Accurate RET Fusion Detection in Solid Tumors Using RNA Sequencing Coverage Imbalance Analysis
Abstract
1. Introduction
2. Results
2.1. RET Coverage Asymmetry Screening
2.2. RET Testing with Targeted NGS Panels and Sanger Sequencing
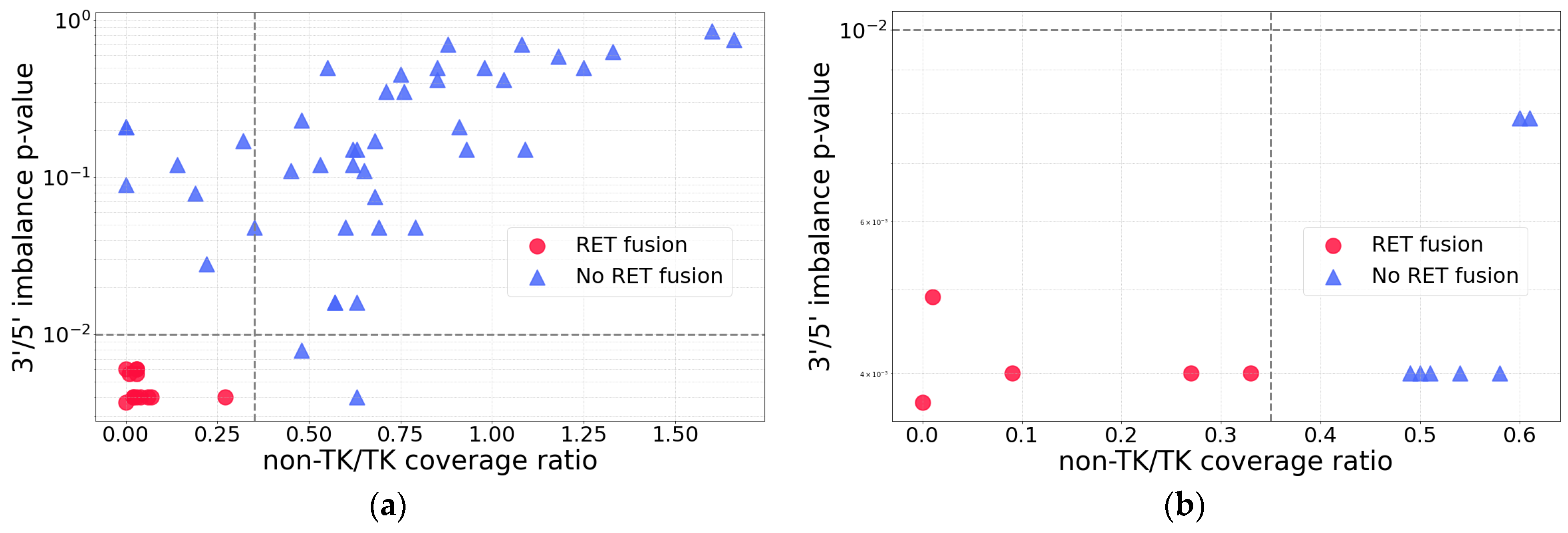
2.3. RNA-Seq Coverage Threshold Values for Detection of RET Fusions
2.4. Rare and Previously Unreported RET Fusions
3. Discussion
4. Materials and Methods
4.1. Biosamples and RNA Sequencing
4.2. Exon Coverage Calculation
4.3. Experimental Validation of RET Fusion Transcripts by Targeted NGS Panels
4.4. Experimental Validation of RET Fusion Transcripts by RT-PCR and Sanger Sequencing
4.5. Bioinformatic Detection of RET Fusion Transcripts
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sorokin, M.; Rabushko, E.; Rozenberg, J.M.; Mohammad, T.; Seryakov, A.; Sekacheva, M.; Buzdin, A. Clinically Relevant Fusion Oncogenes: Detection and Practical Implications. Ther. Adv. Med. Oncol. 2022, 14. [Google Scholar] [CrossRef]
- Drilon, A.; Oxnard, G.R.; Tan, D.S.W.; Loong, H.H.F.; Johnson, M.; Gainor, J.; McCoach, C.E.; Gautschi, O.; Besse, B.; Cho, B.C.; et al. Efficacy of Selpercatinib in RET Fusion–Positive Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 813–824. [Google Scholar] [CrossRef]
- Wirth, L.J.; Sherman, E.; Robinson, B.; Solomon, B.; Kang, H.; Lorch, J.; Worden, F.; Brose, M.; Patel, J.; Leboulleux, S.; et al. Efficacy of Selpercatinib in RET-Altered Thyroid Cancers. N. Engl. J. Med. 2020, 383, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Lopes, G.; Gainor, J.F.; Subbiah, V.; Bowles, D.W.; Doebele, R.C.; Mansfield, A.S.; Baik, C.S.; Gadgeel, S.M.; Kalemkerian, G.P.; Ou, S.-H.I.; et al. Efficacy and Safety of Pralsetinib in Patients with Advanced RET-Fusion-Positive NSCLC: Final Data from the Phase 1/2 ARROW Study. J. Clin. Oncol. 2025, 43, 8644. [Google Scholar] [CrossRef]
- Ke, J.; Huang, S.; Jing, Z.; Duan, M. The Efficacy and Safety of Selective RET Inhibitors in RET Fusion-Positive Non-Small Cell Lung Cancer: A Meta-Analysis. Investig. New Drugs 2023, 41, 768–776. [Google Scholar] [CrossRef]
- Takahashi, M.; Ritz, J.; Cooper, G.M. Activation of a Novel Human Transforming Gene, Ret, by DNA Rearrangement. Cell 1985, 42, 581–588. [Google Scholar] [CrossRef]
- Kawai, K.; Takahashi, M. Intracellular RET Signaling Pathways Activated by GDNF. Cell Tissue Res. 2020, 382, 113–123. [Google Scholar] [CrossRef]
- Regua, A.T.; Najjar, M.; Lo, H.W. RET Signaling Pathway and RET Inhibitors in Human Cancer. Front. Oncol. 2022, 12, 932353. [Google Scholar] [CrossRef]
- Mahato, A.K.; Saarma, M. Biology of RET Receptor and Its Ligands: Focus on the Nervous System. Endocr. Relat. Cancer 2025, 32, e240174. [Google Scholar] [CrossRef] [PubMed]
- Ivanchuk, S.M.; Myers, S.M.; Mulligan, L.M. Expression of RET 3′ Splicing Variants during Human Kidney Development. Oncogene 1998, 16, 991–996. [Google Scholar] [CrossRef]
- Richardson, D.S.; Rodrigues, D.M.; Hyndman, B.D.; Crupi, M.J.F.; Nicolescu, A.C.; Mulligan, L.M. Alternative Splicing Results in RET Isoforms with Distinct Trafficking Properties. Mol. Biol. Cell 2012, 23, 3838–3850. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.S.; Lee, W.-C.; Shin, J.-Y.; Lee, S.; Bleazard, T.; Won, J.-K.; Kim, Y.T.; Kim, J.-I.; Kang, J.-H.; Seo, J.-S. A Transforming KIF5B and RET Gene Fusion in Lung Adenocarcinoma Revealed from Whole-Genome and Transcriptome Sequencing. Genome Res. 2012, 22, 436–445. [Google Scholar] [CrossRef]
- Santoro, M.; Carlomagno, F. Central Role of RET in Thyroid Cancer. Cold Spring Harb. Perspect. Biol. 2013, 5, a009233. [Google Scholar] [CrossRef]
- Bossi, D.; Carlomagno, F.; Pallavicini, I.; Pruneri, G.; Trubia, M.; Raviele, P.R.; Marinelli, A.; Anaganti, S.; Cox, M.C.; Viale, G.; et al. Functional Characterization of a Novel FGFR1OP-RET Rearrangement in Hematopoietic Malignancies. Mol. Oncol. 2014, 8, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Staubitz, J.I.; Schad, A.; Springer, E.; Rajalingam, K.; Lang, H.; Roth, W.; Hartmann, N.; Musholt, T.J. Novel Rearrangements Involving the RET Gene in Papillary Thyroid Carcinoma. Cancer Genet. 2019, 230, 13–20. [Google Scholar] [CrossRef]
- Parimi, V.; Tolba, K.; Danziger, N.; Kuang, Z.; Sun, D.; Lin, D.I.; Hiemenz, M.C.; Schrock, A.B.; Ross, J.S.; Oxnard, G.R.; et al. Genomic Landscape of 891 RET Fusions Detected across Diverse Solid Tumor Types. npj Precis. Oncol. 2023, 7, 10. [Google Scholar] [CrossRef]
- Nagasaka, M.; Brazel, D.; Baca, Y.; Xiu, J.; Al-Hallak, M.N.; Kim, C.; Nieva, J.; Swensen, J.J.; Spetzler, D.; Korn, W.M.; et al. Pan-Tumor Survey of RET Fusions as Detected by next-Generation RNA Sequencing Identified RET Fusion Positive Colorectal Carcinoma as a Unique Molecular Subset. Transl. Oncol. 2023, 36, 101744. [Google Scholar] [CrossRef]
- Pekova, B.B.; Sykorova, V.; Mastnikova, K.; Vaclavikova, E.; Moravcova, J.; Vlcek, P.; Lancova, L.; Lastuvka, P.; Katra, R.; Bavor, P.; et al. RET Fusion Genes in Pediatric and Adult Thyroid Carcinomas: Cohort Characteristics and Prognosis. Endocr. Relat. Cancer 2023, 30, e230117. [Google Scholar] [CrossRef]
- Ou, S.H.I.; Zhu, V.W. Catalog of 5′ Fusion Partners in RET+ NSCLC Circa 2020. JTO Clin. Res. Rep. 2020, 1, 100037. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Chen, G.; He, X.; Shi, Y.; He, C.; Yin, J.C.; Zhang, Z. Characterization of RET Fusions via Integrated DNA and RNA Sequencing in Early-Stage Non-Small Cell Lung Cancer: A Retrospective Study. Transl. Lung Cancer Res. 2025, 14, 4384–4397. [Google Scholar] [CrossRef] [PubMed]
- Mizukami, T.; Shiraishi, K.; Shimada, Y.; Ogiwara, H.; Tsuta, K.; Ichikawa, H.; Sakamoto, H.; Kato, M.; Shibata, T.; Nakano, T.; et al. Molecular Mechanisms Underlying Oncogenic RET Fusion in Lung Adenocarcinoma. J. Thorac. Oncol. 2014, 9, 622–630. [Google Scholar] [CrossRef]
- Goytain, A.; Ng, T. NanoString NCounter Technology: High-Throughput RNA Validation. In Chimeric RNA: Methods and Protocols; Springer: New York, NY, USA, 2020; pp. 125–139. [Google Scholar]
- Reguart, N.; Teixidó, C.; Giménez-Capitán, A.; Paré, L.; Galván, P.; Viteri, S.; Rodríguez, S.; Peg, V.; Aldeguer, E.; Viñolas, N.; et al. Identification of ALK, ROS1, and RET Fusions by a Multiplexed MRNA-Based Assay in Formalin-Fixed, Paraffin-Embedded Samples from Advanced Non–Small-Cell Lung Cancer Patients. Clin. Chem. 2017, 63, 751–760. [Google Scholar] [CrossRef]
- Novaes, L.A.C.; da Silva, L.S.; de Marchi, P.; de Oliveira Cavagna, R.; de Paula, F.E.; Zanon, M.F.; Evangelista, A.F.; da Silva, E.C.A.; da Silva, V.D.; Leal, L.F.; et al. Simultaneous Analysis of ALK, RET, and ROS1 Gene Fusions by NanoString in Brazilian Lung Adenocarcinoma Patients. Transl. Lung Cancer Res. 2021, 10, 292–303. [Google Scholar] [CrossRef]
- Rabushko, E.; Sorokin, M.; Suntsova, M.; Seryakov, A.P.; Kuzmin, D.V.; Poddubskaya, E.; Buzdin, A.A. Experimentally Deduced Criteria for Detection of Clinically Relevant Fusion 3′ Oncogenes from FFPE Bulk RNA Sequencing Data. Biomedicines 2022, 10, 1866. [Google Scholar] [CrossRef]
- Belli, C.; Penault-Llorca, F.; Ladanyi, M.; Normanno, N.; Scoazec, J.-Y.; Lacroix, L.; Reis-Filho, J.S.; Subbiah, V.; Gainor, J.F.; Endris, V.; et al. ESMO Recommendations on the Standard Methods to Detect RET Fusions and Mutations in Daily Practice and Clinical Research. Ann. Oncol. 2021, 32, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-R.; Aypar, U.; Rosen, E.Y.; Mata, D.A.; Benayed, R.; Mullaney, K.; Jayakumaran, G.; Zhang, Y.; Frosina, D.; Drilon, A.; et al. A Performance Comparison of Commonly Used Assays to Detect RET Fusions. Clin. Cancer Res. 2021, 27, 1316–1328. [Google Scholar] [CrossRef] [PubMed]
- Yoh, K.; Seto, T.; Satouchi, M.; Nishio, M.; Yamamoto, N.; Murakami, H.; Nogami, N.; Matsumoto, S.; Kohno, T.; Tsuta, K.; et al. Vandetanib in Patients with Previously Treated RET-Rearranged Advanced Non-Small-Cell Lung Cancer (LURET): An Open-Label, Multicentre Phase 2 Trial. Lancet Respir. Med. 2017, 5, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Fu, S.; Patel, M.R.; Fakih, M.; Wang, D.; Olszanski, A.J.; Morgensztern, D.; Liu, S.V.; Cho, B.C.; Bazhenova, L.; et al. A Phase I/Ib Trial of the VEGFR-Sparing Multikinase RET Inhibitor RXDX-105. Cancer Discov. 2019, 9, 384–395. [Google Scholar] [CrossRef]
- Das, T.K.; Cagan, R.L. KIF5B-RET Oncoprotein Signals through a Multi-Kinase Signaling Hub. Cell Rep. 2017, 20, 2368–2383. [Google Scholar] [CrossRef]
- Mc Leer, A.; Mondet, J.; Magnat, N.; Mersch, M.; Giovannini, D.; Emprou, C.; Toffart, A.C.; Sturm, N.; Lantuéjoul, S.; Benito, D. Rearranged During Transfection Rearrangement Detection by Fluorescence In Situ Hybridization Compared With Other Techniques in NSCLC. JTO Clin. Res. Rep. 2024, 5, 100714. [Google Scholar] [CrossRef]
- Radonic, T.; Geurts-Giele, W.R.R.; Samsom, K.G.; Roemen, G.M.J.M.; von der Thüsen, J.H.; Thunnissen, E.; Meijssen, I.C.; Sleddens, H.F.B.M.; Dinjens, W.N.M.; Boelens, M.C.; et al. RET Fluorescence In Situ Hybridization Analysis Is a Sensitive but Highly Unspecific Screening Method for RET Fusions in Lung Cancer. J. Thorac. Oncol. 2021, 16, 798–806. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, M.; Lyadov, V.; Suntsova, M.; Garipov, M.; Semenova, A.; Popova, N.; Guguchkin, E.; Heydarov, R.; Zolotovskaia, M.; Zhao, X.; et al. Detection of Fusion Events by RNA Sequencing in FFPE versus Freshly Frozen Colorectal Cancer Tissue Samples. Front. Mol. Biosci. 2025, 11, 1448792. [Google Scholar] [CrossRef] [PubMed]
- Zakharova, G.; Suntsova, M.; Rabushko, E.; Mohammad, T.; Drobyshev, A.; Seryakov, A.; Poddubskaya, E.; Moisseev, A.; Smirnova, A.; Sorokin, M.; et al. A New Approach of Detecting ALK Fusion Oncogenes by RNA Sequencing Exon Coverage Analysis. Cancers 2024, 16, 3851. [Google Scholar] [CrossRef]
- Zheng, Z.; Liebers, M.; Zhelyazkova, B.; Cao, Y.; Panditi, D.; Lynch, K.D.; Chen, J.; Robinson, H.E.; Shim, H.S.; Chmielecki, J.; et al. Anchored Multiplex PCR for Targeted Next-Generation Sequencing. Nat. Med. 2014, 20, 1479–1484. [Google Scholar] [CrossRef]
- Beg, S.; Bareja, R.; Ohara, K.; Eng, K.W.; Wilkes, D.C.; Pisapia, D.J.; Al Zoughbi, W.; Kudman, S.; Zhang, W.; Rao, R.; et al. Integration of Whole-Exome and Anchored PCR-Based next Generation Sequencing Significantly Increases Detection of Actionable Alterations in Precision Oncology. Transl. Oncol. 2021, 14, 100944. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, T.L.; Dailey, S.C.; Ottestad, A.L.; Yu, J.; Becker, P.W.; Scaife, S.; Richman, S.D.; Wood, H.M.; Slaney, H.; Bottomley, D.; et al. Adaptor Template Oligo-Mediated Sequencing (ATOM-Seq) Is a New Ultra-Sensitive UMI-Based NGS Library Preparation Technology for Use with CfDNA and CfRNA. Sci. Rep. 2021, 11, 3138. [Google Scholar] [CrossRef]
- Leone, A.; Muscarella, L.A.; Graziano, P.; Tornese, A.; Grillo, L.R.; Di Lorenzo, A.; Bronzini, M.; Scarpino, S.; Sparaneo, A.; Rossi, G. Robust Performance of the Novel Research-Use-Only Idylla GeneFusion Assay Using a Diverse Set of Pathological Samples with a Proposed 1-Day Workflow for Advanced NSCLC Evaluation. Cancers 2022, 15, 292. [Google Scholar] [CrossRef]
- Tong, Y.; Zhao, Z.; Liu, B.; Bao, A.; Zheng, H.; Gu, J.; McGrath, M.; Xia, Y.; Tan, B.; Song, C.; et al. 5′/3′ Imbalance Strategy to Detect ALK Fusion Genes in Circulating Tumor RNA from Patients with Non-Small Cell Lung Cancer. J. Exp. Clin. Cancer Res. 2018, 37, 68. [Google Scholar] [CrossRef]
- Vaughn, C.P.; Costa, J.L.; Feilotter, H.E.; Petraroli, R.; Bagai, V.; Rachiglio, A.M.; Marino, F.Z.; Tops, B.; Kurth, H.M.; Sakai, K.; et al. Simultaneous Detection of Lung Fusions Using a Multiplex RT-PCR next Generation Sequencing-Based Approach: A Multi-Institutional Research Study. BMC Cancer 2018, 18, 828. [Google Scholar] [CrossRef]
- Rogers, T.M.; Arnau, G.M.; Ryland, G.L.; Huang, S.; Lira, M.E.; Emmanuel, Y.; Perez, O.D.; Irwin, D.; Fellowes, A.P.; Wong, S.Q.; et al. Multiplexed Transcriptome Analysis to Detect ALK, ROS1 and RET Rearrangements in Lung Cancer. Sci. Rep. 2017, 7, 42259. [Google Scholar] [CrossRef]
- Goytain, A.; Chang, K.T.E.; Goh, J.Y.; Nielsen, T.O.; Ng, T.L. Diagnosis of Fusion-Associated Sarcomas by Exon Expression Imbalance and Gene Expression. J. Mol. Diagn. 2023, 25, 121–131. [Google Scholar] [CrossRef]
- Sorokin, M.; Garazha, A.; Suntsova, M.; Tkachev, V.; Poddubskaya, E.; Gaifullin, N.; Sushinskaya, T.; Lantsov, D.; Borisov, V.; Naskhletashvili, D.; et al. Prospective Trial of the Oncobox Platform RNA Sequencing Bioinformatic Analysis for Personalized Prescription of Targeted Drugs. Comput. Biol. Med. 2025, 187, 109716. [Google Scholar] [CrossRef] [PubMed]
- Samii, A.; Sorokin, M.; Kar, S.; Makovskaia, L.; Garazha, A.; Hartmann, C.; Moisseev, A.; Kim, E.; Giese, A.; Buzdin, A. Case of Multifocal Glioblastoma with Four Fusion Transcripts of ALK, FGFR2, NTRK2, and NTRK3 Genes Stresses the Need for Tumor Tissue Multisampling for Transcriptomic Analysis. Mol. Case Stud. 2021, 7, a006100. [Google Scholar] [CrossRef] [PubMed]
- Lira, M.E.; Choi, Y.L.; Lim, S.M.; Deng, S.; Huang, D.; Ozeck, M.; Han, J.; Jeong, J.Y.; Shim, H.S.; Cho, B.C.; et al. A Single-Tube Multiplexed Assay for Detecting ALK, ROS1, and RET Fusions in Lung Cancer. J. Mol. Diagn. 2014, 16, 229–243. [Google Scholar] [CrossRef]
- Hofman, V.; Heeke, S.; Bontoux, C.; Chalabreysse, L.; Barritault, M.; Bringuier, P.P.; Fenouil, T.; Benzerdjeb, N.; Begueret, H.; Merlio, J.P.; et al. Ultrafast Gene Fusion Assessment for Nonsquamous NSCLC. JTO Clin. Res. Rep. 2023, 4, 100457. [Google Scholar] [CrossRef] [PubMed]
- Heydt, C.; Wölwer, C.B.; Velazquez Camacho, O.; Wagener-Ryczek, S.; Pappesch, R.; Siemanowski, J.; Rehker, J.; Haller, F.; Agaimy, A.; Worm, K.; et al. Detection of Gene Fusions Using Targeted Next-Generation Sequencing: A Comparative Evaluation. BMC Med. Genom. 2021, 14, 62. [Google Scholar] [CrossRef]
- Sondka, Z.; Dhir, N.B.; Carvalho-Silva, D.; Jupe, S.; Madhumita; McLaren, K.; Starkey, M.; Ward, S.; Wilding, J.; Ahmed, M.; et al. COSMIC: A Curated Database of Somatic Variants and Clinical Data for Cancer. Nucleic Acids Res. 2024, 52, D1210–D1217. [Google Scholar] [CrossRef]
- Kim, P.; Zhou, X. FusionGDB: Fusion Gene Annotation DataBase. Nucleic Acids Res. 2019, 47, D994–D1004. [Google Scholar] [CrossRef]
- Haas, B.J.; Dobin, A.; Li, B.; Stransky, N.; Pochet, N.; Regev, A. Accuracy Assessment of Fusion Transcript Detection via Read-Mapping and de Novo Fusion Transcript Assembly-Based Methods. Genome Biol. 2019, 20, 213. [Google Scholar] [CrossRef]
- Gandhi, M.M.; Ricciuti, B.; Harada, G.; Repetto, M.; Gildenberg, M.S.; Singh, A.; Li, Y.Y.; Gagné, A.; Wang, X.; Aizer, A.; et al. Amplification of Wild-Type RET Represents a Novel Molecular Subtype of Several Cancer Types With Clinical Response to Selpercatinib. JCO Precis. Oncol. 2023, 7, e2300295. [Google Scholar] [CrossRef]
- Zollinger, A.J.; Smith, M.L. Fibronectin, the Extracellular Glue. Matrix Biol. 2017, 60–61, 27–37. [Google Scholar] [CrossRef]
- Char, R.; Pierre, P. The RUFYs, a Family of Effector Proteins Involved in Intracellular Trafficking and Cytoskeleton Dynamics. Front. Cell Dev. Biol. 2020, 8, 779. [Google Scholar] [CrossRef]
- Char, R.; Liu, Z.; Jacqueline, C.; Davieau, M.; Delgado, M.-G.; Soufflet, C.; Fallet, M.; Chasson, L.; Chapuy, R.; Camosseto, V.; et al. RUFY3 Regulates Endolysosomes Perinuclear Positioning, Antigen Presentation and Migration in Activated Phagocytes. Nat. Commun. 2023, 14, 4290. [Google Scholar] [CrossRef] [PubMed]
- Hendrickx, A.; Beullens, M.; Ceulemans, H.; Den Abt, T.; Van Eynde, A.; Nicolaescu, E.; Lesage, B.; Bollen, M. Docking Motif-Guided Mapping of the Interactome of Protein Phosphatase-1. Chem. Biol. 2009, 16, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Amary, F.; Perez-Casanova, L.; Ye, H.; Cottone, L.; Strobl, A.-C.; Cool, P.; Miranda, E.; Berisha, F.; Aston, W.; Rocha, M.; et al. Synovial Chondromatosis and Soft Tissue Chondroma: Extraosseous Cartilaginous Tumor Defined by FN1 Gene Rearrangement. Mod. Pathol. 2019, 32, 1762–1771. [Google Scholar] [CrossRef] [PubMed]
- Kallen, M.E.; Michal, M.; Meyer, A.; Suster, D.I.; Olson, N.J.; Charville, G.W.; Perret, R.; Gross, J.M. Calcified Chondroid Mesenchymal Neoplasm. Am. J. Surg. Pathol. 2023, 47, 725–737. [Google Scholar] [CrossRef]
- Liu, Y.J.; Wang, W.; Yeh, J.; Wu, Y.; Mantilla, J.G.; Fletcher, C.D.M.; Ricciotti, R.W.; Chen, E.Y. Calcified Chondroid Mesenchymal Neoplasms with FN1-Receptor Tyrosine Kinase Gene Fusions Including FGFR2, FGFR1, MERTK, NTRK1, and TEK: A Molecular and Clinicopathologic Analysis. Mod. Pathol. 2021, 34, 1373–1383. [Google Scholar] [CrossRef]
- Bertz, S.; Stöhr, R.; Gaisa, N.T.; Wullich, B.; Hartmann, A.; Agaimy, A. TERT Promoter Mutation Analysis as a Surrogate to Morphology and Immunohistochemistry in Problematic Spindle Cell Lesions of the Urinary Bladder. Histopathology 2020, 77, 949–962. [Google Scholar] [CrossRef]
- Tyurin, V.I.; Preobrazhenskaya, E.V.; Mityushkina, N.V.; Romanko, A.A.; Anuskina, A.A.; Mulkidjan, R.S.; Saitova, E.S.; Shevyakov, M.P.; Aleksakhina, S.N.; Venina, A.R.; et al. Prevalence of RET Translocations in Non-Small Cell Lung Cancer. In Proceedings of the IX St. Petersburg International Oncology Forum “White Nights 2023”, St. Petersburg, Russia, 3–8 July 2023; pp. 343–344. (In Russian). [Google Scholar]
- Zhang, W.; Lin, S.; Wang, Z.; Zhang, W.; Xing, M. Coexisting RET/PTC and TERT Promoter Mutation Predict Poor Prognosis but Effective RET and MEK Targeting in Thyroid Cancer. J. Clin. Endocrinol. Metab. 2024, 109, 3166–3175. [Google Scholar] [CrossRef]
- Buzdin, A.; Sorokin, M.; Garazha, A.; Glusker, A.; Aleshin, A.; Poddubskaya, E.; Sekacheva, M.; Kim, E.; Gaifullin, N.; Giese, A.; et al. RNA Sequencing for Research and Diagnostics in Clinical Oncology. Semin. Cancer Biol. 2020, 60, 311–323. [Google Scholar] [CrossRef]
- Di Grazia, G.; Conti, C.; Nucera, S.; Motta, G.; Martorana, F.; Stella, S.; Massimino, M.; Giuliano, M.; Vigneri, P. REThinking the Role of the RET Oncogene in Breast Cancer. Front. Oncol. 2024, 14, 1427228. [Google Scholar] [CrossRef] [PubMed]
- Jagust, P.; Powell, A.M.; Ola, M.; Watson, L.; de Pablos-Aragoneses, A.; García- Gómez, P.; Fallon, R.; Bane, F.; Heiland, M.; Morris, G.; et al. RET Overexpression Leads to Increased Brain Metastatic Competency in Luminal Breast Cancer. JNCI J. Natl. Cancer Inst. 2024, 116, 1632–1644. [Google Scholar] [CrossRef] [PubMed]
- Spanheimer, P.M.; Lorenzen, A.W.; De Andrade, J.P.; Kulak, M.V.; Carr, J.C.; Woodfield, G.W.; Sugg, S.L.; Weigel, R.J. Receptor Tyrosine Kinase Expression Predicts Response to Sunitinib in Breast Cancer. Ann. Surg. Oncol. 2015, 22, 4287–4294. [Google Scholar] [CrossRef] [PubMed]
- Suntsova, M.; Gaifullin, N.; Allina, D.; Reshetun, A.; Li, X.; Mendeleeva, L.; Surin, V.; Sergeeva, A.; Spirin, P.; Prassolov, V.; et al. Atlas of RNA Sequencing Profiles for Normal Human Tissues. Sci. Data 2019, 6, 36. [Google Scholar] [CrossRef]
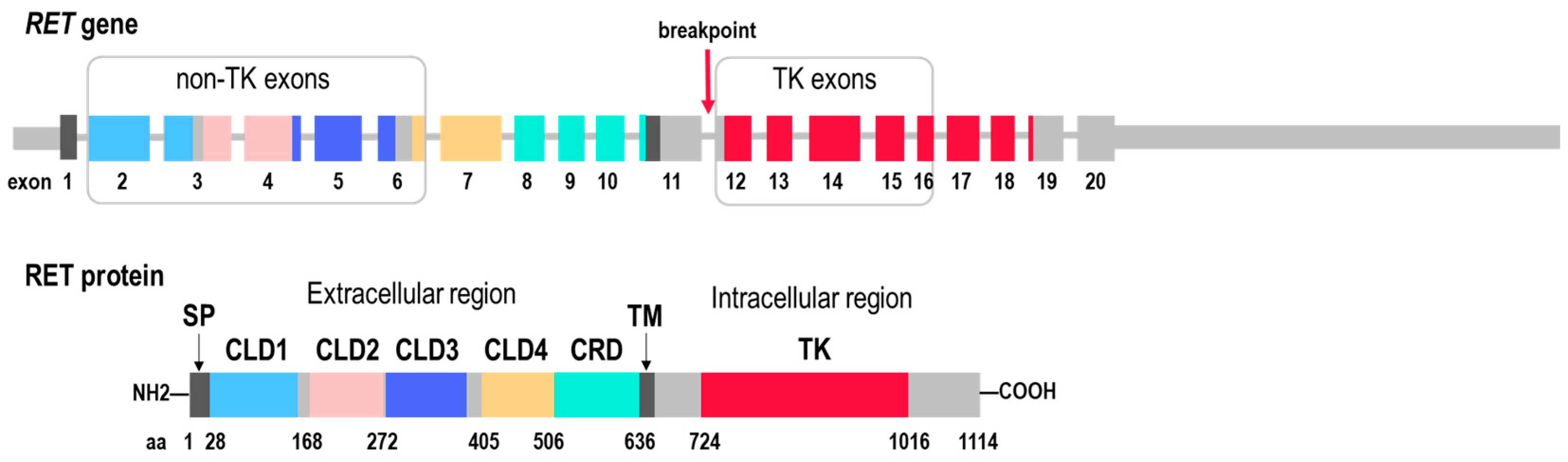

| Tumor Type 1 | Total Cases Sequenced, n (%) | Gender, Male/Female | Age of Onset | ||
|---|---|---|---|---|---|
| Mean | Median | Range | |||
| TC | 221 (16.7%) | 21.3%/74.7% 2 | 42.7 | 44.5 | 6–81 |
| CRC | 164 (12.4%) | 43.9%/56.1% | 59.0 | 59 | 32–87 |
| NSCLC | 154 (11.6%) | 68.2%/31.8% | 60.7 | 62.5 | 29–81 |
| BC | 147 (11.1%) | 0%/100% | 53.0 | 54 | 27–89 |
| CNS | 82 (6.2%) | 57.3%/36.6% 2 | 39.8 | 40 | 3–70 |
| OC | 80 (6.0%) | 0%/100% | 52.3 | 52 | 27–80 |
| SC | 67 (5.0%) | 55.2%/44.8% | 57.4 | 59 | 29–81 |
| PC | 61 (4.6%) | 55.7%/44.3% | 60.2 | 62 | 31–79 |
| MM | 59 (4.2%) | 55.9%/42.4% 2 | 58.4 | 59 | 29–78 |
| AL | 50 (3.8%) | 54.0%/46.0% | 5.8 | 5 | 1–15 |
| KC | 37 (2.6%) | 67.6%/32.4% | 56.0 | 57 | 40–72 |
| Other | 211 (15.9%) | 37.9%/60.7% 2 | 51.6 | 52 | 14–83 |
| Tumor Type | Number of Samples | Number of Samples Tested with | ||
|---|---|---|---|---|
| TruSight NGS | OncoFu NGS | Sanger | ||
| Breast cancer | 4 | 4 | 4 | 0 |
| Cervical cancer | 1 | 1 | 1 | 0 |
| Cholangiocarcinoma | 2 | 0 | 2 | 0 |
| FGF23-producing adenoma | 1 | 0 | 0 | 1 |
| Fibrosarcoma | 1 | 0 | 0 | 1 |
| Glioblastoma, IDH wild type | 2 | 1 | 2 | 0 |
| Hemangioendothelioma | 2 | 0 | 2 | 0 |
| Leukemia | 5 | 5 | 5 | 0 |
| Liposarcoma | 1 | 1 | 1 | 0 |
| Lung cancer | 24 | 22 | 23 | 0 |
| Melanoma | 1 | 1 | 1 | 0 |
| Mesothelioma | 1 | 1 | 1 | 0 |
| Mucoepidermoid carcinoma | 1 | 0 | 1 | 0 |
| Ovarian cancer | 4 | 3 | 4 | 0 |
| Pancreatic cancer | 3 | 3 | 3 | 0 |
| Papillary thyroid cancer | 19 | 2 | 12 | 8 |
| Parathyroid cancer | 1 | 0 | 1 | 0 |
| Prostate cancer | 1 | 0 | 1 | 0 |
| Rectal cancer | 3 | 2 | 3 | 0 |
| Salivary gland cancer | 1 | 1 | 1 | 0 |
| Overall | 78 | 47 | 68 | 10 |
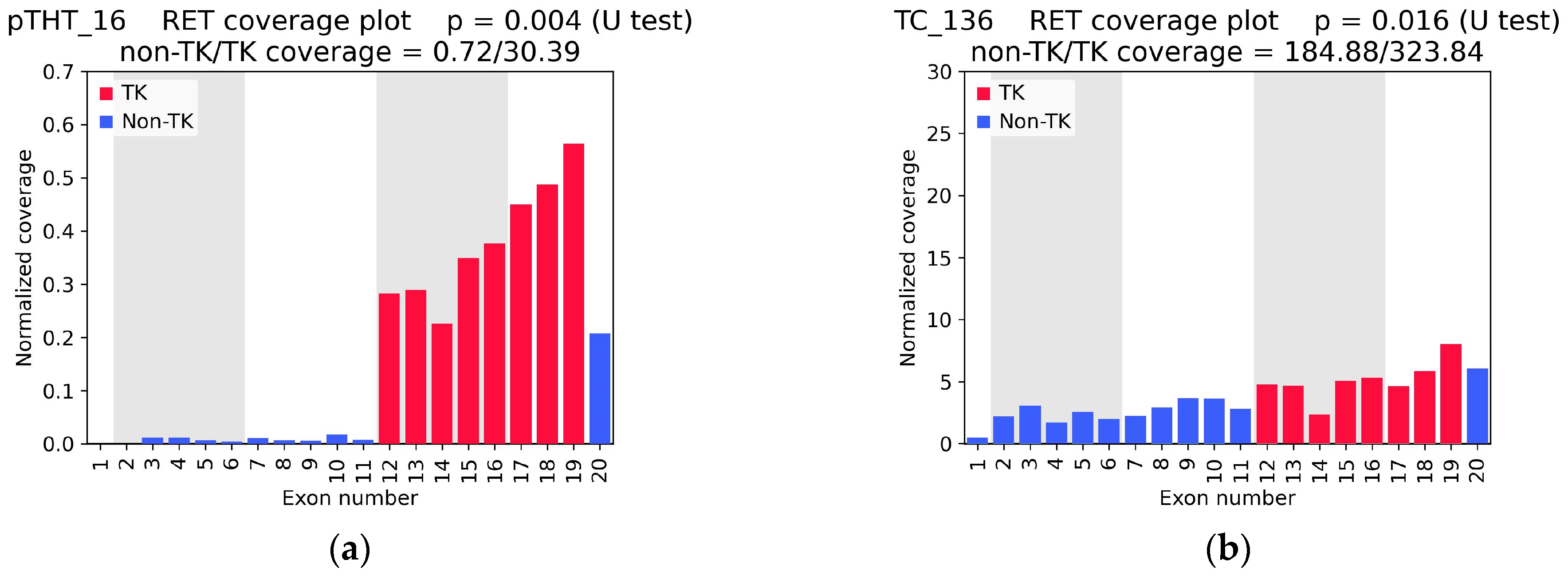
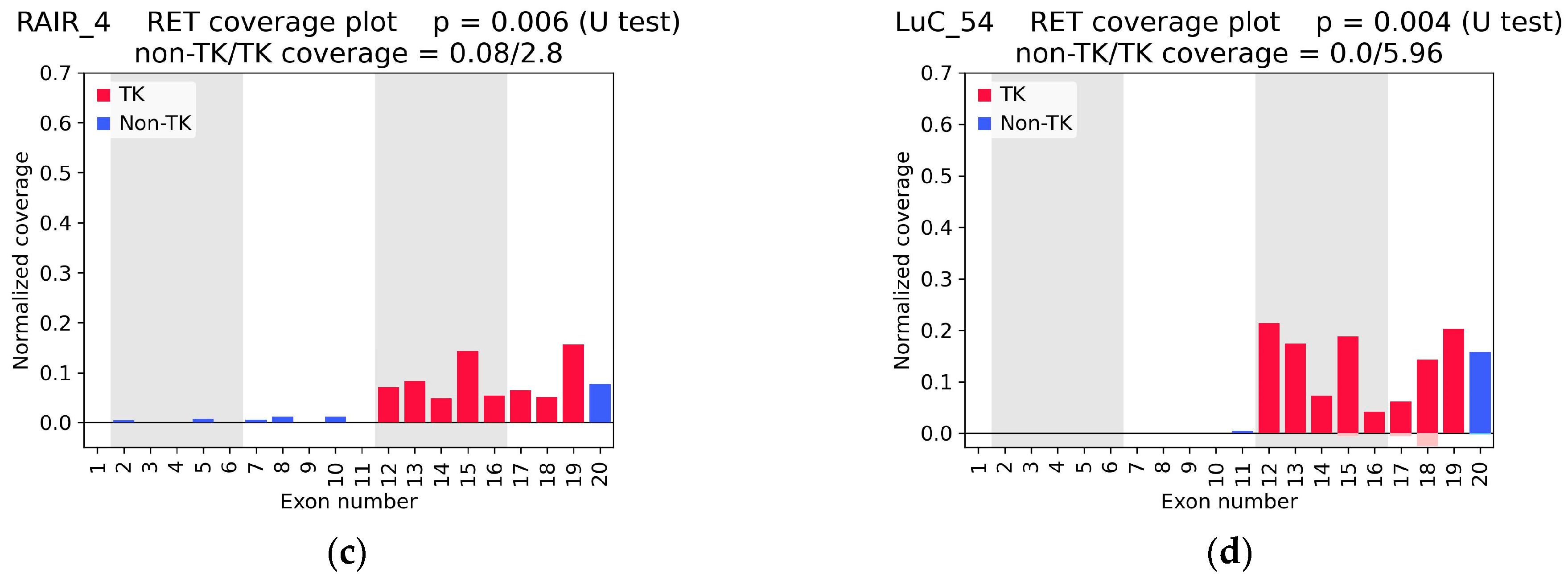
| Sample ID | RNA-Seq Coverage Characteristics | Found RET Fusion (Number of Junction and Spanning Reads) 2 | Confirmed by Sanger Sequencing | ||||
|---|---|---|---|---|---|---|---|
| 5′/3′ Asymmetry p-Value | TK Coverage Depth | Non-TK/TK 1 Coverage Ratio | RNA-Seq | TruSight | OncoFu | ||
| CC_162 | 0.006 | 8.11 | 0.03 | NCOA4(9)::RET(12) (6 + 11) 2 | - | NCOA4(9)::RET(12) (329 + 423) | - |
| FGF_7 | 0.006 | 1 487.74 | 0.00 | FN1(20)::RET(11) (937 + 267) | - | - | yes |
| FS_1 | 0.004 | 8.90 | 0.06 | CCDC6(1)::RET(12) (3 + 0) | - | - | yes |
| LuC_100 | 0.006 | 6.85 | 0.03 | KIF5B(16)::RET(12) (5 + 0) | KIF5B(16)::RET(12) (10 + 0) | KIF5B(16)::RET(12) (93 + 59) | - |
| pTHT_15 | 0.0056 | 29.42 | 0.01 | CCDC6(1)::RET(12) (18 + 4) | - | - | yes |
| pTHT_16 | 0.004 | 30.39 | 0.02 | CCDC6(1)::RET(12) (31 + 5) | - | - | yes |
| pTHT_21 | 0.004 | 12.44 | 0.07 | NCOA4(7)::RET(12) (16 + 1) | - | - | yes |
| pTHT_22 | 0.004 | 18.87 | 0.02 | RUFY3(11)::RET(12) (13 + 2) | - | - | yes |
| pThT_24 | 0.004 | 9.35 | 0.27 | NCOA4(7)::RET(12) (6 + 0) | - | NCOA4(7)::RET(12) (52 + 45) | yes |
| pTHT_25 | 0.004 | 15.83 | 0.04 | CCDC6(1)::RET(12) (10 + 1) | - | - | yes |
| pTHT_26 | 0.004 | 20.39 | 0.03 | TRIM27(3)::RET(12) (15 + 5) | - | - | yes |
| RAIR_4 | 0.0056 | 2.80 | 0.03 | NCOA4(7)::RET(12) (1 + 0) | - | - | yes |
| LuC_54 | 0.0037 | 5.96 | 0.00 | no fusion | KIF5B(15)::RET(12) (68 + 10) | KIF5B(15)::RET(12) (12 + 4) | - |
| BC_100 | 0.048 | 6.94 | 0.79 | no fusion | no fusion | no fusion | - |
| OC_49 | 0.048 | 1.97 | 0.69 | no fusion | no fusion | no fusion | - |
| OC_80 | 0.028 | 1.24 | 0.22 | no fusion | - | no fusion | - |
| pTHT_4 | 0.016 | 7.26 | 0.63 | no fusion | - | no fusion | - |
| pTHT_5 | 0.004 | 3.38 | 0.63 | no fusion | - | no fusion | - |
| pTHT_6 | 0.048 | 3.19 | 0.60 | no fusion | - | no fusion | - |
| pTHT_19 | 0.0079 | 9.82 | 0.48 | no fusion | - | no fusion | - |
| pTHT_32 | 0.016 | 6.77 | 0.57 | no fusion | - | no fusion | - |
| TC_136 | 0.016 | 323.84 | 0.57 | no fusion | - | no fusion | - |
| SgC_2 | 0.048 | 1.04 | 0.35 | no fusion | no fusion | no fusion | - |
| Target Fusion | Forward Primer, 5′–3′ | Reverse Primer, 5′–3′ |
|---|---|---|
| CCDC6(1)::RET(12) | TGGAGACCTACAAACTGAAGTG | CAAGAACCAAGTTCTTCCGAG |
| FN1(20)::RET(11) | CCCAAAGCCACTGGAGTC | GACAGCAGCACCGAGAC |
| NCOA4(7)::RET(12) | ACCTGCCAGTGGTTATCAAG | CAAGAACCAAGTTCTTCCGAG |
| PPP1R21(15)::RET(12) | GTGGATTCATTAGTCCTCTTTCAG | CAAGAACCAAGTTCTTCCGAG |
| RUFY3(11)::RET(12) | GCAGGATGCCCTGGTATC | CAAGAACCAAGTTCTTCCGAG |
| TRIM27(3)::RET(12) | CATCTCCCACCTCAGCAG | CAAGAACCAAGTTCTTCCGAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaziev, I.; Khristichenko, A.; Luppov, D.; Suntsova, M.; Bondarenko, E.; Reinberg, M.; Matrosova, A.; Khilal, N.; Sorokin, M.; Sekacheva, M.; et al. Accurate RET Fusion Detection in Solid Tumors Using RNA Sequencing Coverage Imbalance Analysis. Int. J. Mol. Sci. 2025, 26, 11300. https://doi.org/10.3390/ijms262311300
Gaziev I, Khristichenko A, Luppov D, Suntsova M, Bondarenko E, Reinberg M, Matrosova A, Khilal N, Sorokin M, Sekacheva M, et al. Accurate RET Fusion Detection in Solid Tumors Using RNA Sequencing Coverage Imbalance Analysis. International Journal of Molecular Sciences. 2025; 26(23):11300. https://doi.org/10.3390/ijms262311300
Chicago/Turabian StyleGaziev, Ivan, Anna Khristichenko, Daniil Luppov, Maria Suntsova, Ekaterina Bondarenko, Maria Reinberg, Alina Matrosova, Nadezhda Khilal, Maksim Sorokin, Marina Sekacheva, and et al. 2025. "Accurate RET Fusion Detection in Solid Tumors Using RNA Sequencing Coverage Imbalance Analysis" International Journal of Molecular Sciences 26, no. 23: 11300. https://doi.org/10.3390/ijms262311300
APA StyleGaziev, I., Khristichenko, A., Luppov, D., Suntsova, M., Bondarenko, E., Reinberg, M., Matrosova, A., Khilal, N., Sorokin, M., Sekacheva, M., Poddubskaya, E., Buzdin, A., & Zakharova, G. (2025). Accurate RET Fusion Detection in Solid Tumors Using RNA Sequencing Coverage Imbalance Analysis. International Journal of Molecular Sciences, 26(23), 11300. https://doi.org/10.3390/ijms262311300







