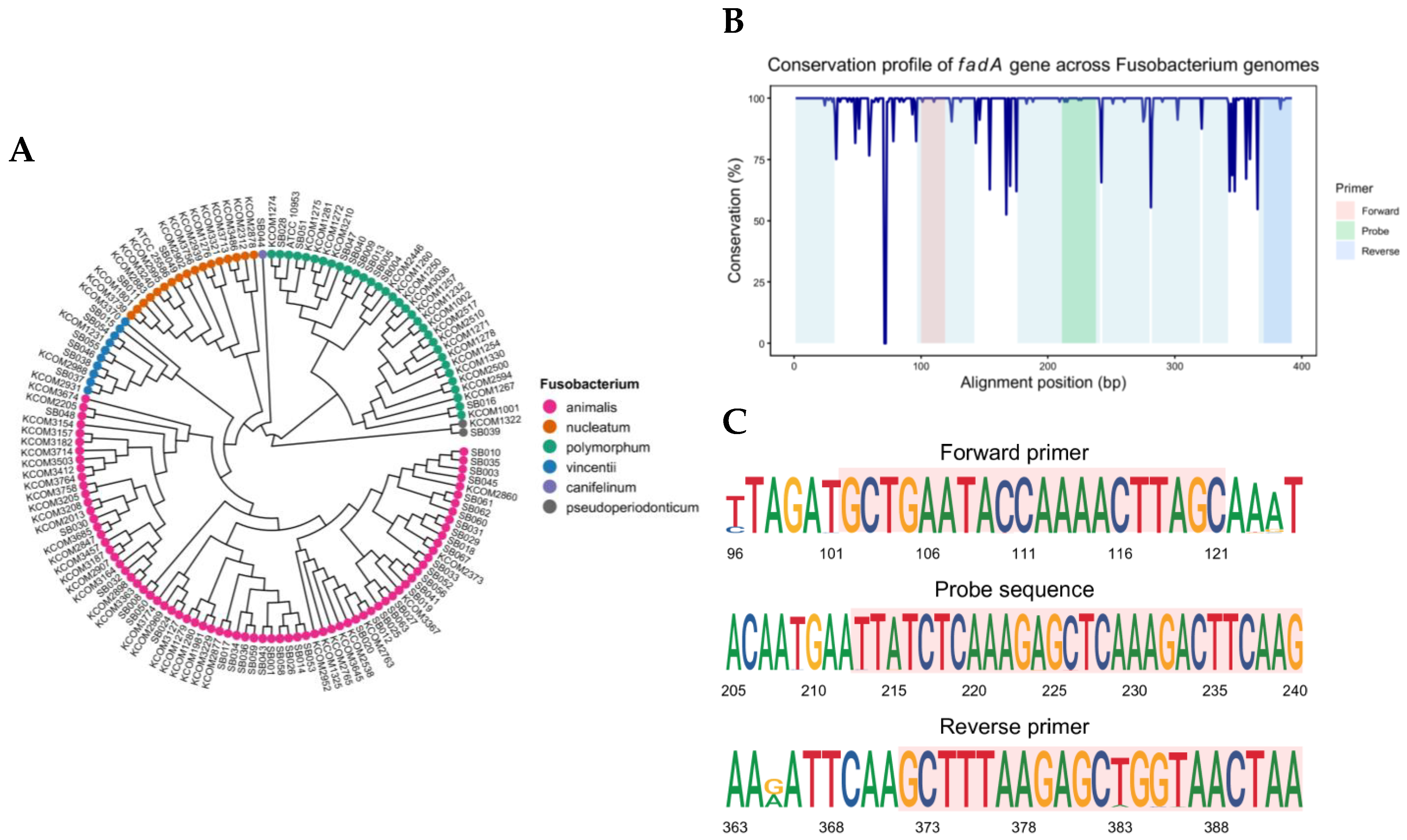
2.1. Identification of Conserved Regions Within the fadA Gene Across Fusobacterium Genomes
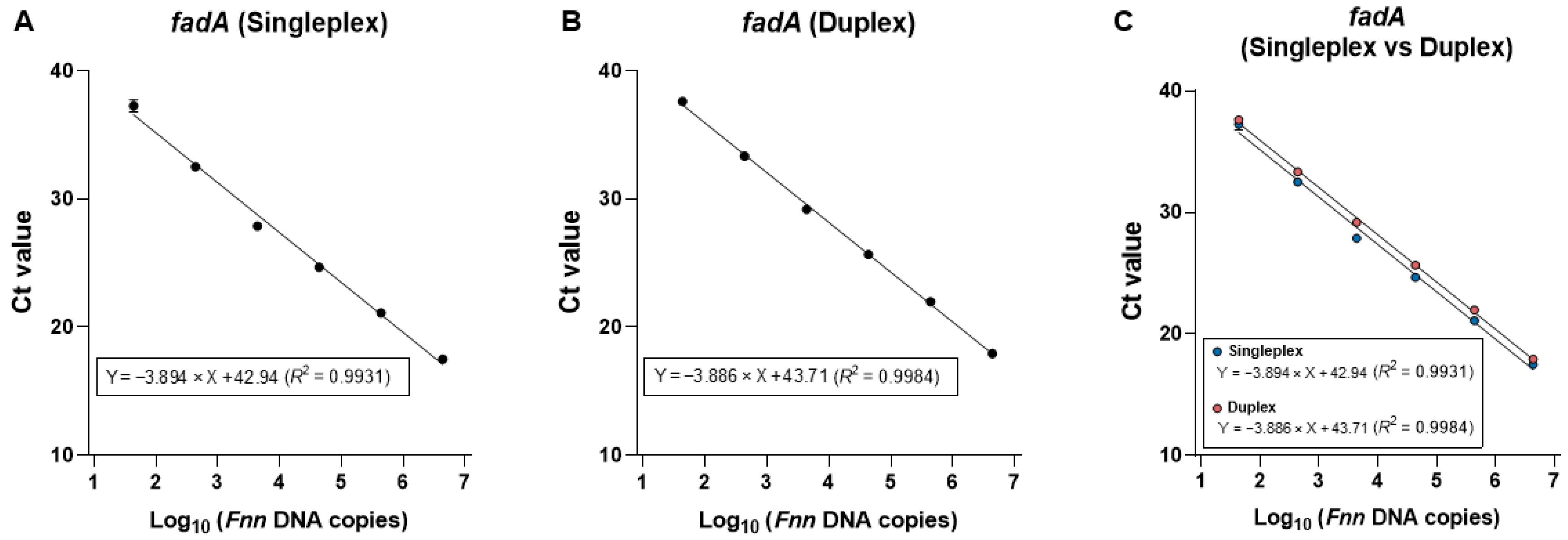
2.2. Assessment of the Duplex qPCR Assay
2.2.1. Cross-Target Interference
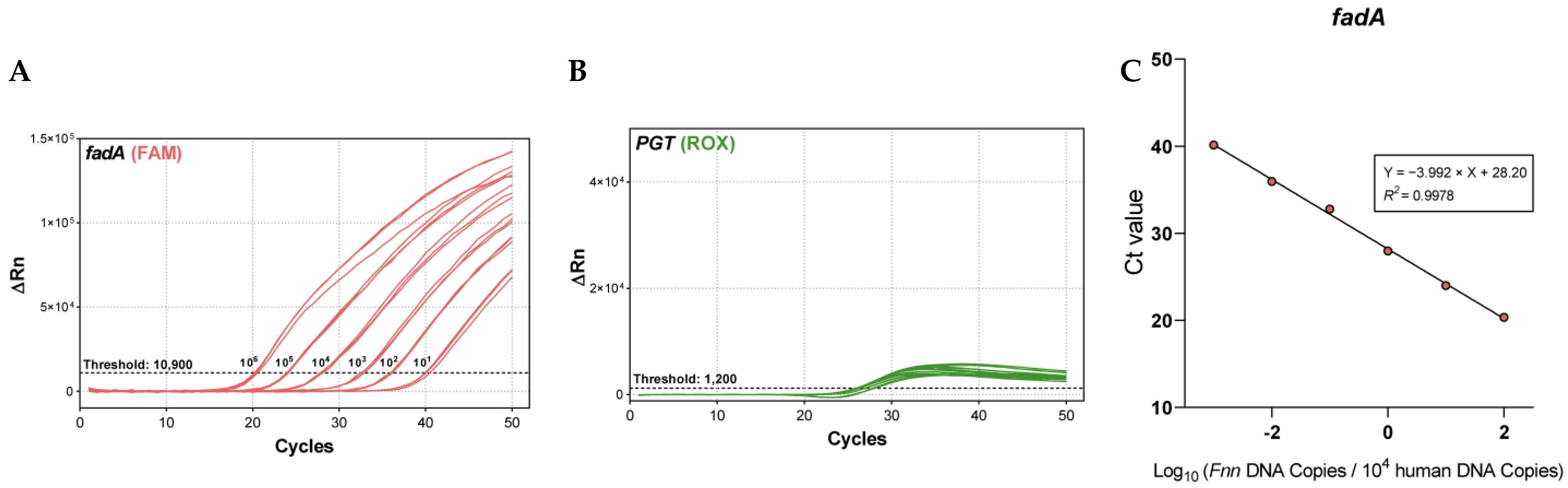
2.2.2. Analytical Sensitivity
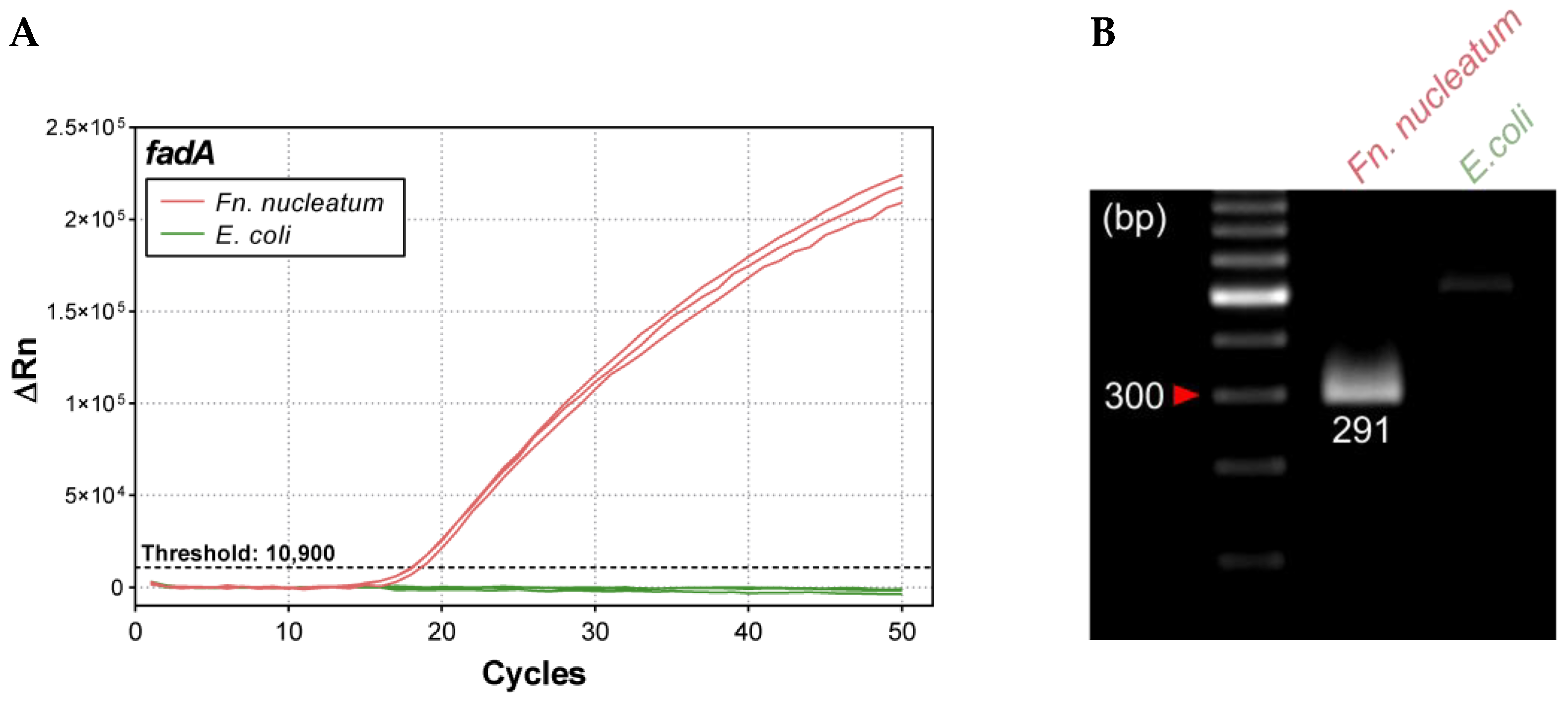
2.2.3. Analytical Specificity
2.2.4. Robust Detection of Fusobacterium DNA in a Human Genomic Background
2.2.5. Precision and Reproducibility
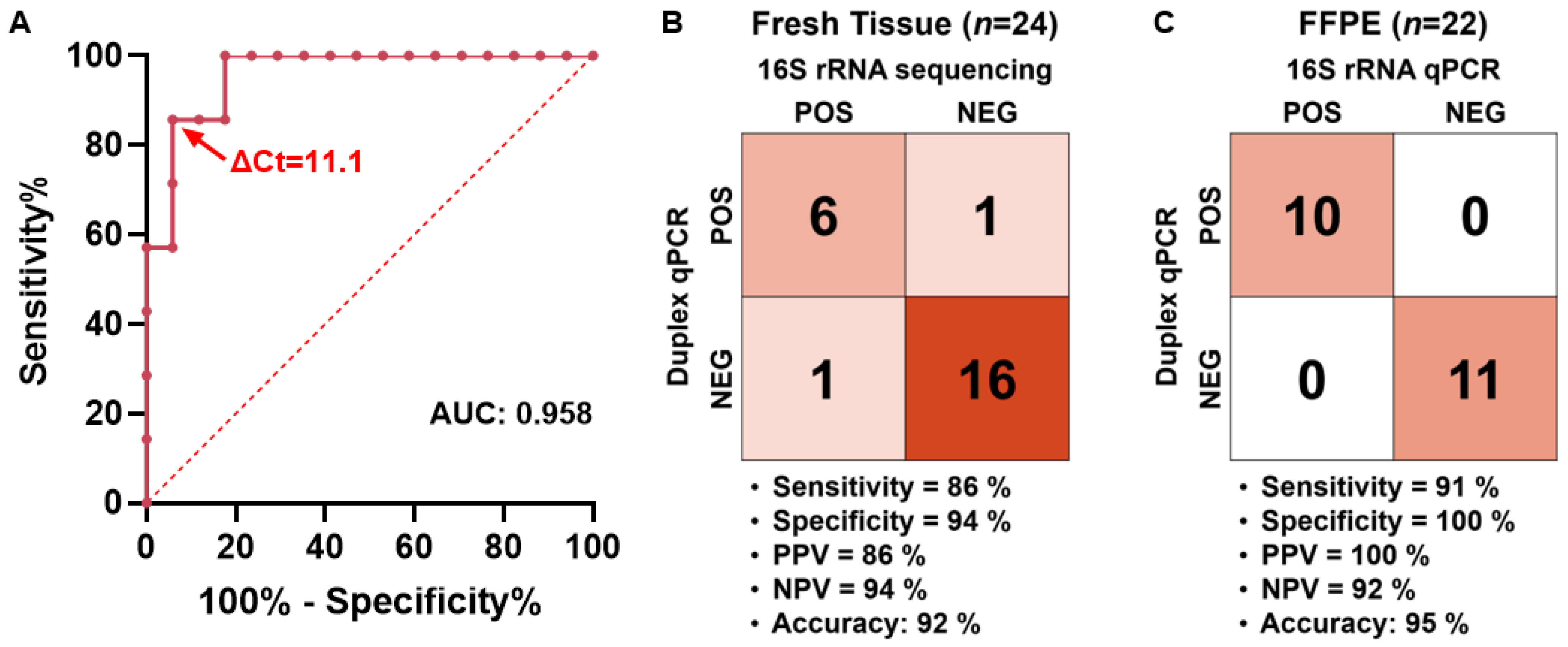
2.3. Clinical Validation and ΔCt Cutoff Determination of the Duplex qPCR
3. Discussion
4. Materials and Methods
4.1. Acquisition of Fusobacterium Genomes
4.2. Primer and Probe Design
4.3. Bacterial Strains
4.4. DNA Extraction Methods
4.5. Quantitative Polymerase Chain Reaction (qPCR)
4.6. 16S rRNA Sequencing
4.7. DNA Gel Electrophoresis
4.8. Determination of Sensitivity and Specificity
4.9. Spike-In Test
4.10. Stability and Reproducibility
4.11. Statistical Analysis
4.12. Ethical Approval
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Segata, N.; Haake, S.K.; Mannon, P.; Lemon, K.P.; Waldron, L.; Gevers, D.; Huttenhower, C.; Izard, J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012, 13, R42. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum—Symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, K.; Baba, Y.; Nakagawa, S.; Mima, K.; Miyake, K.; Nakamura, K.; Sawayama, H.; Kinoshita, K.; Ishimoto, T.; Iwatsuki, M.; et al. Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated with Prognosis. Clin. Cancer Res. 2016, 22, 5574–5581. [Google Scholar] [CrossRef]
- Al-Hebshi, N.N.; Nasher, A.T.; Maryoud, M.Y.; Homeida, H.E.; Chen, T.; Idris, A.M.; Johnson, N.W. Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci. Rep. 2017, 7, 1834. [Google Scholar] [CrossRef]
- Mima, K.; Sukawa, Y.; Nishihara, R.; Qian, Z.R.; Yamauchi, M.; Inamura, K.; Kim, S.A.; Masuda, A.; Nowak, J.A.; Nosho, K. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 2015, 1, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fang, Y.; Liang, W.; Wong, C.C.; Qin, H.; Gao, Y.; Liang, M.; Song, L.; Zhang, Y.; Fan, M.; et al. Fusobacterium nucleatum facilitates anti-PD-1 therapy in microsatellite stable colorectal cancer. Cancer Cell 2024, 42, 1729–1746.E8. [Google Scholar] [CrossRef]
- Zepeda-Rivera, M.; Minot, S.S.; Bouzek, H.; Wu, H.; Blanco-Míguez, A.; Manghi, P.; Jones, D.S.; LaCourse, K.D.; Wu, Y.; McMahon, E.F. A distinct Fusobacterium nucleatum clade dominates the colorectal cancer niche. Nature 2024, 628, 424–432. [Google Scholar] [CrossRef]
- Atout, S.; Shurrab, S.; Loveridge, C. Evaluation of the suitability of RNAscope as a technique to measure gene expression in clinical diagnostics: A systematic review. Mol. Diagn. Ther. 2022, 26, 19–37. [Google Scholar] [CrossRef]
- Han, Y.W. Fusobacterium nucleatum: A commensal-turned pathogen. Curr. Opin. Microbiol. 2015, 23, 141–147. [Google Scholar] [CrossRef]
- Neuzillet, C.; Marchais, M.; Vacher, S.; Hilmi, M.; Schnitzler, A.; Meseure, D.; Leclere, R.; Lecerf, C.; Dubot, C.; Jeannot, E.; et al. Prognostic value of intratumoral Fusobacterium nucleatum and association with immune-related gene expression in oral squamous cell carcinoma patients. Sci. Rep. 2021, 11, 7870. [Google Scholar] [CrossRef]
- Mitsuhashi, K.; Nosho, K.; Sukawa, Y.; Matsunaga, Y.; Ito, M.; Kurihara, H.; Kanno, S.; Igarashi, H.; Naito, T.; Adachi, Y. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget 2015, 6, 7209. [Google Scholar] [CrossRef]
- Datorre, J.G.; de Carvalho, A.C.; Dos Reis, M.B.; Dos Reis, M.; Matsushita, M.; Santos, F.; Guimaraes, D.P.; Reis, R.M. Accuracy and Clinical Relevance of Intra-Tumoral Fusobacterium nucleatum Detection in Formalin-Fixed Paraffin-Embedded (FFPE) Tissue by Droplet Digital PCR (ddPCR) in Colorectal Cancer. Diagnostics 2022, 12, 114. [Google Scholar] [CrossRef]
- Hullar, M.A.; Kahsai, O.J.; Hill, C.; Levy, L.; Malen, R.C.; Curtis, K.R.; Ammar, H.; Sillah, A.; Reedy, A.M.; Lampe, J.W. Highly Sensitive DNA Testing of Fusobacterium nucleatum in Colorectal Tumors. Cancer Epidemiol. Biomark. Prev. 2025, 34, 1377–1385. [Google Scholar] [CrossRef]
- Zuraik, A.A.; Daboul, Y.; Awama, M.A.; Yazigi, H.; Kayasseh, M.A.; Georges, M. Rapid detection of FadA in Fusobacterium nucleatum using the quantitative LAMP colorimetric phenol red method in stool samples from colorectal cancer patients. Sci. Rep. 2024, 14, 13739. [Google Scholar] [CrossRef] [PubMed]
- Aitchison, A.; Pearson, J.F.; Purcell, R.V.; Frizelle, F.A.; Keenan, J.I. Detection of Fusobacterium nucleatum DNA in primary care patient stool samples does not predict progression of colorectal neoplasia. PLoS ONE 2022, 17, e0269541. [Google Scholar] [CrossRef]
- Kurt, M.; Yumuk, Z. Diagnostic accuracy of Fusobacterium nucleatum IgA and IgG ELISA test in colorectal cancer. Sci. Rep. 2021, 11, 1608. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Liu, Y.; Wang, J.; Guo, Y.; Zhang, Y.; Xiao, S. Detection of fusobacterium nucleatum and fadA adhesin gene in patients with orthodontic gingivitis and non-orthodontic periodontal inflammation. PLoS ONE 2014, 9, e85280. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shi, C.; Zheng, J.; Guo, Y.; Fan, T.; Zhao, H.; Jian, D.; Cheng, X.; Tang, H.; Ma, J. Fusobacterium nucleatum predicts a high risk of metastasis for esophageal squamous cell carcinoma. BMC Microbiol. 2021, 21, 301. [Google Scholar] [CrossRef] [PubMed]
- Dregelies, T.; Haumaier, F.; Sterlacci, W.; Backert, S.; Vieth, M. Detection of Fusobacterium nucleatum in Patients with Colitis-Associated Colorectal Cancer. Curr. Microbiol. 2023, 80, 293. [Google Scholar] [CrossRef] [PubMed]
- Kashani, N.; Abadi, A.B.; Rahimi, F.; Forootan, M. FadA-positive Fusobacterium nucleatum is prevalent in biopsy specimens of Iranian patients with colorectal cancer. New Microbes New Infect. 2020, 34, 100651. [Google Scholar] [CrossRef]
- Meng, Q.; Gao, Q.; Mehrazarin, S.; Tangwanichgapong, K.; Wang, Y.; Huang, Y.; Pan, Y.; Robinson, S.; Liu, Z.; Zangiabadi, A.; et al. Fusobacterium nucleatum secretes amyloid-like FadA to enhance pathogenicity. EMBO Rep. 2021, 22, e52891. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lee, S.-T.; Choi, S.; Lee, H.; Kwon, S.S.; Byun, J.H.; Kim, Y.A.; Rhee, K.-J.; Choi, J.R.; Kim, T.I. Fusobacterium nucleatum in biopsied tissues from colorectal cancer patients and alcohol consumption in Korea. Sci. Rep. 2020, 10, 19915. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Chen, Z.; Wong, M.C.; Hui, M.; Yu, J.; Ng, S.C.; Sung, J.J.; Chan, F.K.; Chan, P.K. Southern Chinese populations harbour non-nucleatum Fusobacteria possessing homologues of the colorectal cancer-associated FadA virulence factor. Gut 2020, 69, 1998–2007. [Google Scholar] [CrossRef]
- Wong, S.H.; Kwong, T.N.Y.; Chow, T.C.; Luk, A.K.C.; Dai, R.Z.W.; Nakatsu, G.; Lam, T.Y.T.; Zhang, L.; Wu, J.C.Y.; Chan, F.K.L.; et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 2017, 66, 1441–1448. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Gui, X.; Zhang, Y.; Zhang, Z.; Chen, W.; Zhang, X.; Wang, Y.; Zhang, M.; Shang, Z.; et al. Salivary Fusobacterium nucleatum serves as a potential biomarker for colorectal cancer. iScience 2022, 25, 104203. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Wilson, G.M.; Flibotte, S.; Chopra, V.; Melnyk, B.L.; Honer, W.G.; Holt, R.A. DNA copy-number analysis in bipolar disorder and schizophrenia reveals aberrations in genes involved in glutamate signaling. Human. Mol. Genet. 2006, 15, 743–749. [Google Scholar] [CrossRef]
- Lee, J.B.; Kim, K.-A.; Cho, H.Y.; Kim, D.; Kim, W.K.; Yong, D.; Lee, H.; Yoon, S.S.; Han, D.H.; Han, Y.D. Association between Fusobacterium nucleatum and patient prognosis in metastatic colon cancer. Sci. Rep. 2021, 11, 20263. [Google Scholar] [CrossRef] [PubMed]
- Repass, J. Replication study: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. eLife 2018, 7, e25801. [Google Scholar] [CrossRef]
- Boutaga, K.; van Winkelhoff, A.J.; Vandenbroucke-Grauls, C.M.J.E.; Savelkoul, P.H.M. Periodontal pathogens: A quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunol. Med. Microbiol. 2005, 45, 191–199. [Google Scholar] [CrossRef]
- Choi, I.; Kim, K.-A.; Kim, S.C.; Park, D.; Nam, K.T.; Cha, J.H.; Baek, S.; Cha, J.; Jo, H.-Y.; Jung, M. Secretory IgA dysfunction underlies poor prognosis in Fusobacterium-infected colorectal cancer. Gut Microbes 2025, 17, 2528428. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Fu, L.; Niu, B.; Wu, S.; Wooley, J. Ultrafast clustering algorithms for metagenomic sequence analysis. Brief. Bioinform. 2012, 13, 656–668. [Google Scholar] [CrossRef] [PubMed]
- CLSI EP06; Evaluation of Linearity of Quantitative Measurement Procedures. Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2020.
- CLSI EP15-A3; User Verification of Precision and Estimation of Bias. Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2014.

| Detection Target | Fnn DNA (ng) | Intra-Day | Inter-Day | Inter-Instrument | Inter-Operator | ||||
|---|---|---|---|---|---|---|---|---|---|
| Ct | CV (%) * | Ct | CV (%) * | Ct | CV (%) * | Ct | CV (%) * | ||
| fadA | 101 | 18.0 ± 0.1 | 0.8 | 17.8 ± 0.2 | 1.3 | 17.5 ± 0.6 | 3.3 | 17.5 ± 0.4 | 2.5 |
| 100 | 21.9 ± 0.1 | 0.6 | 21.6 ± 0.7 | 3.1 | 21.3 ± 1.0 | 4.5 | 21.0 ± 0.2 | 1.1 | |
| 10−1 | 25.6 ± 0.1 | 0.2 | 25.2 ± 0.7 | 2.7 | 25.9 ± 0.3 | 1.1 | 24.8 ± 0.4 | 1.6 | |
| 10−2 | 29.1 ± 0.1 | 0.5 | 28.7 ± 0.6 | 2.2 | 30.2 ± 1.4 | 4.6 | 28.4 ± 0.5 | 1.7 | |
| 10−3 | 33.2 ± 0.2 | 0.5 | 32.7 ± 0.7 | 2.1 | 34.1 ± 1.1 | 3.2 | 32.3 ± 0.3 | 1.0 | |
| 10−4 | 37.0 ± 0.5 | 1.4 | 37.2 ± 1.3 | 3.5 | 38.4 ± 1.0 | 2.7 | 36.0 ± 0.3 | 0.7 | |
| Target Gene | Name | Sequence (5′-3′) | Reference |
|---|---|---|---|
| fadA | fadA-Forward | GCTGAATACCAAAACTTAGC | this study |
| fadA-Reverse | TTAGTTACCAGCTCTTAAAGC | ||
| fadA-Probe | (FAM)-TTATCTCAAAGAGCTCAAAGACTTCAAG-(BHQ1) | ||
| PGT | PGT-Forward | ATCCCCAAAGCACCTGGTTT | [4] |
| PGT-Reverse | AGAGGCCAAGATAGTCCTGGTAA | ||
| PGT-Probe-ROX | (ROX)-CCATCCATGTCCTCATCTC-(BHQ2) | ||
| PGT-Probe-FAM | (FAM)-CCATCCATGTCCTCATCTC-(TAMRA) | ||
| 16S rRNA | 16S-Forward | GGATTTATTGGGCGTAAAGC | [35] |
| 16S-Reverse | GGCATTCCTACAAATATCTACGAA | ||
| 16S-Probe | (FAM)-CTCTACACTTGTAGTTCCG-(TAMRA) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, Y.; Kim, K.-A.; Lee, S.; Lim, Y.H.; Seo, Y.; Kim, T.K.; Kim, C.H.; Kim, Y.; Jereis, F.; Kim, W.K.; et al. A Duplex qPCR Assay Targeting the fadA Gene Enables Robust Detection of Fusobacterium in Clinical Samples. Int. J. Mol. Sci. 2025, 26, 11319. https://doi.org/10.3390/ijms262311319
Seo Y, Kim K-A, Lee S, Lim YH, Seo Y, Kim TK, Kim CH, Kim Y, Jereis F, Kim WK, et al. A Duplex qPCR Assay Targeting the fadA Gene Enables Robust Detection of Fusobacterium in Clinical Samples. International Journal of Molecular Sciences. 2025; 26(23):11319. https://doi.org/10.3390/ijms262311319
Chicago/Turabian StyleSeo, Yurin, Kyung-A Kim, Suho Lee, Yujin H. Lim, Yura Seo, Taeyul K. Kim, Chae Hyun Kim, Yeleem Kim, Francesca Jereis, Won Kyu Kim, and et al. 2025. "A Duplex qPCR Assay Targeting the fadA Gene Enables Robust Detection of Fusobacterium in Clinical Samples" International Journal of Molecular Sciences 26, no. 23: 11319. https://doi.org/10.3390/ijms262311319
APA StyleSeo, Y., Kim, K.-A., Lee, S., Lim, Y. H., Seo, Y., Kim, T. K., Kim, C. H., Kim, Y., Jereis, F., Kim, W. K., Han, Y. D., Jung, M., Lee, H., Lee, K., Ahn, J. B., Yoon, J. G., & Kim, H. S. (2025). A Duplex qPCR Assay Targeting the fadA Gene Enables Robust Detection of Fusobacterium in Clinical Samples. International Journal of Molecular Sciences, 26(23), 11319. https://doi.org/10.3390/ijms262311319






