FBXW7 Gene Mutation and Expression in Colorectal Cancer (CRC): A Systematic Review from Molecular Mechanisms to Clinical Translation
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Study Selection Criteria and Data Extraction
3. Results
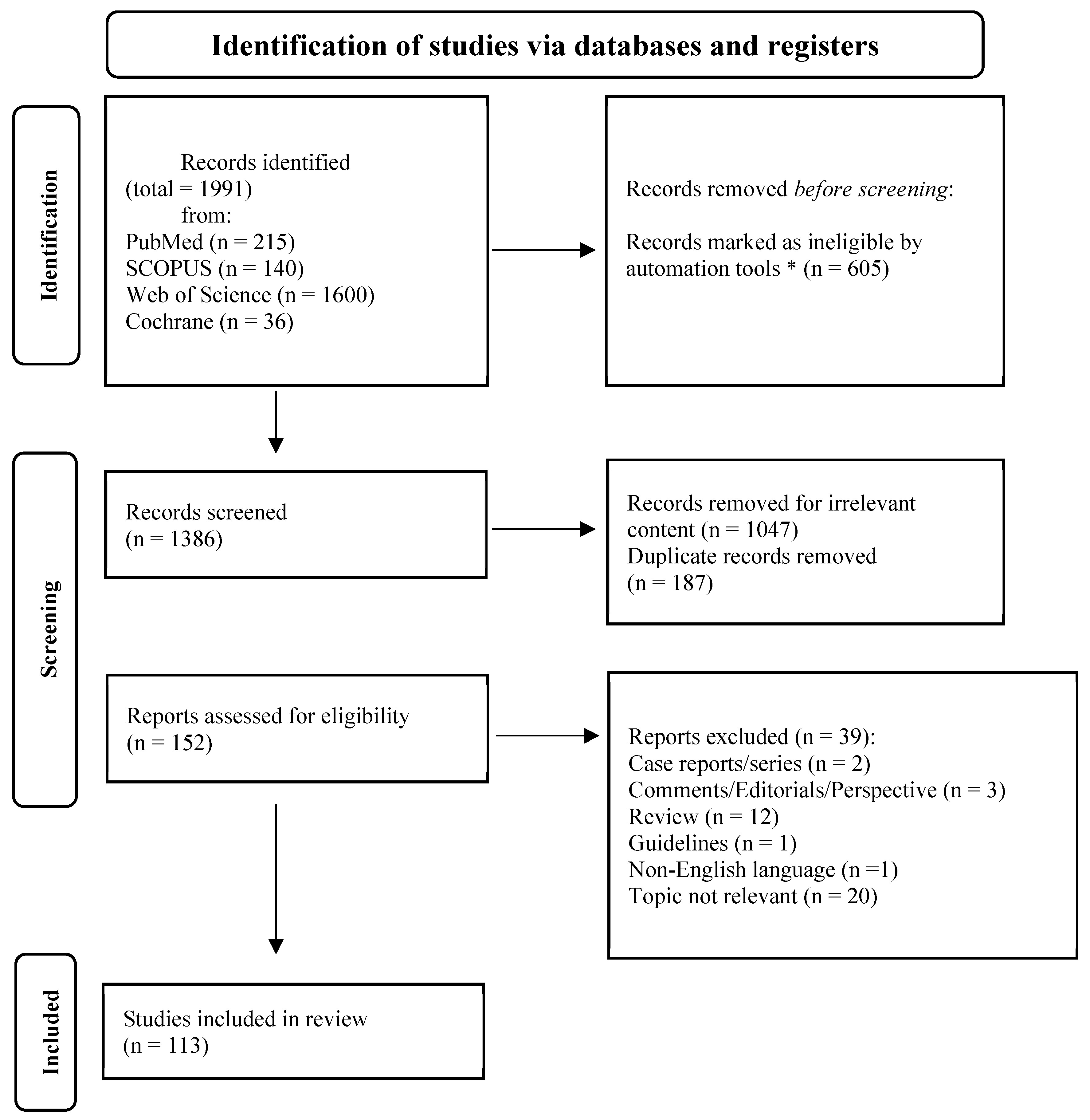
3.1. Literature Search and Included Studies
3.2. Preclinical Studies
3.2.1. FBXW7 in CRC and Tumorigenesis
| Author, Year | Type of Study | Mechanism of FBXW7 Studied | Biological Effect on Tumorigenesis Mediated by FBXW7 |
|---|---|---|---|
| Bialkowska AB 2014 [32] | Genomic analysis of CRC cell lines | Oncogenic role for the P301S KLF5 mutant through interaction with FBXW7α | KLF5 P301S mutation blocks FBXW7-mediated degradation, increasing its stability and activity. |
| Chan DKH 2023 [33] | WT and mutant FBXW7 CRC cells cocultured using a Transwell system Mass spectrometry | AKAP8 mediates DNA damage transfer from FBXW7-mutant to neighboring wild-type cells, affecting local subclonal populations. | AKAP8 is secreted in the microenvironment of FBXW7 mutant cells |
| Luan RG 2025 [34] | Clinical observational study analyzing protein expression in 38 colon cancer tissue samples. | FBXW7 mediates ubiquitination and degradation of cyclin E1, correlated with phosphorylation at Ser73 and Thr395. | Promotes degradation of cyclin E1, potentially inhibiting cell cycle progression and tumor growth in colon cancer. |
| Huber AL 2016 [35] | Experimental study using primary mouse fibroblasts, genetic knockout mice (Cry2−/−), and human cancer cell lines to investigate CRY2 function. | Although FBXW7 is a standard SCF adaptor, the study shows that CRY2–FBXL3 forms its own SCF complex that degrades c-MYC independently of FBXW7. | FBXW7 normally limits MYC-driven proliferation by degrading c-MYC, but CRY2–FBXL3 provides a separate MYC-degradation pathway; losing CRY2 therefore increases MYC activity and promotes tumorigenesis. |
| Duhamel S 2016 [36] | Comprehensive study using epithelial cells, imaging, gene silencing, drug inhibition, and a transgenic mouse model. | ERK1/2 signaling downregulates Fbxw7β, preventing Aurora A’s ubiquitin-mediated degradation and causing its accumulation. | Loss of Fbxw7β leads to Aurora A buildup, causing cytokinesis failure, polyploidy, and chromosomal instability that promote genomic instability and cancer progression. |
| Mu Y 2017 [37] | Study combining CRC patient tissues with functional assays in HCT116 and SW480 cell lines. | FBXW7 suppresses Notch1 signaling, and FAM83D knockdown increases FBXW7 levels, thereby reducing Notch1 activity. | Higher FBXW7 levels inhibit CRC progression by reducing proliferation, migration, invasion, and promoting apoptosis. |
| Grim JE 2012 [25] | Mouse model study examining the effects of jointly deleting FBXW7 and p53 in intestinal epithelium. | FBXW7 acts as a tumor suppressor by degrading multiple oncogenic proteins, helping control proliferation, differentiation, and genomic stability. | FBXW7 loss alone alters differentiation and oncogene levels, but combined with p53 loss drives aggressive, metastatic intestinal cancer. |
| Li QG 2018 [38] | Study of 276 CRC tissues with in vitro validation using IHC, molecular assays, and functional analyses. | FBXW7 functions as an E3 ubiquitin ligase targeting HIF1α for degradation, thereby reducing CEACAM5 (CEA) expression in a HIF1α-dependent manner. | FBXW7 suppresses CRC progression by inhibiting tumor cell migration and metastasis, leading to improved overall survival and disease-free survival. |
| Zhan P 2015 [39] | Experimental study of colon cancer using proteomics and molecular assays. | FBXW7 targets ENO1 for GSK3β-dependent ubiquitin-proteasomal degradation. | FBXW7 inhibits colorectal cancer by downregulating ENO1, reducing CCL20, lactate, proliferation, and migration. |
| Lu HR 2019 [40] | Experimental study of CRC using clinical tissues, cell lines, and mouse xenografts. | Circ-FBXW7 acts as a tumor-suppressive RNA by regulating NEK2, mTOR, and PTEN pathways. | Circ-FBXW7 suppresses colorectal cancer by inhibiting proliferation, migration, invasion, and tumor growth. |
| Babaei-Jadidi R 2011 [18] | Conditional knockout mouse study using ApcMin/+ crosses with histological, molecular, and proteomic analyses. | FBXW7, as part of the SCFFBXW7 complex, degrades oncogenic proteins; its loss causes aberrant signaling and oncogene accumulation. | FBXW7 loss promotes intestinal tumorigenesis by enhancing proliferation, disrupting differentiation, causing DEK accumulation, altering TPM splicing, and accelerating adenomas with APC deficiency. |
| Li, N 2015 [41] | Experimental study of colorectal cancer using cell lines, mouse models, and human tissue validation. | FBXW7 indirectly modulates p53 phosphorylation at Ser15/Ser18 via upstream kinases like CK1α. | FBXW7 loss causes phospho-p53(Ser15) accumulation, alters TP53 gene expression, and drives chemoresistance, promoting colorectal cancer progression. |
| Wei W 2023 [27] | Experimental study of colorectal cancer using cell lines, organoids, and mouse/xenograft models with molecular, biochemical, and metabolic analyses. | FBXW7β promotes FASN degradation via K48-linked ubiquitination, while CSN6 stabilizes FASN by degrading FBXW7β; GSK3β phosphorylation of FASN enables FBXW7β recognition. | FBXW7β suppresses colorectal cancer by reducing FASN, lipid accumulation, and tumor growth; its loss promotes FASN stability and tumor progression. |
| Long, YP 2019 [42] | In vitro co-culture study of colon cancer cells and macrophages, validated by RNA-seq and qRT-PCR. | FBXW7α controls macrophage polarization through the miR-205/SMAD1 axis; its loss shifts M1/M2 balance. | FBXW7α suppresses TAM M1 polarization; its knockdown boosts inflammatory cytokines, potentially inhibiting colorectal cancer. |
| Iwatsuki M 2010 [43] | Clinical observational study combined with in vitro functional assays. | FBXW7 regulates degradation of oncogenic proteins c-MYC and cyclin E via ubiquitin-mediated proteolysis; its suppression leads to accumulation of these proteins. | Loss of FBXW7 promotes tumor progression by enhancing cell proliferation, invasion, and poor prognosis in colorectal cancer, acting as a tumor suppressor. |
| Liu Z 2021 [19] | Study integrating bioinformatics (TCGA, GTEx), CRC tissues, and in vitro experiments in HCT116 cells. | miR-223 directly inhibits FBXW7 by binding its 3′UTR, reducing mRNA and protein levels. | FBXW7 downregulation enhances CRC cell proliferation and inhibits apoptosis through Notch and Akt/mTOR activation. |
| Chen Y 2024 [44] | Study integrating GEO dataset analysis, CRC tissues, and in vitro experiments in HCT116 and Caco-2 cells. | miR-25-3p directly targets FBXW7 3′UTR, decreasing its mRNA and protein levels. | FBXW7 loss promotes CRC cell proliferation and survival via oncogenic protein accumulation, while its overexpression suppresses growth and induces apoptosis. |
| Gong L 2018 (abstract) [21] | In vitro study to analyze the effect on CRC invasion of miR-92b-3p, a target of FBXW7 | miR-92b-3p downregulates FBXW7 in CRC, and blocking it or overexpressing FBXW7 limits tumor growth and spread. | Inhibiting miR-92b-3p suppresses CRC proliferation, invasion, and migration by upregulating FBXW7, highlighting its role in carcinogenesis and metastasis. |
| Ou BC 2016 [45] | Study integrating clinical tissue analysis with in vitro and in vivo functional experiments. | Plk2 binds to FBXW7 and promotes its proteasome-mediated degradation, leading to stabilization and accumulation of Cyclin E. | Degradation of FBXW7 by Plk2 increases Cyclin E levels, promoting cell proliferation, inhibiting apoptosis, and enhancing colorectal tumor growth. |
| Wang YL 2013 [31] | In vitro study using FBXW7 knockout colon cancer cell lines. | FBXW7 regulates EMT, migration, invasion, and stemness via mTOR signaling. | FBXW7 loss promotes EMT, migration, invasion, and stemness, which are counteracted by mTOR inhibition. |
| Guo Z 2012 [46] | In vitro study using human CRC cell lines SW620, HT29 and HCT116 | Rictor associates with FBXW7 to form an E3 complex participating in the regulation of c-Myc and cyclin E degradation. | Rictor, within the FBXW7 complex, drives c-Myc and cyclin E degradation, and impaired signaling leads to their accumulation in CRC cells |
| Li L 2014 * [24] | Study of human colon tissues, CRC cell lines, and knockout models using molecular and histological analyses. | Sequential upregulation of miR-182 and miR-503 promotes colon adenoma-to-adenocarcinoma progression by jointly suppressing FBXW7. | Blocking both miR-182 and miR-503 in HCT116 colon cancer cells resulted in increased FBXW7 expression and significantly reduced tumor size in xenograft models |
| Khan OM 2018 [47] | In vitro and in vivo study identifying FBXW7 interactors via proteomics in CRC cells, validated by Usp9x deletion in mouse intestines. | The deubiquitinase USP9X antagonizes FBXW7 ubiquitylation, thereby stabilizing FBXW7 protein and regulating c-MYC degradation. | Usp9x inactivation reduced secretory cell differentiation, increased progenitor proliferation and tumor burden, while c-Myc heterozygosity mitigated tumor formation in Usp9x-deficient mice |
| Lee YS * [48] | Analysis of 232 CRC patient tissues for tumor-associated macrophage subtypes, complemented by in vitro, in vivo, and ex vivo studies on PI3Kγ regulation | M1 and M2 macrophages exerted opposite effects on CRC progression, mediated via the FBXW7-MCL-1 axis, while PI3Kγ inhibition in macrophages influenced EMT and cytotoxicity in tumor cells. | Patients with a lower M2/M1 ratio (<3) experienced significantly better progression-free and overall survival; targeting macrophage PI3Kγ suppressed EMT features and enhanced colon cancer cell death, supporting its value as an immunotherapeutic target. |
| Lin L 2020 [49] | Experimental study in CRC and normal epithelial cells assessing Trametinib and TRAIL effects using siRNA, transfection, viability, colony formation, apoptosis, Western blot, qPCR, and co-immunoprecipitation. | FBXW7 functions as an E3 ligase that promotes proteasomal degradation of Mcl-1 phosphorylated by GSK-3β. | Trametinib enhances TRAIL-mediated apoptosis in CRC cells by promoting FBXW7-dependent Mcl-1 degradation, thereby boosting its anticancer effect. |
| Bengoechea-Alonso MT 2010 [50] | In vitro study examining TGIF1 phosphorylation and degradation under altered Fbxw7 activity. | FBXW7 targets phosphorylated TGIF1 for ubiquitin-mediated proteasomal degradation. | Loss of FBXW7 function leads to accumulation of phosphorylated TGIF1 and suppression of TGFβ-dependent transcriptional activity. |
| Kumar Y 2016 [22] | In vitro study of FBXW7-mediated CDX2 ubiquitination using coimmunoprecipitation and phosphodegron mutations in colon cancer and HEK293T cells. | FBXW7, in a GSK3β-dependent manner, binds to two phosphodegron motifs on CDX2 via its WD domain, promoting CDX2 ubiquitination and proteasomal degradation | FBXW7 and GSK3β overexpression reduces CDX2 levels and function, causing growth arrest in colon cancer cells; disruption of both CDX2 phosphodegrons prevents FBXW7-mediated degradation. |
| Bajpai S 2022 [51] | Study combining molecular and cellular experiments with protein interaction, ubiquitination assays, and conditional knockout mice. | FBXW7 functions as an E3 ubiquitin ligase that recognizes phosphorylated c-Myc at T58/S62, mediating its polyubiquitination and proteasomal degradation. | FBXW7 limits c-Myc accumulation, thereby suppressing uncontrolled cell proliferation and acting as a tumor suppressor. |
| Diefenbacher ME 2015 [52] | In vivo and in vitro study using MEFs, intestinal epithelial cells, and double knockout mouse models. | FBXW7 targets oncogenic substrates for degradation, while USP28 opposes this by deubiquitinating them, including in FBXW7-deficient cells. | USP28 loss in FBXW7-deficient mice slows proliferation, restores differentiation, and partially reverses tumorigenesis, showing USP28 promotes tumor growth without FBXW7. |
| Eun JC et al., 2021 [29] | Translational ex vivo study of 87 patient-derived CRC organoids combining multi-omic profiling and drug sensitivity testing. | FBXW7 loss in CRC associates with CD8+ T-cell exhaustion, UPR activation, WNT/β-catenin dependency, and heightened sensitivity to PPAR inhibition, revealing potential therapeutic vulnerabilities. | FBXW7 loss promotes tumorigenesis by driving immune evasion (CD8+ T-cell exhaustion) and proteostatic stress (UPR), while creating WNT/CTNNB1-dependent vulnerabilities (e.g., FH535 sensitivity). |
| Boretto et al., 2024 [30] | Study using engineered human CRC organoids with CRISPR base editing, proteomic and transcriptomic analyses, and drug response testing. | FBXW7 normally degrades phosphorylated proteins like EGFR; mutations impair this, causing EGFR and other oncogene accumulation. | FBXW7 mutations reduce EGF dependency, enhance MAPK/EGFR signaling, promote proliferation, and confer resistance to anti-EGFR therapies, thereby driving tumor progression. |
| Gao et al., 2025 [28] | Preclinical experimental study combining bioinformatics analyses, in vitro assays, and in vivo mouse models of CRC. | FBXW7 promotes c-Myc degradation, while SET stabilizes c-Myc by competing for FBXW7 binding. | FBXW7 suppresses MSS CRC tumorigenesis by degrading c-Myc, reducing mismatch repair proteins, immune evasion, and the immunosuppressive microenvironment. |
3.2.2. FBXW7 and Drug Resistance
3.3. Clinical Studies in CRC
3.3.1. Correlation Between FBXW7 Alterations and Demographic/Clinic-Pathological Features of CRC
3.3.2. FBXW7 Alterations in Localized CRC
3.3.3. FBXW7 Alterations in mCRC
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mardis, E.R. A decade’s perspective on DNA sequencing technology. Nature 2011, 470, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Sidoli, S.; Kulej, K.; Garcia, B.A. Why proteomics is not the new genomics and the future of mass spectrometry in cell biology. J. Cell Biol. 2017, 216, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Komander, D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009, 37, 937–953. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.-H.; Bellon, M.; Nicot, C. FBXW7: A critical tumor suppressor of human cancers. Mol. Cancer 2018, 17, 115. [Google Scholar] [CrossRef]
- O’NEil, J.; Grim, J.; Strack, P.; Rao, S.; Tibbitts, D.; Winter, C.; Hardwick, J.; Welcker, M.; Meijerink, J.P.; Pieters, R.; et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J. Exp. Med. 2007, 204, 1813–1824. [Google Scholar] [CrossRef]
- Inuzuka, H.; Shaik, S.; Onoyama, I.; Gao, D.; Tseng, A.; Maser, R.S.; Zhai, B.; Wan, L.; Gutierrez, A.; Lau, A.W.; et al. SCFFBW7 regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 2011, 471, 104–109. [Google Scholar] [CrossRef]
- Jin, J.; Cardozo, T.; Lovering, R.C.; Elledge, S.J.; Pagano, M.; Harper, J.W. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004, 18, 2573–2580. [Google Scholar] [CrossRef]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017, 45, D777–D783. [Google Scholar] [CrossRef]
- Davis, H.; Lewis, A.; Behrens, A.; Tomlinson, I. Investigation of the atypical FBXW7 mutation spectrum in human tumours by conditional expression of a heterozygous propellor tip missense allele in the mouse intestines. Gut 2014, 63, 792–799. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, H.-H.; Lin, J.-K.; Lin, C.-C.; Lan, Y.-T.; Wang, H.-S.; Yang, S.-H.; Chen, W.-S.; Lin, T.-C.; Jiang, J.-K.; et al. FBXW7 mutation analysis and its correlation with clinicopathological features and prognosis in colorectal cancer patients. Int. J. Biol. Markers 2015, 30, 88–95. [Google Scholar] [CrossRef]
- Fan, J.; Bellon, M.; Ju, M.; Zhao, L.; Wei, M.; Fu, L.; Nicot, C. Clinical significance of FBXW7 loss of function in human cancers. Mol. Cancer 2022, 21, 87. [Google Scholar] [CrossRef]
- Wang, H.-W.; Yan, X.-L.; Wang, L.-J.; Zhang, M.-H.; Yang, C.-H.; Liu, W.; Jin, K.-M.; Bao, Q.; Li, J.; Wang, K.; et al. Characterization of genomic alterations in Chinese colorectal cancer patients with liver metastases. J. Transl. Med. 2021, 19, 313. [Google Scholar] [CrossRef]
- Stein, M.K.; Williard, F.W.; Xiu, J.; Tsao, M.W.; Martin, M.G.; Deschner, B.W.; Dickson, P.V.; Glazer, E.S.; Yakoub, D.; Shibata, D.; et al. Comprehensive tumor profiling reveals unique molecular differences between peritoneal metastases and primary colorectal adenocarcinoma. J. Surg. Oncol. 2020, 121, 1320–1328. [Google Scholar] [CrossRef]
- Chen, S.; Lin, J.; Zhao, J.; Lin, Q.; Liu, J.; Wang, Q.; Mui, R.; Ma, L. FBXW7 attenuates tumor drug resistance and enhances the efficacy of immunotherapy. Front. Oncol. 2023, 13, 1147239. [Google Scholar] [CrossRef]
- Uroog, L.; Zeya, B.; Imtiyaz, K.; Wani, R.A.; Alam Rizvi, M.M. FBXW7 polymorphism asserts susceptibility to colorectal cancer. Gene 2024, 901, 148181. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.L.; Singh, T.; Mero, P.; Moffat, J.; Hieter, P. Dependence of Human Colorectal Cells Lacking the FBW7 Tumor Suppressor on the Spindle Assembly Checkpoint. Genetics 2015, 201, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Babaei-Jadidi, R.; Li, N.; Saadeddin, A.; Spencer-Dene, B.; Jandke, A.; Muhammad, B.; Ibrahim, E.E.; Muraleedharan, R.; Abuzinadah, M.; Davis, H.; et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J. Exp. Med. 2011, 208, 295–312. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, T.; Duan, J.; Liu, X.; Liu, L. MicroRNA-223-induced inhibition of the FBXW7 gene affects the proliferation and apoptosis of colorectal cancer cells via the Notch and Akt/mTOR pathways. Mol. Med. Rep. 2021, 23, 154. [Google Scholar] [CrossRef]
- Jahid, S.; Sun, J.; Edwards, R.A.; Dizon, D.; Panarelli, N.C.; Milsom, J.W.; Sikandar, S.S.; Gümüş, Z.H.; Lipkin, S.M. miR-23a Promotes the Transition from Indolent to Invasive Colorectal Cancer. Cancer Discov. 2012, 2, 540–553. [Google Scholar] [CrossRef]
- Gong, L.; Ren, M.; Lv, Z.; Yang, Y.; Wang, Z. miR-92b-3p Promotes Colorectal Carcinoma Cell Proliferation, Invasion, and Migration by Inhibiting FBXW7 In Vitro and In Vivo. DNA Cell Biol. 2018, 37, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.; Shukla, N.; Thacker, G.; Kapoor, I.; Lochab, S.; Bhatt, M.L.B.; Chattopadhyay, N.; Sanyal, S.; Trivedi, A.K. Ubiquitin Ligase, Fbw7, Targets CDX2 for Degradation via Two Phosphodegron Motifs in a GSK3β-Dependent Manner. Mol. Cancer Res. 2016, 14, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Babaei-Jadidi, R.; Lorenzi, F.; Spencer-Dene, B.; Clarke, P.; Domingo, E.; Tulchinsky, E.; Vries, R.G.J.; Kerr, D.; Pan, Y.; et al. An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis 2019, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sarver, A.L.; Khatri, R.; Hajeri, P.B.; Kamenev, I.; French, A.J.; Thibodeau, S.N.; Steer, C.J.; Subramanian, S. Sequential expression of miR-182 and miR-503 cooperatively targets FBXW7, contributing to the malignant transformation of colon adenoma to adenocarcinoma. J. Pathol. 2014, 234, 488–501. [Google Scholar] [CrossRef]
- Grim, J.E.; Knoblaugh, S.E.; Guthrie, K.A.; Hagar, A.; Swanger, J.; Hespelt, J.; Delrow, J.J.; Small, T.; Grady, W.M.; Nakayama, K.I.; et al. Fbw7 and p53 cooperatively suppress advanced and chromosomally unstable intestinal cancer. Mol. Cell. Biol. 2012, 32, 2160–2167. [Google Scholar] [CrossRef]
- Takada, M.; Zhang, W.; Suzuki, A.; Kuroda, T.S.; Yu, Z.; Inuzuka, H.; Gao, D.; Wan, L.; Zhuang, M.; Hu, L.; et al. FBW7 Loss Promotes Chromosomal Instability and Tumorigenesis via Cyclin E1/CDK2–Mediated Phosphorylation of CENP-A. Cancer Res. 2017, 77, 4881–4893. [Google Scholar] [CrossRef]
- Wei, W.; Qin, B.; Wen, W.; Zhang, B.; Luo, H.; Wang, Y.; Xu, H.; Xie, X.; Liu, S.; Jiang, X.; et al. FBXW7β loss-of-function enhances FASN-mediated lipogenesis and promotes colorectal cancer growth. Signal Transduct. Target. Ther. 2023, 8, 187. [Google Scholar] [CrossRef]
- Gao, L.; Li, Y.; Wang, H.; Liu, J.; Zhang, R.; Shan, W.; Zeng, L.; Zhao, Q.; Li, Y.; Liu, J. SET facilitates immune escape of microsatellite stability colorectal cancer by inhibiting c-Myc degradation. Cancer Sci. 2025, 116, 29–43. [Google Scholar] [CrossRef]
- Cho, E.J.; Kim, M.; Jo, D.; Kim, J.; Oh, J.-H.; Chung, H.C.; Lee, S.-H.; Kim, D.; Chun, S.-M.; Kim, J.; et al. Immuno-genomic classification of colorectal cancer organoids reveals cancer cells with intrinsic immunogenic properties associated with patient survival. J. Exp. Clin. Cancer Res. 2021, 40, 230. [Google Scholar] [CrossRef]
- Boretto, M.; Geurts, M.H.; Gandhi, S.; Ma, Z.; Staliarova, N.; Celotti, M.; Lim, S.; He, G.-W.; Millen, R.; Driehuis, E.; et al. Epidermal growth factor receptor (EGFR) is a target of the tumor-suppressor E3 ligase FBXW7. Proc. Natl. Acad. Sci. USA 2024, 121, e2309902121. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Lu, J.; Zhang, P.; Wang, Y.; Xu, Y.; Wang, Z.; Mao, J.-H.; Wei, G. Rapamycin inhibits FBXW7 loss-induced epithelial–mesenchymal transition and cancer stem cell-like characteristics in colorectal cancer cells. Biochem. Biophys. Res. Commun. 2013, 434, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Bialkowska, A.B.; Liu, Y.; Nandan, M.O.; Yang, V.W. A Colon Cancer-derived mutant of Krüppel-like factor 5 (KLF5) is resistant to degradation by glycogen synthase kinase 3β (GSK3β) and the E3 ubiquitin ligase F-box and WD repeat domain-containing 7α (FBW7α). J. Biol. Chem. 2014, 289, 5997–6005. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.K.H.; Mandal, A.; Hester, S.; Yu, Z.; Higgins, G.S.; Kessler, B.M.; Fischer, R. Biallelic FBXW7 knockout induces AKAP8-mediated DNA damage in neighbouring wildtype cells. Cell Death Discov. 2025, 9, 200. [Google Scholar] [CrossRef] [PubMed]
- Luan, R.-G.; Liu, M.-D.; Deng, Z.-F.; Lu, C.-L.; Yu, M.-L.; Zhang, M.-Y.; Liu, R.; An, R.; Yao, Y.-L.; Guo, D.-B.; et al. Correlations of the expression of Cx43, SCFFBXW7, p-cyclin E1 (Ser73), p-cyclin E1 (Thr77) and p-cyclin E1 (Thr395) in colon cancer tissues. World J. Gastrointest. Oncol. 2025, 17, 98410. [Google Scholar] [CrossRef]
- Huber, A.-L.; Papp, S.J.; Chan, A.B.; Henriksson, E.; Jordan, S.D.; Kriebs, A.; Nguyen, M.; Wallace, M.; Li, Z.; Metallo, C.M.; et al. CRY2 and FBXL3 Cooperatively Degrade c-MYC. Mol. Cell 2016, 64, 774–789. [Google Scholar] [CrossRef]
- Duhamel, S.; Girondel, C.; Dorn, J.F.; Tanguay, P.-L.; Voisin, L.; Smits, R.; Maddox, P.S.; Meloche, S. Deregulated ERK1/2 MAP kinase signaling promotes aneuploidy by a Fbxw7β-Aurora A pathway. Cell Cycle 2016, 15, 1631–1642. [Google Scholar] [CrossRef]
- Mu, Y.; Zou, H.; Chen, B.; Fan, Y.; Luo, S. FAM83D knockdown regulates proliferation, migration and invasion of colorectal cancer through inhibiting FBXW7/Notch-1 signalling pathway. Biomed. Pharmacother. 2017, 90, 548–554. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Li, J.; Ma, Y.; Dai, W.; Mo, S.; Xu, Y.; Li, X.; Cai, S. FBW7 suppresses metastasis of colorectal cancer by inhibiting HIF1α/CEACAM5 functional axis. Int. J. Biol. Sci. 2018, 14, 726–735. [Google Scholar] [CrossRef]
- Zhan, P.; Wang, Y.; Zhao, S.; Liu, C.; Wang, Y.; Wen, M.; Mao, J.-H.; Wei, G.; Zhang, P. FBXW7 negatively regulates ENO1 expression and function in colorectal cancer. Mod. Pathol. 2015, 95, 995–1004. [Google Scholar] [CrossRef]
- Lu, H.; Yao, B.; Wen, X.; Jia, B. FBXW7 circular RNA regulates proliferation, migration and invasion of colorectal carcinoma through NEK2, mTOR, and PTEN signaling pathways in vitro and in vivo. BMC Cancer 2019, 19, 918. [Google Scholar] [CrossRef]
- Li, N.; Lorenzi, F.; Kalakouti, E.; Normatova, M.; Babaei-Jadidi, R.; Tomlinson, I.; Nateri, A.S. FBXW7-mutated colorectal cancer cells exhibit aberrant expression of phosphorylated-p53 at Serine-15. Oncotarget 2015, 6, 9240–9256. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Zhu, Y. Identification of FBXW7α-regulated genes in M1-polarized macrophages in colorectal cancer by RNA sequencing. Saudi Med. J. 2019, 40, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki, M.; Mimori, K.; Ishii, H.; Yokobori, T.; Takatsuno, Y.; Sato, T.; Toh, H.; Onoyama, I.; Nakayama, K.I.; Baba, H.; et al. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: Clinical significance. Int. J. Cancer 2010, 126, 1828–1837. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, B.; Tu, S.; Yuan, H. miR-25-3p serves as an oncogenic in colorectal cancer cells by regulating the ubiquitin ligase FBXW7 function. Oncol. Rep. 2024, 52, 153. [Google Scholar] [CrossRef]
- Ou, B.; Zhao, J.; Guan, S.; Wangpu, X.; Zhu, C.; Zong, Y.; Ma, J.; Sun, J.; Zheng, M.; Feng, H.; et al. Plk2 promotes tumor growth and inhibits apoptosis by targeting Fbxw7/Cyclin E in colorectal cancer. Cancer Lett. 2016, 380, 457–466. [Google Scholar] [CrossRef]
- Guo, Z.; Zhou, Y.; Evers, B.M.; Wang, Q. Rictor regulates FBXW7-dependent c-Myc and cyclin E degradation in colorectal cancer cells. Biochem. Biophys. Res. Commun. 2012, 418, 426–432. [Google Scholar] [CrossRef]
- Khan, O.M.; Carvalho, J.; Spencer-Dene, B.; Mitter, R.; Frith, D.; Snijders, A.P.; Wood, S.A.; Behrens, A. The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer. J. Clin. Investig. 2018, 128, 1326–1337. [Google Scholar] [CrossRef]
- Lee, Y.S.; Song, S.J.; Hong, H.K.; Oh, B.Y.; Lee, W.Y.; Cho, Y.B. The FBW7-MCL-1 axis is key in M1 and M2 macrophage-related colon cancer cell progression: Validating the immunotherapeutic value of targeting PI3Kγ. Exp. Mol. Med. 2020, 52, 815–831. [Google Scholar] [CrossRef]
- Lin, L.; Ding, D.; Xiao, X.; Li, B.; Cao, P.; Li, S. Trametinib potentiates TRAIL-induced apoptosis via FBW7-dependent Mcl-1 degradation in colorectal cancer cells. J. Cell. Mol. Med. 2020, 24, 6822–6832. [Google Scholar] [CrossRef]
- Bengoechea-Alonso, M.T.; Ericsson, J. Tumor suppressor Fbxw7 regulates TGFβ signaling by targeting TGIF1 for degradation. Oncogene 2010, 29, 5322–5328. [Google Scholar] [CrossRef]
- Bajpai, S.; Jin, H.R.; Mucha, B.; Diehl, J.A. Ubiquitylation of unphosphorylated c-myc by novel E3 ligase SCFFbxl8. Cancer Biol. Ther. 2022, 23, 348–357. [Google Scholar] [CrossRef]
- Diefenbacher, M.E.; Chakraborty, A.; Blake, S.M.; Mitter, R.; Popov, N.; Eilers, M.; Behrens, A. Usp28 counteracts Fbw7 in intestinal homeostasis and cancer. Cancer Res. 2015, 75, 1181–1186. [Google Scholar] [CrossRef]
- Fang, L.; Yang, Z.; Zhou, J.; Tung, J.-Y.; Hsiao, C.-D.; Wang, L.; Deng, Y.; Wang, P.; Wang, J.; Lee, M.-H. Circadian clock gene CRY2 degradation is involved in chemoresistance of colorectal cancer. Mol. Cancer Ther. 2015, 14, 1476–1487. [Google Scholar] [CrossRef]
- Xu, Y.; Qiu, A.; Peng, F.; Tan, X.; Wang, J.; Gong, X. Exosomal transfer of circular RNA FBXW7 ameliorates the chemoresistance to oxaliplatin in colorectal cancer by sponging miR-18b-5p. Neoplasma 2021, 68, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, F.; Babaei-Jadidi, R.; Sheard, J.; Spencer-Dene, B.; Nateri, A.S. Fbxw7-associated drug resistance is reversed by induction of terminal differentiation in murine intestinal organoid culture. Mol. Ther. Methods Clin. Dev. 2016, 3, 16024. [Google Scholar] [CrossRef] [PubMed]
- Fiore, D.; Piscopo, C.; Proto, M.C.; Vasaturo, M.; Dal Piaz, F.; Fusco, B.M.; Pagano, C.; Laezza, C.; Bifulco, M.; Gazzerro, P. N6-Isopentenyladenosine Inhibits Colorectal Cancer and Improves Sensitivity to 5-Fluorouracil-Targeting FBXW7 Tumor Suppressor. Cancers 2019, 11, 1456. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Ikoma, T.; Yamamura, S.; Miura, K.; Tsuduki, T.; Watanabe, T.; Nagai, H.; Takatani, M.; Yasui, H. Regorafenib is suitable for advanced colorectal cancer patients who have previously received trifluridine/tipiracil plus bevacizumab. Sci. Rep. 2023, 13, 2433, Erratum in Sci. Rep. 2023, 13, 11935. [Google Scholar] [CrossRef]
- Tong, J.; Tan, S.; Zou, F.; Yu, J.; Zhang, L. FBW7 mutations mediate resistance of colorectal cancer to targeted therapies by blocking Mcl-1 degradation. Oncogene 2017, 36, 787–796. [Google Scholar] [CrossRef]
- Wang, H.-P.; Chen, W.-J.; Shen, J.-M.; Ye, T.; Xie, H.-W. Attenuating glucose metabolism by Fbxw7 promotes Taxol sensitivity of colon cancer cells through downregulating NADPH oxidase 1 (Nox1). Ann. Transl. Med. 2021, 9, 886. [Google Scholar] [CrossRef]
- Jiménez-Izquierdo, R.; Morrugares, R.; Suanes-Cobos, L.; Correa-Sáez, A.; Garrido-Rodríguez, M.; Cerero-Tejero, L.; Khan, O.M.; de la Luna, S.; Sancho, R. FBXW7 tumor suppressor regulation by dualspecificity tyrosine-regulated kinase 2. Cell Death Dis. 2023, 14, 202. [Google Scholar] [CrossRef]
- Ding, J.; Zhao, Z.; Song, J.; Luo, B.; Huang, L. MiR-223 promotes the doxorubicin resistance of colorectal cancer cells via regulating epithelial–mesenchymal transition by targeting FBXW7. Acta Biochim. Biophys. Sin. 2018, 50, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Tan, S.; Nikolovska-Coleska, Z.; Yu, J.; Zou, F.; Zhang, L. FBW7-Dependent Mcl-1 Degradation Mediates the Anticancer Effect of Hsp90 Inhibitors. Mol. Cancer Ther. 2017, 16, 1979–1988. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Zhang, H.; Li, H.; Xu, Q.; Sun, W.; Zhang, S.; Zhang, X.; Zhu, S.; Wang, H. Prognostic risk analysis related to radioresistance genes in colorectal cancer. Front. Oncol. 2023, 12, 1100481. [Google Scholar] [CrossRef]
- Sun, T.; Yin, Y.; Jin, H.; Liu, H.; Tian, W. Exosomal microRNA-19b targets FBXW7 to promote colorectal cancer stem cell stemness and induce resistance to radiotherapy. Kaohsiung J. Med. Sci. 2022, 38, 108–119. [Google Scholar] [CrossRef]
- Huang, G.; Long, K. Sensitization of colon cancer cells to cisplatin by Fbxw7 via negative regulation of the Nox1-mTOR pathway. Pathol. Res. Pr. 2023, 247, 154479. [Google Scholar] [CrossRef]
- Honma, S.; Hisamori, S.; Nishiuchi, A.; Itatani, Y.; Obama, K.; Shimono, Y.; Sakai, Y. F-Box/WD Repeat Domain-Containing 7 Induces Chemotherapy Resistance in Colorectal Cancer Stem Cells. Cancers 2019, 11, 635. [Google Scholar] [CrossRef]
- Izumi, D.; Ishimoto, T.; Miyake, K.; Eto, T.; Arima, K.; Kiyozumi, Y.; Uchihara, T.; Kurashige, J.; Iwatsuki, M.; Baba, Y.; et al. Colorectal Cancer Stem Cells Acquire Chemoresistance Through the Upregulation of F-Box/WD Repeat-Containing Protein 7 and the Consequent Degradation of c-Myc. Stem Cells 2017, 35, 2027–2036. [Google Scholar] [CrossRef]
- Ruan, F.; Ruan, Y.; Gu, H.; Sun, J.; Chen, Q. Clitocine enhances the drug sensitivity of colon cancer cells by promoting FBXW7-mediated MCL-1 degradation via inhibiting the A2B/cAMP/ERK axis. Am. J. Physiol. Physiol. 2024, 327, C884–C900. [Google Scholar] [CrossRef]
- Belmonte-Fernández, A.; Herrero-Ruíz, J.; Galindo-Moreno, M.; Limón-Mortés, M.C.; Mora-Santos, M.; Sáez, C.; Japón, M.Á.; Tortolero, M.; Romero, F. Cisplatin-induced cell death increases the degradation of the MRE11-RAD50-NBS1 complex through the autophagy/lysosomal pathway. Cell Death Differ. 2023, 30, 488–499. [Google Scholar] [CrossRef]
- Baxter, J.S.; Brough, R.; Krastev, D.B.; Song, F.; Sridhar, S.; Gulati, A.; Alexander, J.; Roumeliotis, T.I.; Kozik, Z.; Choudhary, J.S.; et al. Cancer-associated FBXW7 loss is synthetic lethal with pharmacological targeting of CDC7. Mol. Oncol. 2024, 18, 369–385. [Google Scholar] [CrossRef]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef]
- Sanchez-Burgos, L.; Navarro-González, B.; García-Martín, S.; Sirozh, O.; Mota-Pino, J.; Fueyo-Marcos, E.; Tejero, H.; Antón, M.E.; Murga, M.; Al-Shahrour, F.; et al. Activation of the integrated stress response is a vulnerability for multidrug-resistant FBXW7 -deficient cells. EMBO Mol. Med. 2022, 14, e15855. [Google Scholar] [CrossRef]
- Pfohl, U.; Loskutov, J.; Bashir, S.; Kühn, R.; Herter, P.; Templin, M.; Mamlouk, S.; Belanov, S.; Linnebacher, M.; Bürtin, F.; et al. A RAS-Independent Biomarker Panel to Reliably Predict Response to MEK Inhibition in Colorectal Cancer. Cancers 2022, 14, 3252. [Google Scholar] [CrossRef]
- Escobar, D.; Bushara, O.; Sun, L.; Liao, J.; Yang, G.-Y. Clinicopathologic characteristics of FBXW7-mutated colorectal adenocarcinoma and association with aberrant beta-catenin localization. Hum. Pathol. 2022, 119, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Tricoli, J.V.; Boardman, L.A.; Patidar, R.; Sindiri, S.; Jang, J.S.; Walsh, W.D.; McGregor, P.M.; Camalier, C.E.; Ma, M.G.M.; Furman, W.L.; et al. A mutational comparison of adult and adolescent and young adult (AYA) colon cancer. Cancer 2018, 124, 1070–1082. [Google Scholar] [CrossRef] [PubMed]
- Kothari, N.; Teer, J.K.; Abbott, A.M.; Srikumar, T.; Zhang, Y.; Yoder, S.J.; Brohl, A.S.; Kim, R.D.; Reed, D.R.; Shibata, D. Increased incidence of FBXW7 and POLE proofreading domain mutations in young adult colorectal cancers. Cancer 2016, 122, 2828–2835. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Peng, W.; Tian, C.; Zhang, Y.; Ji, D.; Wang, L.; Jin, K.; Wang, F.; Shao, Y.; Wang, X.; et al. Molecular characteristics of early-onset compared with late-onset colorectal cancer: A case controlled study. Int. J. Surg. 2024, 110, 4559–4570. [Google Scholar] [CrossRef]
- Mouradov, D.; Greenfield, P.; Li, S.; In, E.-J.; Storey, C.; Sakthianandeswaren, A.; Georgeson, P.; Buchanan, D.D.; Ward, R.L.; Hawkins, N.J.; et al. Oncomicrobial Community Profiling Identifies Clinicomolecular and Prognostic Subtypes of Colorectal Cancer. Gastroenterology 2023, 165, 104–120. [Google Scholar] [CrossRef]
- Holowatyj, A.N.; Wen, W.; Gibbs, T.; Seagle, H.M.; Keller, S.R.; Edwards, D.R.V.; Washington, M.K.; Eng, C.; Perea, J.; Zheng, W.; et al. Racial/Ethnic and Sex Differences in Somatic Cancer Gene Mutations among Patients with Early-Onset Colorectal Cancer. Cancer Discov. 2023, 13, 570–579. [Google Scholar] [CrossRef]
- Roberto, M.; Arrivi, G.; Pilozzi, E.; Montori, A.; Balducci, G.; Mercantini, P.; Laghi, A.; Ierinò, D.; Panebianco, M.; Marinelli, D.; et al. The Potential Role of Genomic Signature in Stage II Relapsed Colorectal Cancer (CRC) Patients: A Mono-Institutional Study. Cancer Manag. Res. 2022, 14, 1353–1369. [Google Scholar] [CrossRef]
- Miyaki, M.; Yamaguchi, T.; Iijima, T.; Takahashi, K.; Matsumoto, H.; Mori, T. Somatic Mutations of the CDC4 (FBXW7) Gene in Hereditary Colorectal Tumors. Oncology 2009, 76, 430–434. [Google Scholar] [CrossRef]
- Intarajak, T.; Udomchaiprasertkul, W.; Bunyoo, C.; Yimnoon, J.; Soonklang, K.; Wiriyaukaradecha, K.; Lamlertthon, W.; Sricharunrat, T.; Chaiwiriyawong, W.; Siriphongpreeda, B.; et al. Genetic Aberration Analysis in Thai Colorectal Adenoma and Early-Stage Adenocarcinoma Patients by Whole-Exome Sequencing. Cancers 2019, 11, 977. [Google Scholar] [CrossRef]
- Ling, C.; Wang, L.; Wang, Z.; Xu, L.; Sun, L.; Yang, H.; Li, W.-D.; Wang, K. A pathway-centric survey of somatic mutations in chinese patients with colorectal carcinomas. PLoS ONE 2015, 10, e0116753. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Jiang, S.; Zhou, X.; Si, C.; Shao, P.; Jiang, Q.; Zhu, L.; Shen, L.; Meng, Q.; Yin, J.C.; et al. FBXW7 and Its Downstream NOTCH Pathway Could be Potential Indicators of Organ-Free Metastasis in Colorectal Cancer. Front. Oncol. 2021, 11, 783564. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.H.; Yu, G.Y.; Hong, Y.G.; Lian, W.; Chouhan, H.; Xu, Y.; Liu, L.J.; Bai, C.G.; Zhang, W. Clinical significance of multiple gene detection with a 22-gene panel in formalin-fixed paraffin-embedded specimens of 207 colorectal cancer patients. Int. J. Clin. Oncol. 2019, 24, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Gao, J.; Mao, Z.; Wang, J.; Li, J.; Li, W.; Lei, Y.; Li, S.; Wu, Z.; Tang, C.; et al. Genetic mutations in human rectal cancers detected by targeted sequencing. J. Hum. Genet. 2015, 60, 589–596. [Google Scholar] [CrossRef]
- Korphaisarn, K.; Pongpaibul, A.; Roothumnong, E.; Pongsuktavorn, K.; Thamlikitkul, L.; Anekpuritanang, T.; Poungvarin, N.; Thongnoppakhun, W.; Pithukpakorn, M. High Frequency of KRAS Codon 146 and FBXW7 Mutations in Thai Patients with Stage II-III Colon Cancer. Asian Pac. J. Cancer Prev. 2019, 20, 2319–2326. [Google Scholar] [CrossRef]
- Jauhri, M.; Bhatnagar, A.; Gupta, S.; Shokeen, Y.; Minhas, S. Targeted molecular profiling of rare genetic alterations in colorectal cancer using next-generation sequencing. Med. Oncol. 2016, 33, 106. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, H.; Bao, H.; Zhang, J.; Wu, R.; Zhu, L. Comprehensive characterization of FBXW7 mutational and clinicopathological profiles in human colorectal cancers. Front. Oncol. 2023, 13, 1154432. [Google Scholar] [CrossRef]
- Heczko, L.; Liška, V.; Vyčítal, O.; Fiala, O.; Šůsová, S.; Hlaváč, V.; Souček, P. Targeted panel sequencing of pharmacogenes and oncodrivers in colorectal cancer patients reveals genes with prognostic significance. Hum. Genom. 2024, 18, 83. [Google Scholar] [CrossRef]
- Mouradov, D.; Domingo, E.; Gibbs, P.; Jorissen, R.N.; Li, S.; Soo, P.Y.; Lipton, L.; Desai, J.; E Danielsen, H.; Oukrif, D.; et al. Survival in stage II/III colorectal cancer is independently predicted by chromosomal and microsatellite instability, but not by specific driver mutations. Am. J. Gastroenterol. 2013, 108, 1785–1793. [Google Scholar] [CrossRef]
- Yeşilkaya, F. Examination of the expression levels of MACC1, Filamin A and FBXW7 genes in colorectal cancer patients. North. Clin. Istanb. 2020, 7, 1–5. [Google Scholar] [CrossRef]
- Lee, C.S.; Song, I.H.; Lee, A.; Kang, J.; Lee, Y.S.; Lee, I.K.; Song, Y.S.; Lee, S.H. Enhancing the landscape of colorectal cancer using targeted deep sequencing. Sci. Rep. 2021, 11, 8154. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhou, W.; Li, Q.; Li, P.; Yang, L.; Xia, X.; Yi, X.; Wan, D. Tumor-derived mutations in postoperative plasma of colorectal cancer with microsatellite instability. Transl. Oncol. 2021, 14, 100945. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Xu, L.; Zhang, Y.; Yang, H.; Liu, S.; Ding, S.; Zhao, H.; Sui, Y.; Wang, C.; Quan, L.; et al. Correlation between NGS panel-based mutation results and clinical information in colorectal cancer patients. Heliyon 2024, 10, e29299. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, R.; Li, J.; Yi, Y.; Liu, X.; Chen, J.; Zhang, H.; Lu, J.; Li, C.; Wu, H.; et al. Comprehensive analysis of oncogenic fusions in mismatch repair deficient colorectal carcinomas by sequential DNA and RNA next generation sequencing. J. Transl. Med. 2021, 19, 433. [Google Scholar] [CrossRef]
- Su, H.; Chang, C.; Hao, J.; Xu, X.; Bao, M.; Luo, S.; Zhao, C.; Liu, Q.; Wang, X.; Zhou, Z.; et al. Identification of Genomic Alterations of Perineural Invasion in Patients with Stage II Colorectal Cancer. OncoTargets Ther. 2020, 13, 11571–11582. [Google Scholar] [CrossRef]
- Liu, H.; Wang, K.; Fu, H.; Song, J. Low expression of the ubiquitin ligase FBXW7 correlates with poor prognosis of patients with colorectal cancer. Int. J. Clin. Exp. Pathol. 2018, 11, 413–419. [Google Scholar]
- Lee, C.S.; Kim, H.S.; Schageman, J.; Lee, I.K.; Kim, M.; Kim, Y. Postoperative Circulating Tumor DNA Can Predict High Risk Patients with Colorectal Cancer Based on Next-Generation Sequencing. Cancers 2021, 13, 4190. [Google Scholar] [CrossRef]
- Armengol, G.; Sarhadi, V.K.; Ghanbari, R.; Doghaei-Moghaddam, M.; Ansari, R.; Sotoudeh, M.; Puolakkainen, P.; Kokkola, A.; Malekzadeh, R.; Knuutila, S. Driver Gene Mutations in Stools of Colorectal Carcinoma Patients Detected by Targeted Next-Generation Sequencing. J. Mol. Diagn. 2016, 18, 471–479. [Google Scholar] [CrossRef]
- Kawashita, Y.; Morine, Y.; Ikemoto, T.; Saito, Y.; Iwahashi, S.; Yamada, S.; Higashijima, J.; Imura, S.; Ogawa, H.; Yagi, T.; et al. Loss of Fbxw7 expression is a predictor of recurrence in colorectal liver metastasis. J. Hepato-Biliary-Pancreat. Sci. 2017, 24, 576–583. [Google Scholar] [CrossRef]
- Ye, J.; Lin, M.; Zhang, C.; Zhu, X.; Li, S.; Liu, H.; Yin, J.; Yu, H.; Zhu, K. Tissue gene mutation profiles in patients with colorectal cancer and their clinical implications. Biomed. Rep. 2020, 13, 43–48. [Google Scholar] [CrossRef] [PubMed]
- De Nicola, F.; Goeman, F.; Pallocca, M.; Sperati, F.; Pizzuti, L.; Melucci, E.; Casini, B.; Amoreo, C.A.; Gallo, E.; Diodoro, M.G.; et al. Deep sequencing and pathway-focused analysis revealed multigene oncodriver signatures predicting survival outcomes in advanced colorectal cancer. Oncogenesis 2018, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Kovaleva, V.; Geissler, A.-L.; Lutz, L.; Fritsch, R.; Makowiec, F.; Wiesemann, S.; Hopt, U.T.; Passlick, B.; Werner, M.; Lassmann, S. Spatio-temporal mutation profiles of case-matched colorectal carcinomas and their metastases reveal unique de novo mutations in metachronous lung metastases by targeted next generation sequencing. Mol. Cancer 2016, 15, 63. [Google Scholar] [CrossRef] [PubMed]
- Nemecek, R.; Berkovcova, J.; Radova, L.; Kazda, T.; Mlcochova, J.; Vychytilova-Faltejskova, P.; Slaby, O.; Svoboda, M. Mutational analysis of primary and metastatic colorectal cancer samples underlying the resistance to cetuximab-based therapy. Onco Targets Ther. 2016, 9, 4695–4703. [Google Scholar] [CrossRef]
- Jesinghaus, M.; Wolf, T.; Pfarr, N.; Muckenhuber, A.; Ahadova, A.; Warth, A.; Goeppert, B.; Sers, C.; Kloor, M.; Endris, V.; et al. Distinctive Spatiotemporal Stability of Somatic Mutations in Metastasized Microsatellite-stable Colorectal Cancer. Am. J. Surg. Pathol. 2015, 39, 1140–1147. [Google Scholar] [CrossRef]
- Ciepiela, I.; Szczepaniak, M.; Ciepiela, P.; Hińcza-Nowak, K.; Kopczyński, J.; Macek, P.; Kubicka, K.; Chrapek, M.; Tyka, M.; Góźdź, S.; et al. Tumor location matters, next generation sequencing mutation profiling of left-sided, rectal, and right-sided colorectal tumors in 552 patients. Sci. Rep. 2024, 14, 4619. [Google Scholar] [CrossRef]
- Li, W.; Qiu, T.; Guo, L.; Ying, J.; Zhou, A. NGS-based oncogenic mutations analysis in advanced colorectal cancer patients improves targeted therapy prediction. Pathol. Res. Pr. 2019, 215, 483–489. [Google Scholar] [CrossRef]
- Antoniotti, C.; Germani, M.M.; Rossini, D.; Lonardi, S.; Pietrantonio, F.; Santini, D.; Marmorino, F.; Allegrini, G.; Daniel, F.; Raimondi, A.; et al. FOLFOXIRI and bevacizumab in patients with early-onset metastatic colorectal cancer. A pooled analysis of TRIBE and TRIBE2 studies. Eur. J. Cancer 2022, 167, 23–31. [Google Scholar] [CrossRef]
- Dallol, A.; Buhmeida, A.; Al-Ahwal, M.S.; Al-Maghrabi, J.; Bajouh, O.; Al-Khayyat, S.; Alam, R.; Abusanad, A.; Turki, R.; Elaimi, A.; et al. Clinical significance of frequent somatic mutations detected by high-throughput targeted sequencing in archived colorectal cancer samples. J. Transl. Med. 2016, 14, 118. [Google Scholar] [CrossRef]
- Braxton, D.R.; Zhang, R.; Morrissette, J.D.; Loaiza-Bonilla, A.; Furth, E.E. Clinicopathogenomic analysis of mismatch repair proficient colorectal adenocarcinoma uncovers novel prognostic subgroups with differing patterns of genetic evolution. Int. J. Cancer 2016, 139, 1546–1556. [Google Scholar] [CrossRef]
- Lavacchi, D.; Gelmini, S.; Calabri, A.; Rossi, G.; Simi, L.; Caliman, E.; Mancini, I.; Salvianti, F.; Petroni, G.; Guidolin, A.; et al. Early changes in circulating tumor DNA (ctDNA) predict treatment response in metastatic KRAS-mutated colorectal cancer (mCRC) patients. Heliyon 2023, 9, e21853. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Kageyama, S.-I.; Seki, M.; Suzuki, A.; Okumura, M.; Hojo, H.; Motegi, A.; Akimoto, T. Liquid Biopsy Cell-free DNA Biomarkers in Patients with Oligometastatic Colorectal Cancer Treated by Ablative Radiotherapy. Anticancer. Res. 2021, 41, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Adua, D.; Di Fabio, F.; Ercolani, G.; Fiorentino, M.; Gruppioni, E.; Altimari, A.; Limpe, F.L.R.; Normanno, N.; Pinna, A.D.; Pinto, C. Heterogeneity in the colorectal primary tumor and the synchronous resected liver metastases prior to and after treatment with an anti-EGFR monoclonal antibody. Mol. Clin. Oncol. 2017, 7, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Cho, Y.B.; Wang, K.; Huang, D.; Hong, H.K.; Choi, Y.-L.; Ko, Y.H.; Nam, D.-H.; Jin, J.; Yang, H.; et al. Patterns of somatic alterations between matched primary and metastatic colorectal tumors characterized by whole-genome sequencing. Genomics 2014, 104, 234–241. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Newhook, T.E.; Cao, H.S.T.; Tzeng, C.-W.D.; Chun, Y.S.; Aloia, T.A.; Dasari, A.; Kopetz, S.; Vauthey, J.-N. Alteration of FBXW7 is Associated with Worse Survival in Patients Undergoing Resection of Colorectal Liver Metastases. J. Gastrointest. Surg. 2021, 25, 186–194. [Google Scholar] [CrossRef]
- Korphaisarn, K.; Morris, V.K.; Overman, M.J.; Fogelman, D.R.; Kee, B.K.; Raghav, K.P.S.; Manuel, S.; Shureiqi, I.; Wolff, R.A.; Eng, C.; et al. FBXW7 missense mutation: A novel negative prognostic factor in metastatic colorectal adenocarcinoma. Oncotarget 2017, 8, 39268–39279. [Google Scholar] [CrossRef]
- Haan, J.C.; Labots, M.; Rausch, C.; Koopman, M.; Tol, J.; Mekenkamp, L.J.M.; van de Wiel, M.A.; Israeli, D.; van Essen, H.F.; van Grieken, N.C.T.; et al. Genomic landscape of metastatic colorectal cancer. Nat. Commun. 2014, 5, 5457. [Google Scholar] [CrossRef]
- Koopman, M.; Antonini, N.F.; Douma, J.; Wals, J.; Honkoop, A.H.; Erdkamp, F.L.; de Jong, R.S.; Rodenburg, C.J.; Vreugdenhil, G.; Loosveld, O.J.; et al. Sequential versus combination chemotherapy with capecitabine, irinotecan, and oxaliplatin in advanced colorectal cancer (CAIRO): A phase III randomised controlled trial. Lancet 2007, 370, 135–142. [Google Scholar] [CrossRef]
- Tol, J.; Koopman, M.; Cats, A.; Rodenburg, C.J.; Creemers, G.J.; Schrama, J.G.; Erdkamp, F.L.; Vos, A.H.; van Groeningen, C.J.; Sinnige, H.A.; et al. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N. Engl. J. Med. 2009, 360, 563–572, Erratum in N. Engl. J. Med. 2010, 363, 2573. [Google Scholar] [CrossRef]
- Broek, E.v.D.; Dijkstra, M.J.J.; Krijgsman, O.; Sie, D.; Haan, J.C.; Traets, J.J.H.; van de Wiel, M.A.; Nagtegaal, I.D.; Punt, C.J.A.; Carvalho, B.; et al. High Prevalence and Clinical Relevance of Genes Affected by Chromosomal Breaks in Colorectal Cancer. PLoS ONE 2015, 10, e0138141. [Google Scholar] [CrossRef]
- Okamoto, W.; Sakai, K.; Makiyama, A.; Yamamoto, Y.; Shitara, K.; Denda, T.; Izawa, N.; Nakano, Y.; Nishina, T.; Esaki, T.; et al. A phase II study to explore biomarkers for the use of mFOLFOX6/XELOX plus bevacizumab as a first-line chemotherapy in patients with metastatic colorectal cancer (WJOG7612GTR). ESMO Open 2022, 7, 100592. [Google Scholar] [CrossRef]
- Malapelle, U.; Pisapia, P.; Sgariglia, R.; Vigliar, E.; Biglietto, M.; Carlomagno, C.; Giuffrè, G.; Bellevicine, C.; Troncone, G. Less frequently mutated genes in colorectal cancer: Evidences from next-generation sequencing of 653 routine cases. J. Clin. Pathol. 2016, 69, 767–771. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; El Hage, M.; Linnebacher, M. Mutation patterns in colorectal cancer and their relationship with prognosis. Heliyon 2024, 10, e36550. [Google Scholar] [CrossRef] [PubMed]
- Lahoz, S.; Rodríguez, A.; Fernández, L.; Gorría, T.; Moreno, R.; Esposito, F.; Oliveres, H.; Albiol, S.; Saurí, T.; Pesantez, D.; et al. Mutational Status of SMAD4 and FBXW7 Affects Clinical Outcome in TP53–Mutated Metastatic Colorectal Cancer. Cancers 2022, 14, 5921. [Google Scholar] [CrossRef] [PubMed]
- Shida, D.; Kuchiba, A.; Shibata, T.; Hamaguchi, T.; Yamasaki, S.; Ito, M.; Kobatake, T.; Tonooka, T.; Masaki, T.; Shiozawa, M.; et al. Genomic landscape and its prognostic significance in stage III colorectal cancer: JCOG1506A1, an ancillary of JCOG0910. Cancer Sci. 2023, 114, 3352–3363. [Google Scholar] [CrossRef]
- Huang, W.; Li, H.; Shi, X.; Lin, M.; Liao, C.; Zhang, S.; Shi, W.; Zhang, L.; Zhang, X.; Gan, J. Characterization of genomic alterations in Chinese colorectal cancer patients. Ultrasound Med. Biol. 2021, 51, 120–129. [Google Scholar] [CrossRef]
- Crobach, S.; Ruano, D.; van Eijk, R.; Schrumpf, M.; PALGA Group; Fleuren, G.; van Wezel, T.; Morreau, H. Somatic mutation profiles in primary colorectal cancers and matching ovarian metastases: Identification of driver and passenger mutations. J. Pathol. Clin. Res. 2016, 2, 166–174. [Google Scholar] [CrossRef]
- Lupini, L.; Bassi, C.; Mlcochova, J.; Musa, G.; Russo, M.; Vychytilova-Faltejskova, P.; Svoboda, M.; Sabbioni, S.; Nemecek, R.; Slaby, O.; et al. Prediction of response to anti-EGFR antibody-based therapies by multigene sequencing in colorectal cancer patients. BMC Cancer 2015, 15, 808. [Google Scholar] [CrossRef]
- Liu, J.; Wei, L.; Hu, N.; Wang, D.; Ni, J.; Zhang, S.; Liu, H.; Lv, T.; Yin, J.; Ye, M.; et al. FBW7-mediated ubiquitination and destruction of PD-1 protein primes sensitivity to anti-PD-1 immunotherapy in non-small cell lung cancer. J. Immunother. Cancer 2022, 10, e005116. [Google Scholar] [CrossRef]
- Gstalder, C.; Liu, D.; Miao, D.; Lutterbach, B.; DeVine, A.L.; Lin, C.; Shettigar, M.; Pancholi, P.; Buchbinder, E.I.; Carter, S.L.; et al. Inactivation of Fbxw7 impairs dsRNA sensing and confers resistance to PD-1 blockade. Cancer Discov. 2025, 10, 1296–1311. [Google Scholar] [CrossRef]
| Author, Year | Type of Study | Mechanism of FBXW7 Studied | Therapy Resistance Mechanisms Mediated by FBXW7 |
|---|---|---|---|
| Fang L 2015 [53] | In vitro | CRY2 and FBXW7 expression are inversely correlated, influencing chemoresistance. | FBXW7 promotes CRY2 degradation, increasing CRC cells’ sensitivity to oxaliplatin. |
| Tong J 2017 [58] | In vitro/in vivo (Mice) | Role of GSK3β/FBXW7-dependent Mcl-1 degradation in killing of CRC cells by Hsp90 inhibitors | GSK3β/FBXW7-driven Mcl-1 degradation is essential for Hsp90 inhibitor–induced apoptosis in CRC; FBXW7 status and Mcl-1 stability determine drug response and guide patient stratification and combination therapies. |
| Ding J 2018 [61] | In vitro | Role of miR-223/FBXW7 pathway in chemosensitivity in different CRC cell lines | High FBXW7 enhances doxorubicin sensitivity in CRC, and miR-223 reduces FBXW7 levels, decreasing drug sensitivity. |
| Fiore D 2019 [56] | In vitro In vivo (Mice) | N6-isopentenyladenosine role in CRC proliferation with different mutational status of FBXW7 and TP53 genes | In FBXW7- and TP53-wild type CRC cells, N6-isopentenyladenosine synergizes with 5-Fluorouracil, suggesting FBXW7 restoration as a strategy for personalized therapy. |
| Xu Y 2021 [54] | In vitro/ In vivo (Mice) | Role of Circular RNA F-box and WD repeat domain containing 7 (circ-FBXW7) in CRC oxaliplatin resistance | Circ-FBXW7 is downregulated in oxaliplatin-resistant CRC, and exosomal circ-FBXW7 can reduce resistance by targeting miR-128-3p. |
| Wang HP 2021 [59] | In vitro | Biological roles and mechanisms of FBXW7 in taxol resistance | FBXW7 sensitizes CRC cells to Taxol by suppressing Nox1 and glucose metabolism, and its downregulation drives resistance, highlighting the FBXW7–Nox1–metabolism axis as a therapeutic target |
| Sun T 2022 [64] | In vitro In vivo (Mice) | Roles of exosomal microRNA-19b (miR-19b) in CRC radioresistance. | CRC-derived exosomes promote radioresistance and stemness via miR-19b, which targets and downregulates FBXW7. |
| Huang G 2023 [65] | In vivo (pts 30) in vitro | FBXW7-Nox1-mTOR pathway and Cisplatin chemoresistance of CRC | FBXW7 was negatively associated with CRC tumor recurrence and cisplatin resistance and positively associated with CRC patient survival rates. |
| Homma S 2019 [66] | In vivo (55 CRC patients receiving neoadjuvant therapy) | The role of FBXW7 as a cell cycle mediator was investigated, focusing on its expression in colorectal cancer stem cell populations and its impact on cell cycle arrest and chemoresistance | High FBXW7 expression correlated with poor pathological response to treatment, increased chemoresistance in cancer stem cell subsets, and FBXW7 knockdown enhanced sensitivity to anticancer drugs both in vitro and in vivo. |
| Izumi D 2017 [67] | In vitro | Upregulation of FBXW7 leads to c-Myc degradation, promoting colorectal CSC resistance to chemotherapy-induced cell cycle arrest. | FBXW7 is specifically upregulated in CSCs after drug treatment, decreasing c-Myc and conferring drug resistance; FBXW7 knockdown increases drug sensitivity, with findings confirmed in liver metastases from CRC patients post-chemotherapy. |
| Ruan F 2024 [68] | Only abstract In vitro/in vivo | Clitocine upregulates FBXW7 by inhibiting A2B receptor/cAMP/ERK signaling, enhancing FBXW7-mediated MCL-1 degradation. | Clitocine decreases intracellular cAMP via competitive binding to A2B receptor, enhancing FBXW7 expression through reduced promoter DNA methylation, leading to increased MCL-1 degradation and improved drug sensitivity; A2B knockout or cAMP analog treatment reverses these effects. |
| Belmonte-Fernández A 2023 [69] | In vitro | FBXW7 regulates DNA damage response and cisplatin sensitivity by ubiquitinating the MRN complex. | Cisplatin induces FBXW7-dependent MRN degradation through the autophagy-lysosome pathway, promoting tumor cell death, while lysosome inhibition blocks this effect, suggesting a strategy to enhance chemosensitivity. |
| Baxter JS 2024 [70] | In vitro | BXW7 loss creates a RIF1-dependent synthetic lethality to CDC7 inhibition. | FBXW7-deficient cells show selective sensitivity to CDC7 inhibition; RIF1 silencing rescues this effect, identifying a potential therapeutic vulnerability in FBXW7-mutant cancers. |
| Hu JL 2019 [71] | In vitro/in vivo (40 cases of CRC tissues and corresponding normal mucosa) | CAF-derived exosomal miR-92a-3p suppresses FBXW7 and MOAP1, activating Wnt/β-catenin signaling to promote EMT, stemness, metastasis, and chemoresistance in CRC. | CAF-derived exosomes enhance CRC metastasis and resistance to 5-FU/L-OHP via miR-92a-3p–mediated suppression of FBXW7 and MOAP1; high serum miR-92a-3p correlates clinically with poor prognosis and chemoresistance in CRC patients. |
| Li N 2019 [23] | Combined in vitro, ex vivo, and In vivo study (mice) | FBXW7 degrades ZEB2 to suppress EMT, CSC traits, and chemoresistance; its loss stabilizes ZEB2, promoting these processes. | FBXW7 suppresses EMT and CSC formation by degrading ZEB2; its loss enhances chemoresistance and metastasis, while ZEB2 knockdown reverses these effects, highlighting the FBXW7–ZEB2 axis as a key modulator of drug resistance and tumor progression. |
| Sanchez Burgos L 2022 [72] | In vitro and in vivo (mice) | FBXW7 loss drives multidrug resistance, making cells dependent on the integrated stress response for survival. | FBXW7-deficient, multidrug-resistant cells show hyperactivation of the ISR, making them selectively vulnerable to ISR inhibition both in vitro and in vivo, revealing a therapeutic opportunity to target chemoresistant tumors with defective FBXW7. |
| Pfohl, U 2022 [73] | In vitro (organoids) | Molecular pattern of CRC and sensitivity to MEK inhibitors | SMAD4 loss and the SFAB signature (SMAD4, FBXW7, ARID1A, BMPR2) predict MEK-inhibitor sensitivity in PDOs, regardless of RAS or BRAF status. |
| Molecular Mechanism/Substrate Affected | Biologic Effect | Clinical Correlate/ Observed Finding |
|---|---|---|
| EGFR stabilization (impaired ubiquitination) | Increased proliferative signaling; reduced EGF dependency | ↓ Sensitivity to anti-EGFR therapy; increased adaptive MAPK/PI3K signaling (organoid and clinical evidence) |
| Cyclin E1 accumulation | Cell-cycle acceleration; genomic instability | More aggressive biology in some FBXW7-mutant CRCs; worse DFS/OS when co-mutated with TP53 |
| MCL-1 stabilization (impaired degradation) | Anti-apoptotic signaling; survival of stressed clones | Resistance to chemotherapy; MCL-1 inhibition restores drugs sensitivity (preclinical) |
| NOTCH1/NOTCH signaling dysregulation | Progenitor cell expansion; impaired differentiation | Higher mutation frequency in early-stage, organ-confined CRC; enrichment in AYA CRC and NOTCH-altered tumors |
| c-MYC stabilization (via SET–FBXW7 interaction | Immunosuppressive microenvironment via CCL5 repression | Immune-excluded MSS CRC phenotype; potential reduced benefit from immunotherapy (preclinical evidence) |
| HIF-1α stabilization | Upregulation of VEGF-A; angiogenic shift | Possible modulation of responsiveness to anti-angiogenic therapy (clinical data limited) |
| ZEB2 degradation failure | EMT induction; stemness, migration, invasion | Greater metastatic potential, intrapatient heterogeneity, site-specific differences between primaries/metastases |
| Interaction with miRNAs (miR-223, miR-27a, miR-182, miR-92b) | Post-transcriptional downregulation of FBXW7 | ↑ Proliferation, ↑ invasion, ↑ FOXO/Notch/Akt signaling; contribution to chemo- and radio-resistance |
| circ-FBXW7/exosomal regulation | Modulates FBXW7 expression; miRNA sponging | Exosomal circ-FBXW7reverses oxaliplatin resistance (preclinical) |
| FBXW7β hotspot mutations (e.g., R465C/H, R479Q) | Variant-specific substrate affinity loss | MSI-H association; better prognosis in specific hotspots; supports variant-level reporting |
| Loss in colonic stem/progenitor cells | Expansion of undifferentiated cells | Higher prevalence in early-onset CRC; association with right-sided, high-grade tumors |
| Clonal evolution under treatment (ctDNA emergence) | Resistant clone expansion | Early detection of progression on EGFR therapy; rising FBXW7-mutant ctDNA predicts shorter PFS |
| High intratumoral heterogeneity | Discordant mutation detection depending on sampling site | Need of re-biopsy of metastatic lesions; stool DNA and ctDNA improve detection |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arrivi, G.; Gentile, G.; Roberto, M.; Delle Cave, D. FBXW7 Gene Mutation and Expression in Colorectal Cancer (CRC): A Systematic Review from Molecular Mechanisms to Clinical Translation. Int. J. Mol. Sci. 2025, 26, 11318. https://doi.org/10.3390/ijms262311318
Arrivi G, Gentile G, Roberto M, Delle Cave D. FBXW7 Gene Mutation and Expression in Colorectal Cancer (CRC): A Systematic Review from Molecular Mechanisms to Clinical Translation. International Journal of Molecular Sciences. 2025; 26(23):11318. https://doi.org/10.3390/ijms262311318
Chicago/Turabian StyleArrivi, Giulia, Gabriella Gentile, Michela Roberto, and Donatella Delle Cave. 2025. "FBXW7 Gene Mutation and Expression in Colorectal Cancer (CRC): A Systematic Review from Molecular Mechanisms to Clinical Translation" International Journal of Molecular Sciences 26, no. 23: 11318. https://doi.org/10.3390/ijms262311318
APA StyleArrivi, G., Gentile, G., Roberto, M., & Delle Cave, D. (2025). FBXW7 Gene Mutation and Expression in Colorectal Cancer (CRC): A Systematic Review from Molecular Mechanisms to Clinical Translation. International Journal of Molecular Sciences, 26(23), 11318. https://doi.org/10.3390/ijms262311318






