During the Formation of Vasculogenic Mimicry by Melanoma Cells, the Silencing of Two Sets of Developmental Genes Is Coupled Either with an Increase or a Decrease in Contacts with the Nucleoli
Abstract
1. Introduction
2. Results
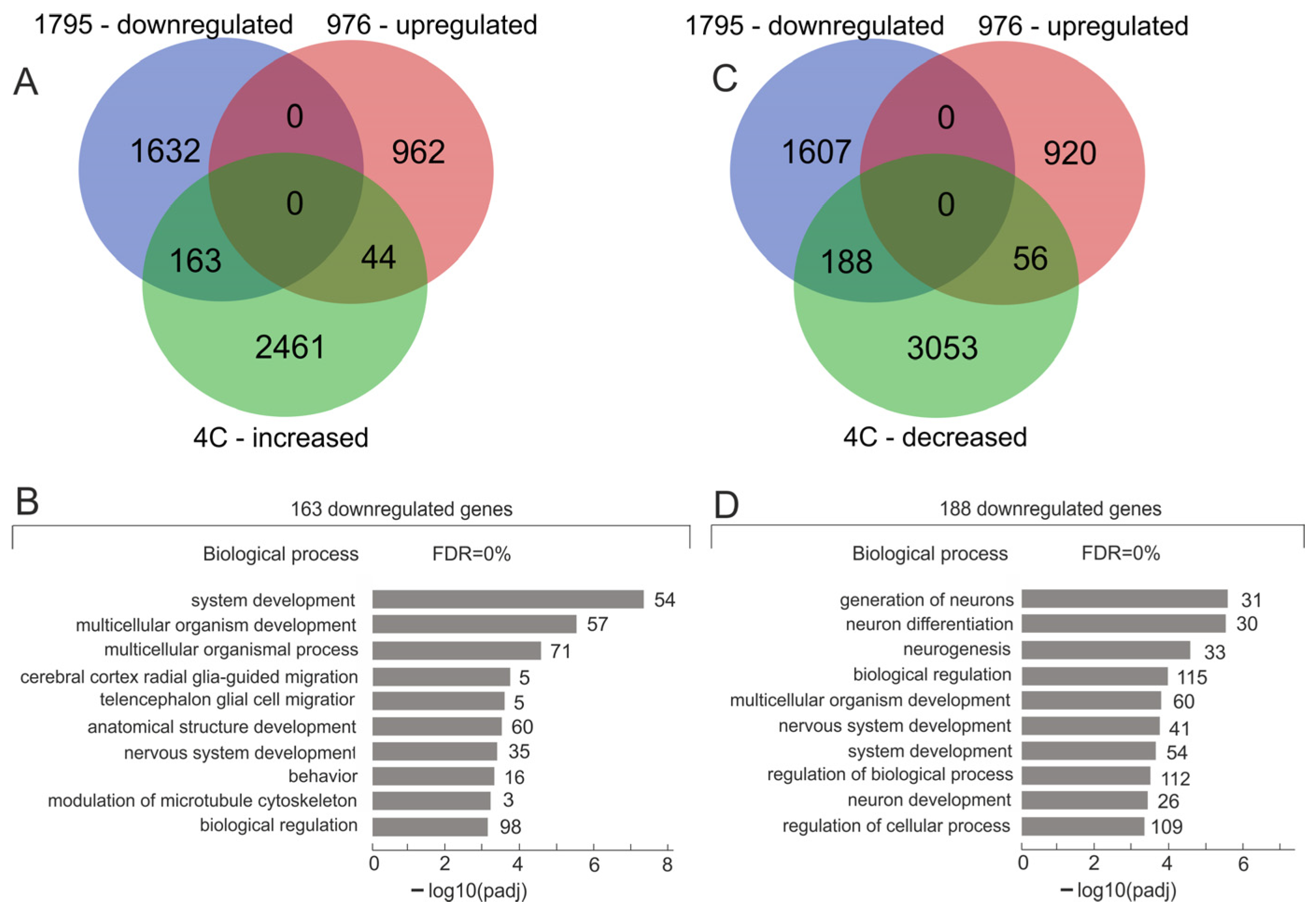
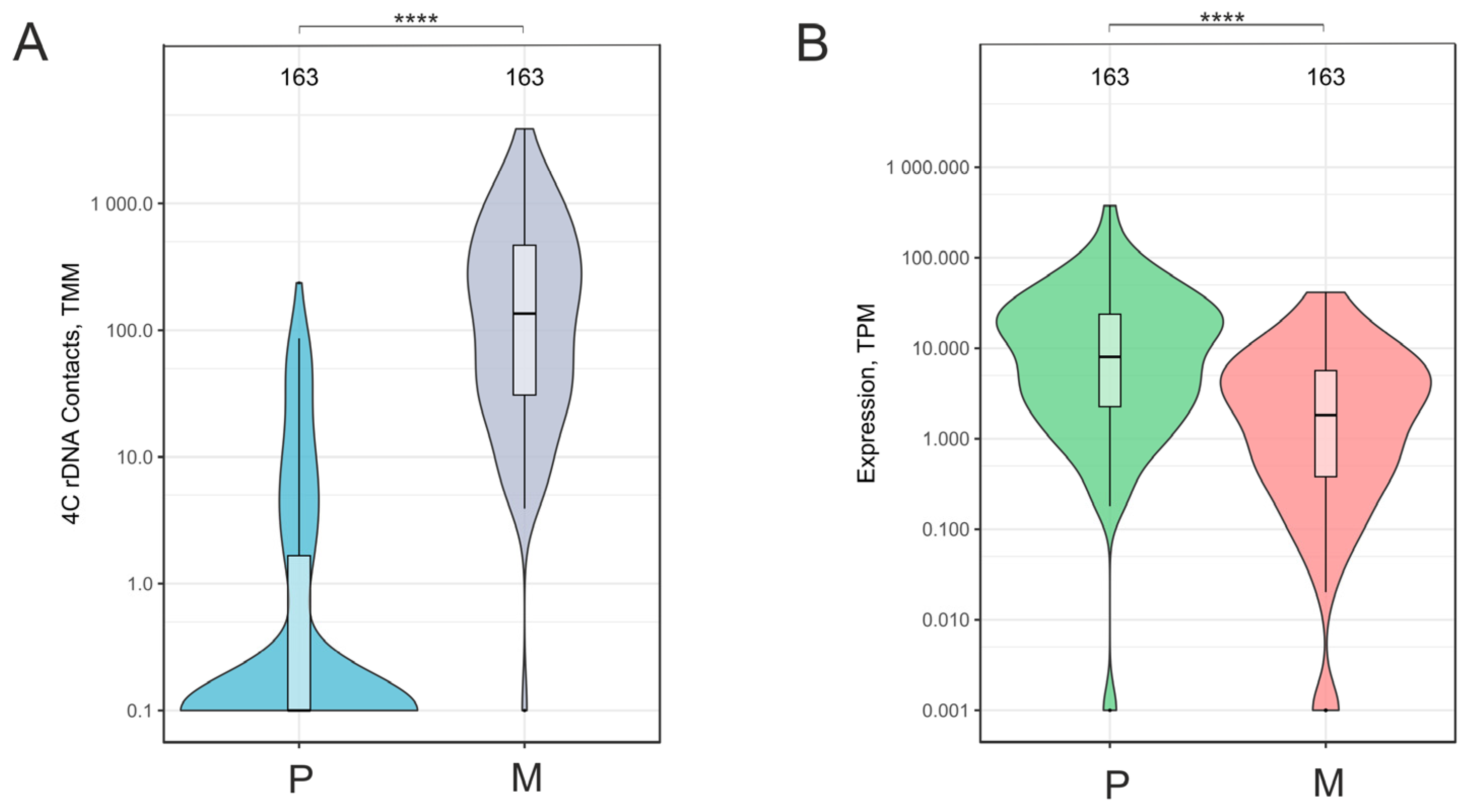
2.1. A Total of 163 Downregulated Genes Had an Increased Number of Contacts with Nucleoli During the Formation of the VM Phenotype
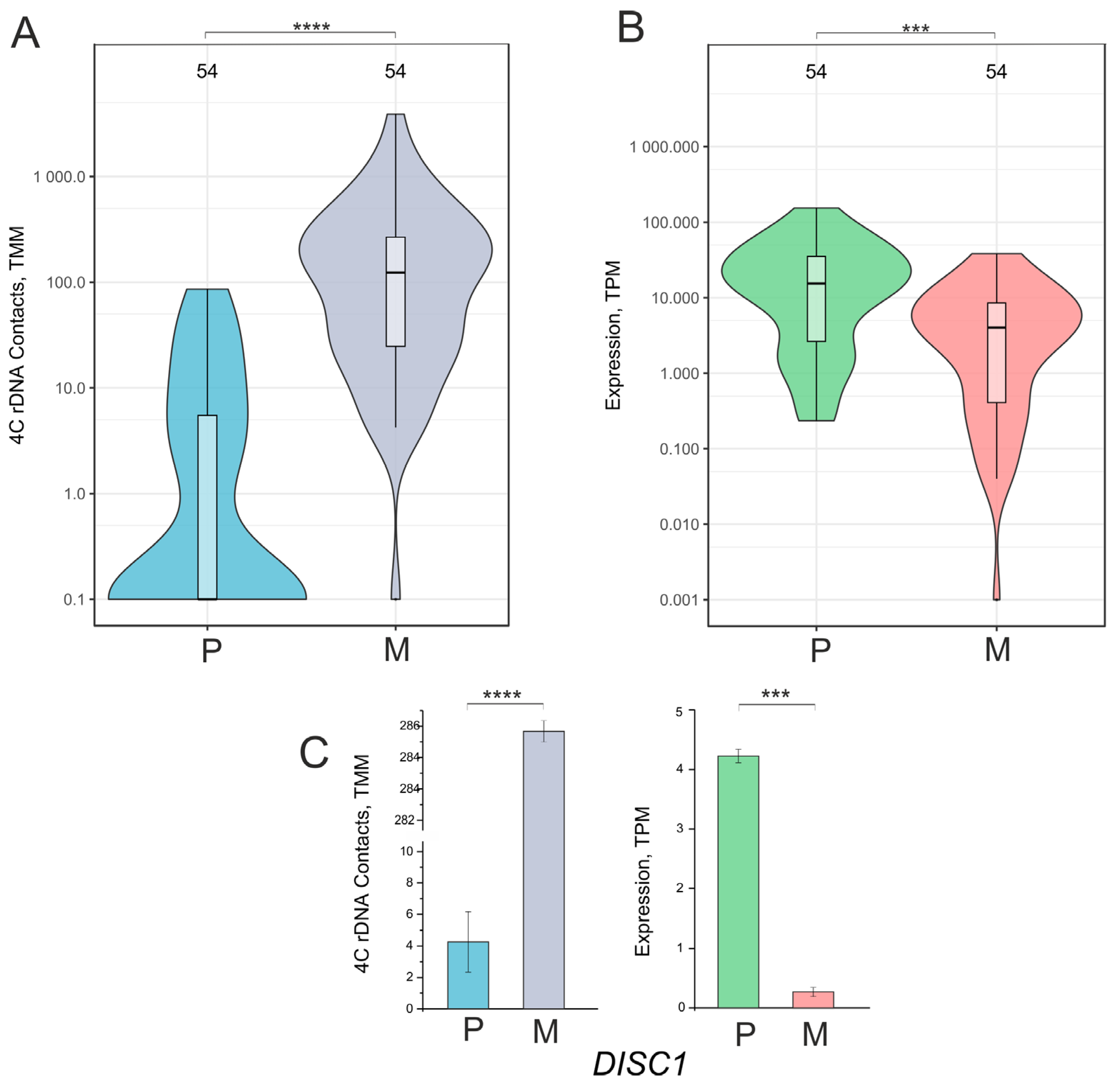
2.2. Downregulated Genes Controlling System Development Had a Decreased Number of Contacts with Nucleoli During the Formation of the VM Phenotype
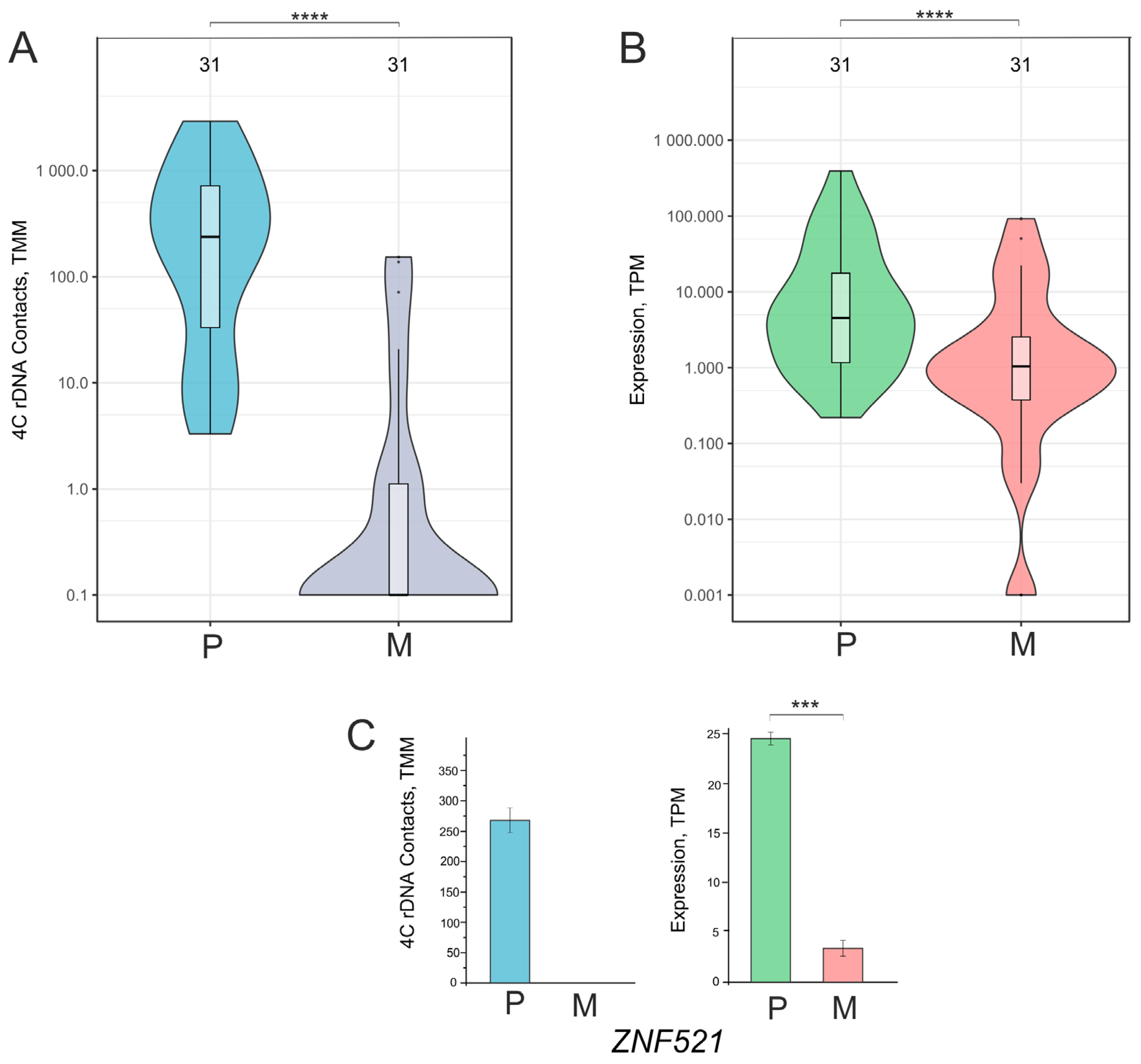
2.3. A Total of 188 Downregulated Genes Controlling the Generation of Neurons Had a Decreased Number of Contacts with Nucleoli During the Formation of VM Phenotype
2.4. Downregulated Genes Controlling the Generation of Neurons Had a Decreased Number of Contacts with Nucleoli During the Formation of the VM Phenotype
2.5. Concerted Downregulation of Two Big Groups of Developmental Genes Was Performed Simultaneously Using Different Mechanisms and Resulted in the Formation of Less Differentiated Cancer Cells
3. Discussion
3.1. Matrigel Induces an Epigenetic Switch in Mel Z Cells
3.2. How Could Nucleoli Downregulate or Upregulate Developmental Genes?
3.3. How Could Dynamic Inter-Chromosomal Contacts with Nucleoli Be Formed?
4. Materials and Methods
4.1. Mel Z Cell Culture
4.2. 4C-rDNA Procedure
4.3. 4C Mapping and Processing
4.4. Violin Plots for 4C-rDNA Data
4.5. Violin Plots for Gene Expression DATA
4.6. Code Accessibility
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Latonen, L. Phase-to-Phase with Nucleoli—Stress Responses, Protein Aggregation and Novel Roles of RNA. Front. Cell. Neurosci. 2019, 13, 151. [Google Scholar] [CrossRef]
- Gupta, S.; Santoro, R. Regulation and Roles of the Nucleolus in Embryonic Stem Cells: From Ribosome Biogenesis to Genome Organization. Stem Cell Rep. 2020, 15, 1206–1219. [Google Scholar] [CrossRef] [PubMed]
- Tchurikov, N.A.; Kravatsky, Y.V. The Role of rDNA Clusters in Global Epigenetic Gene Regulation. Front. Genet. 2021, 12, 730633. [Google Scholar] [CrossRef]
- Tchurikov, N.A.; Fedoseeva, D.M.; Sosin, D.V.; Snezhkina, A.V.; Melnikova, N.V.; Kudryavtseva, A.V.; Kravatsky, Y.V.; Kretova, O.V. Hot spots of DNA double-strand breaks and genomic contacts of human rDNA units are involved in epigenetic regulation. J. Mol. Cell Biol. 2015, 7, 366–382. [Google Scholar] [CrossRef]
- Diesch, J.; Bywater, M.J.; Sanij, E.; Cameron, D.P.; Schierding, W.; Brajanovski, N.; Son, J.; Sornkom, J.; Hein, N.; Evers, M.; et al. Changes in long-range rDNA-genomic interactions associate with altered RNA polymerase II. gene programs during malignant transformation. Commun. Biol. 2019, 2, 39. [Google Scholar] [CrossRef]
- Tchurikov, N.A.; Fedoseeva, D.M.; Klushevskaya, E.S.; Slovohotov, I.Y.; Chechetkin, V.R.; Kravatsky, Y.V.; Kretova, O.V. rDNA Clusters Make Contact with Genes that Are Involved in Differentiation and Cancer and Change Contacts after Heat Shock Treatment. Cells 2019, 8, 1393. [Google Scholar] [CrossRef]
- Tchurikov, N.A.; Klushevskaya, E.S.; Alembekov, I.R.; Kretova, A.N.; Chechetkin, V.R.; Kravatskaya, G.I.; Kravatsky, Y.V. Induction of the Erythroid Differentiation of K562 Cells Is Coupled with Changes in the Inter-Chromosomal Contacts of rDNA Clusters. Int. J. Mol. Sci. 2023, 24, 9842. [Google Scholar] [CrossRef]
- Yu, S.; Lemos, B. The long-range interaction map of ribosomal DNA arrays. PLoS Genet. 2018, 14, e1007258. [Google Scholar] [CrossRef]
- Tchurikov, N.A.; Klushevskaya, E.S.; Fedoseeva, D.M.; Alembekov, I.R.; Kravatskaya, G.I.; Chechetkin, V.R.; Kravatsky, Y.V.; Kretova, O.V. Dynamics of Whole-Genome Contacts of Nucleoli in Drosophila Cells Suggests a Role for rDNA Genes in Global Epigenetic Regulation. Cells 2020, 9, 2587. [Google Scholar] [CrossRef]
- Tchurikov, N.A.; Vartanian, A.A.; Klushevskaya, E.S.; Alembekov, I.R.; Kretova, A.N.; Lukicheva, V.N.; Chechetkin, V.R.; Kravatskaya, G.I.; Kosorukov, V.S.; Kravatsky, Y.V. Strong Activation of ID1, ID2, and ID3 Genes Is Coupled with the Formation of Vasculogenic Mimicry Phenotype in Melanoma Cells. Int. J. Mol. Sci. 2024, 25, 9291. [Google Scholar] [CrossRef] [PubMed]
- Tchurikov, N.A.; Klushevskaya, E.S.; Lukicheva, V.N.; Kretova, A.N.; Poperekova, E.N.; Chechetkin, V.R.; Kravatskaya, G.I.; Vartanian, A.A.; Kosorukov, V.S.; Alembekov, I.R.; et al. Formation of the Vasculogenic Mimicry Phenotype in Melanoma Mel Z Cells Is Coupled with Changes in Inter-Chromosomal Contacts of Developmental Genes with rDNA Clusters. Int. J. Mol. Sci. 2025, 26, 8085. [Google Scholar] [CrossRef] [PubMed]
- Maniotis, A.J.; Folberg, R.; Hess, A.; Seftor, E.A.; Gardner, L.M.; Pe’er, J.; Trent, J.M.; Meltzer, P.S.; Hendrix, M.J. Vascular channel formation by human melanoma cells in vivo and in vitro: Vasculogenic mimicry. Am. J. Pathol. 1999, 155, 739–752. [Google Scholar] [CrossRef]
- Folberg, R.; Hendrix, M.J.; Maniotis, A.J. Vasculogenic mimicry and tumor angiogenesis. Am. J. Pathol. 2000, 156, 361–381. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, M.J.; Seftor, E.A.; Seftor, R.E.; Chao, J.T.; Chien, D.S.; Chu, Y.W. Tumor cell vascular mimicry: Novel targeting opportunity in melanoma. Pharmacol. Ther. 2016, 159, 83–92. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Ikari, R.; Mukaisho, K.I.; Kageyama, S.; Nagasawa, M.; Kubota, S.; Nakayama, T.; Murakami, S.; Taniura, N.; Tanaka, H.; Kushima, R.P.; et al. Differences in the central energy metabolism of cancer cells between conventional 2d and novel 3d culture systems. Int. J. Mol. Sci. 2021, 22, 1805. [Google Scholar] [CrossRef]
- Price, K.J.; Tsykin, A.; Giles, K.M.; Sladic, R.T.; Epis, M.R.; Ganss, R.; Goodall, G.J.; Leedman, P.J. Matrigel basement membrane matrix influences expression of microRNAs in cancer cell lines. Biochem. Biophys. Res. Commun. 2012, 427, 343–348. [Google Scholar] [CrossRef]
- Liu, H.; Wang, H.; Chen, D.; Gu, C.; Huang, J.; Mi, K. Endoplasmic reticulum stress inhibits 3D Matrigel-induced vasculogenic mimicry of breast cancer cells via TGF-β1/Smad2/3 and β-catenin signaling. FEBS Open Bio 2021, 11, 2607–2618. [Google Scholar] [CrossRef]
- Andonegui-Elguera, M.A.; Alfaro-Mora, Y.; Cáceres-Gutiérrez, R.; Caro-Sánchez, C.H.S.; Herrera, L.A.; Díaz-Chávez, J. An Overview of Vasculogenic Mimicry in Breast Cancer. Front. Oncol. 2020, 10, 220. [Google Scholar] [CrossRef] [PubMed]
- Francescone, R.; Vendramini-Costa, D.B. In Vitro Tube Formation Assays in Matrigel. In Vasculogenic Mimicry: Methods and Protocols; Springer: NewYork, NY, USA, 2022; Volume 2514, pp. 31–38. [Google Scholar] [CrossRef]
- Mingo, G.; Valdivia, A.; Santander, G.N.; Bobbitt, N.; Aldana, V.; Pradenas, J.; González, P.; Canales, C.; Toledo, J.A.; Ibáñez, C.; et al. Extracellular matrix protein signaling promotes multi-step cancer vasculogenic mimicry formation. Cell Commun. Signal. 2025, 23, 422. [Google Scholar] [CrossRef]
- Xie, J.; Tang, J.; Li, Y.; Kong, X.; Wang, W.; Wu, H. ATM Activation is Key in Vasculogenic Mimicry Formation by Glioma Stem-like Cells. Biomed. Environ. Sci. 2024, 37, 834–849. [Google Scholar] [CrossRef]
- Aure, M.R.; Fleischer, T.; Bjørklund, S.; Ankil, J.; Castro-Mondragon, J.A.; OSBREAC; Børresen-Dale, A.-L.; Tost, J.; Sahlberg, K.K.; Mathelier, A.; et al. Crosstalk between microRNA expression and DNA methylation drives the hormone-dependent phenotype of breast cancer. Genome Med. 2021, 13, 72. [Google Scholar] [CrossRef]
- Tchurikov, N.A.; Klushevskaya, E.S.; Kravatsky, Y.V.; Kravatskaya, G.I.; Fedoseeva, D.M. Interchromosomal Contacts of rDNA Clusters in Three Human Cell Lines Are Associated with Silencing of Genes Controlling Morphogenesis. Dokl. Biochem. Biophys. 2021, 496, 22–26. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Villa, T.; Porrua, O. Pervasive transcription: A controlled risk. FEBS J. 2023, 290, 3723–3736. [Google Scholar] [CrossRef] [PubMed]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Genet. 2013, 9, e1003569. [Google Scholar] [CrossRef]
- Clark, M.B.; Amaral, P.P.; Schlesinger, F.J.; Dinger, M.E.; Taft, R.J.; Rinn, J.L.; Ponting, C.P.; Stadler, P.F.; Morris, K.V.; Morillon, A.; et al. The Reality of Pervasive Transcription. PLoS Biol. 2011, 9, e1000625. [Google Scholar] [CrossRef] [PubMed]
- Tchurikov, N.A.; Alembekov, I.R.; Klushevskaya, E.S.; Kretova, A.N.; Lukicheva, V.N.; Chechetkin, V.R.; Kravatskaya, G.I.; Kravatsky, Y.V. Preferential Co-Expression and Colocalization of rDNA-Contacting Genes with LincRNAs Suggest Their Involvement in Shaping Inter-Chromosomal Interactions with Nucleoli. Int. J. Mol. Sci. 2024, 25, 6333. [Google Scholar] [CrossRef] [PubMed]
- Begnis, M.; Duc, J.; Offner, S.; Grun, D.; Sheppard, S.; Rosspopoff, O.; Trono, D. Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments. Sci. Adv. 2024, 10, eado1662. [Google Scholar] [CrossRef]
- Frietze, S.; O’Geen, H.; Blahnik, K.R.; Jin, V.X.; Farnham, P.J. ZNF274 recruits the histone methyltransferase SETD B1 to the 3′ ends of ZNF genes. PLoS ONE 2010, 5, e15082. [Google Scholar] [CrossRef]
- Bushnell, B. BBTools: A Suite of Fast, Multithreaded Bioinformatics Tools Designed for Analysis of DNA and RNA Sequence Data. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 13 November 2025).
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Storer, J.; Hubley, R.; Rosen, J.; Wheeler, T.J.; Smit, A.F. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob. DNA 2021, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, F.; Ryan, D.P.; Gruning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dundar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Dowle, M.; Srinivasan, A.; Gorecki, J.; Chirico, M.; Stetsenko, P. Data. Table: Extension of “Data. Frame.”; The Comprehensive R Archive Network: Vienna, Austria, 2024. [Google Scholar]
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. Dplyr: A Grammar of Data Manipulation; Posit Software, PBC: Boston, MA, USA, 2023. [Google Scholar]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. In Use R! 2nd ed.; Springer: Cham, Switzerland, 2016; p. 1. [Google Scholar]
- Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Wickham, H.; Vaughan, D.; Girlich, M. Tidyr: Tidy Messy Data; Posit Software, PBC: Boston, MA, USA, 2024. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tchurikov, N.A.; Klushevskaya, E.S.; Lukicheva, V.N.; Kretova, A.N.; Poperekova, E.N.; Chechetkin, V.R.; Kravatskaya, G.I.; Vartanian, A.A.; Alembekov, I.R.; Kravatsky, Y.V. During the Formation of Vasculogenic Mimicry by Melanoma Cells, the Silencing of Two Sets of Developmental Genes Is Coupled Either with an Increase or a Decrease in Contacts with the Nucleoli. Int. J. Mol. Sci. 2025, 26, 11289. https://doi.org/10.3390/ijms262311289
Tchurikov NA, Klushevskaya ES, Lukicheva VN, Kretova AN, Poperekova EN, Chechetkin VR, Kravatskaya GI, Vartanian AA, Alembekov IR, Kravatsky YV. During the Formation of Vasculogenic Mimicry by Melanoma Cells, the Silencing of Two Sets of Developmental Genes Is Coupled Either with an Increase or a Decrease in Contacts with the Nucleoli. International Journal of Molecular Sciences. 2025; 26(23):11289. https://doi.org/10.3390/ijms262311289
Chicago/Turabian StyleTchurikov, Nickolai A., Elena S. Klushevskaya, Viktoriya N. Lukicheva, Antonina N. Kretova, Elizaveta N. Poperekova, Vladimir R. Chechetkin, Galina I. Kravatskaya, Amalia A. Vartanian, Ildar R. Alembekov, and Yuri V. Kravatsky. 2025. "During the Formation of Vasculogenic Mimicry by Melanoma Cells, the Silencing of Two Sets of Developmental Genes Is Coupled Either with an Increase or a Decrease in Contacts with the Nucleoli" International Journal of Molecular Sciences 26, no. 23: 11289. https://doi.org/10.3390/ijms262311289
APA StyleTchurikov, N. A., Klushevskaya, E. S., Lukicheva, V. N., Kretova, A. N., Poperekova, E. N., Chechetkin, V. R., Kravatskaya, G. I., Vartanian, A. A., Alembekov, I. R., & Kravatsky, Y. V. (2025). During the Formation of Vasculogenic Mimicry by Melanoma Cells, the Silencing of Two Sets of Developmental Genes Is Coupled Either with an Increase or a Decrease in Contacts with the Nucleoli. International Journal of Molecular Sciences, 26(23), 11289. https://doi.org/10.3390/ijms262311289




