Genome-Wide Identification and Hormone-Induced Expression Analysis of the Anthocyanidin Reductase Gene Family in Sainfoin (Onobrychis viciifolia Scop.)
Abstract
1. Introduction
2. Results
2.1. Identification of the OvANR Gene Family in Sainfoin
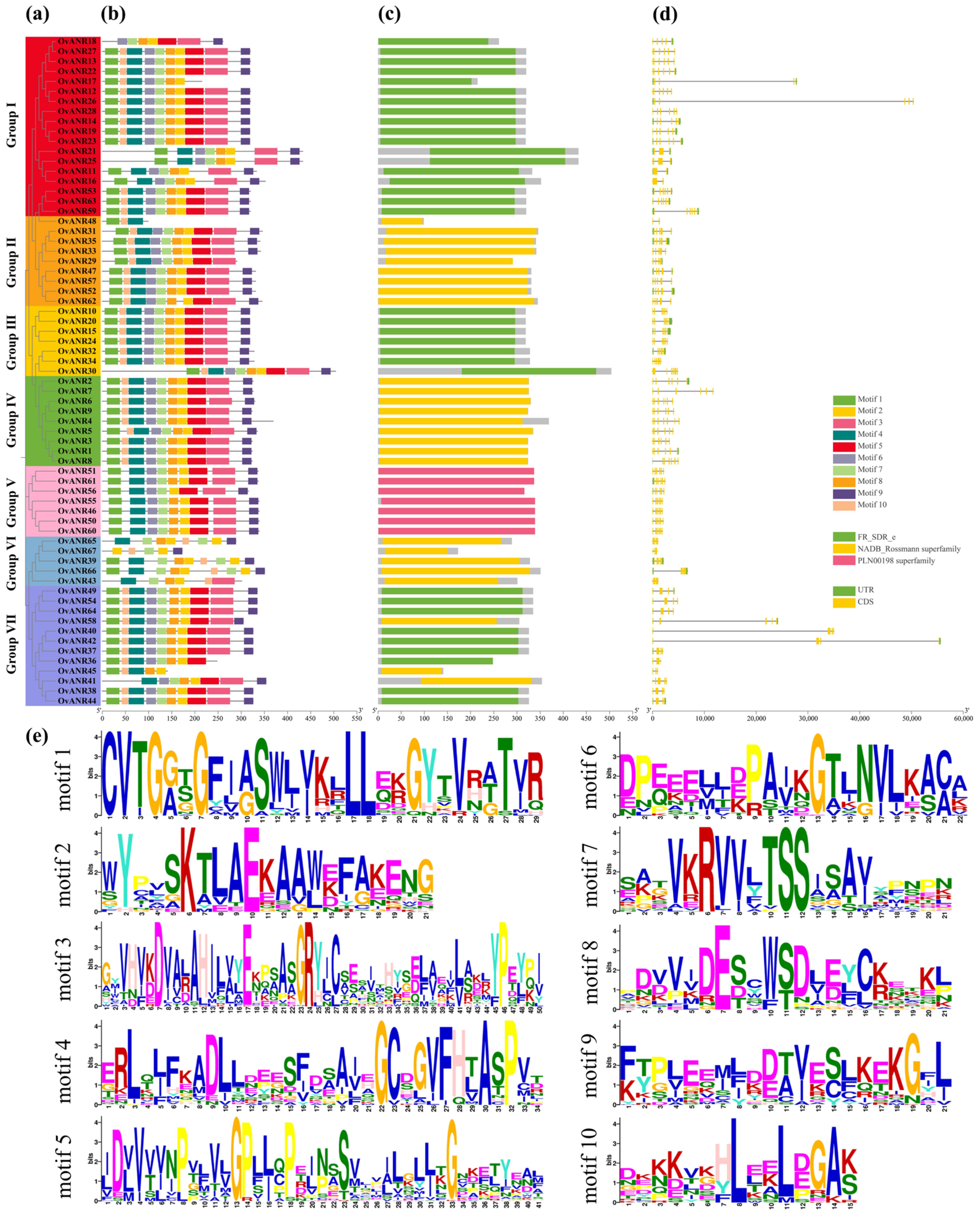
2.2. Evolutionary Relationship, Conserved Domains, Motif Composition, and Gene Structure of OvANRs in Sainfoin
2.3. Multiple Sequence Alignments, Secondary and Tertiary Structures of the OvANR Proteins in Sainfoin
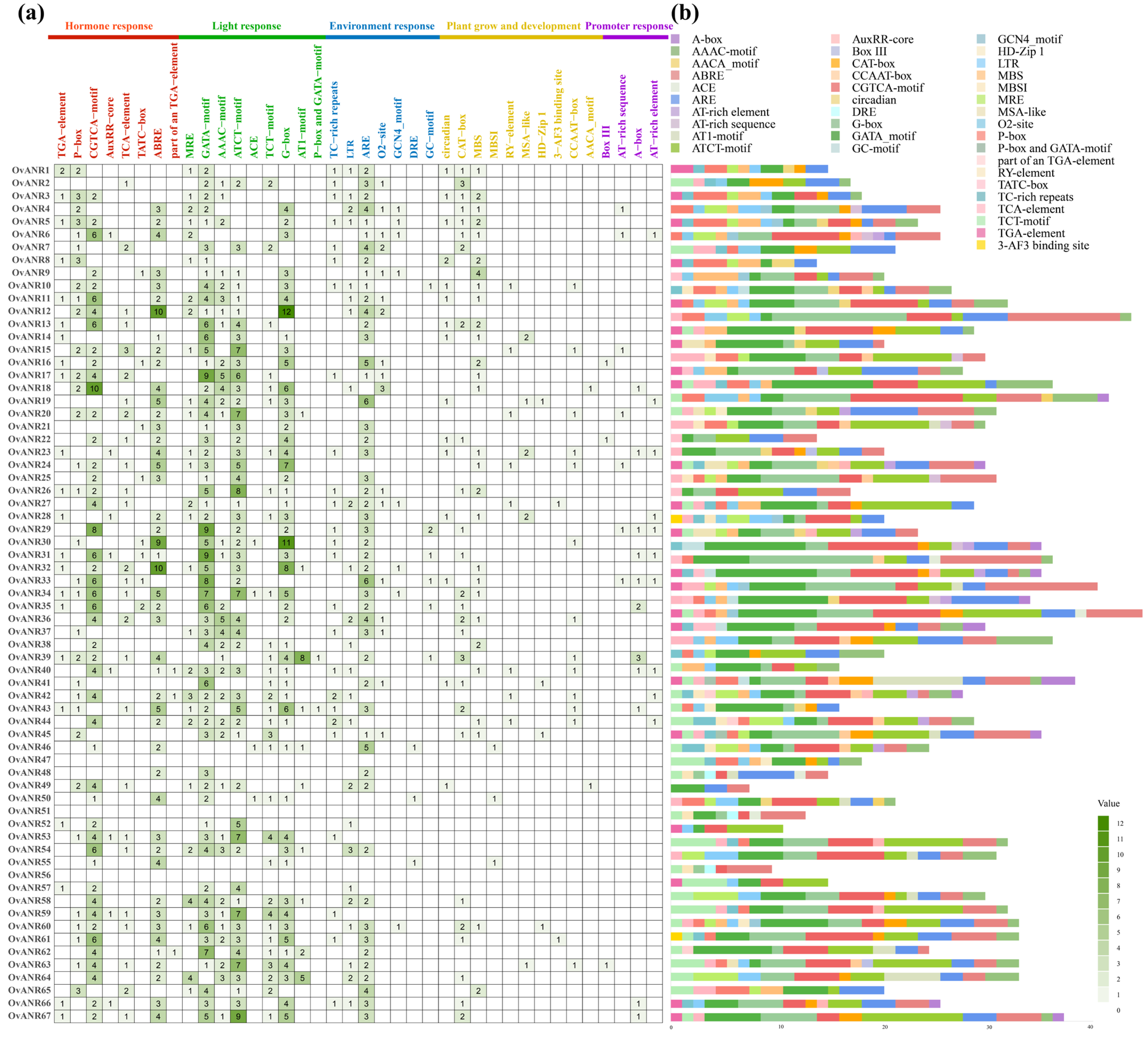
2.4. Cis-Acting Element Analysis of OvANR Gene Family in Sainfoin
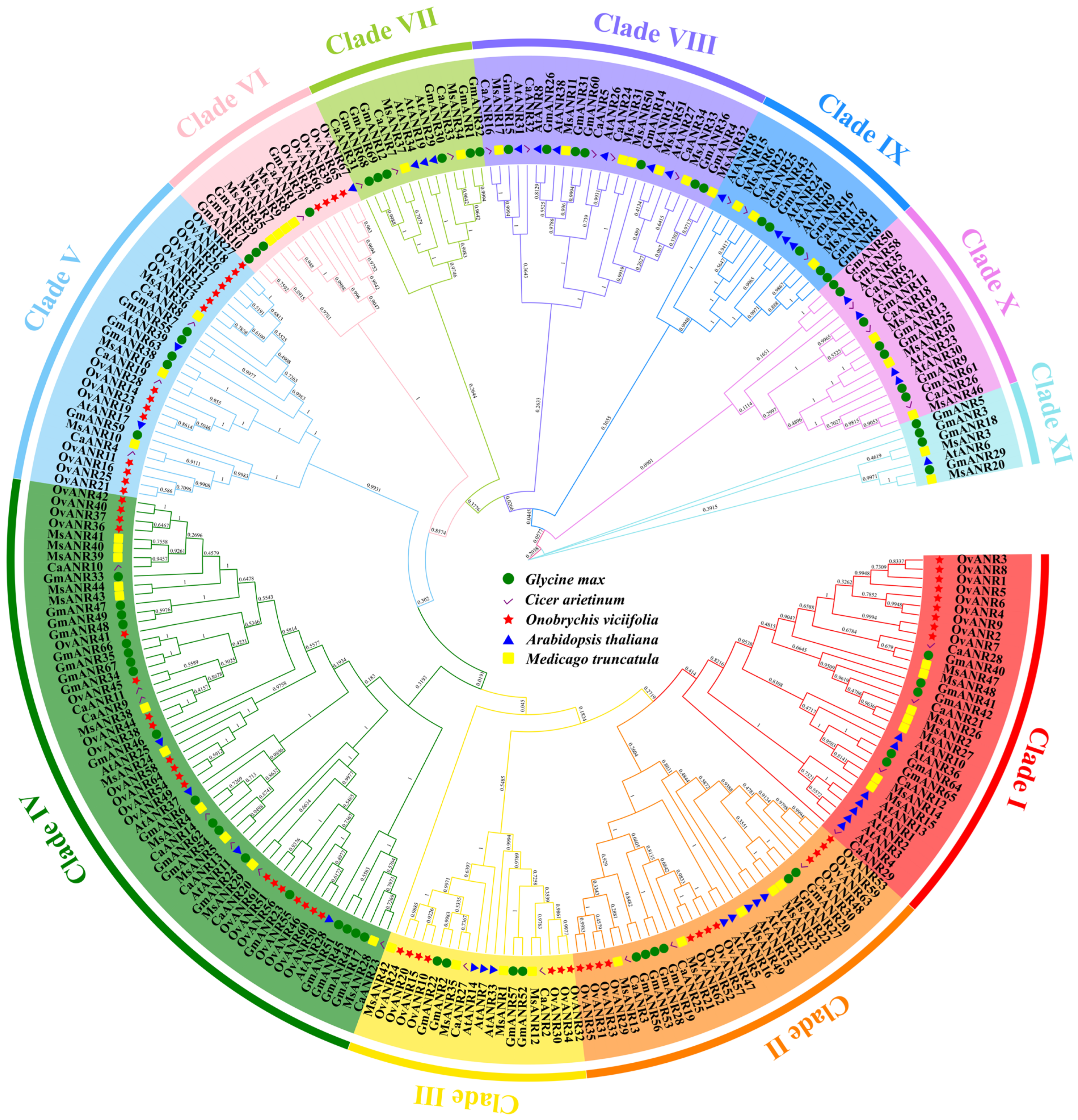
2.5. Evolutionary Analysis of OvANR Proteins
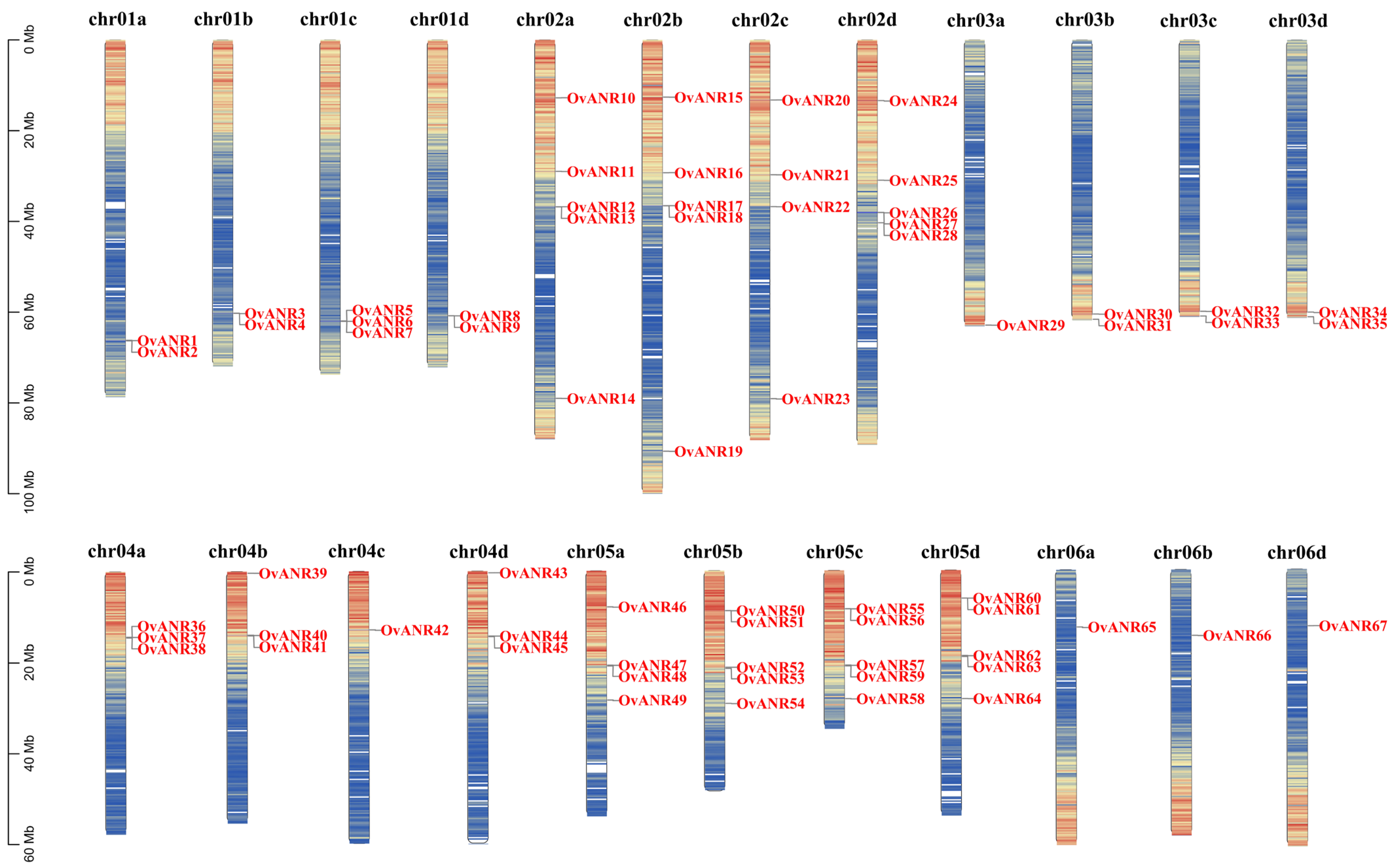
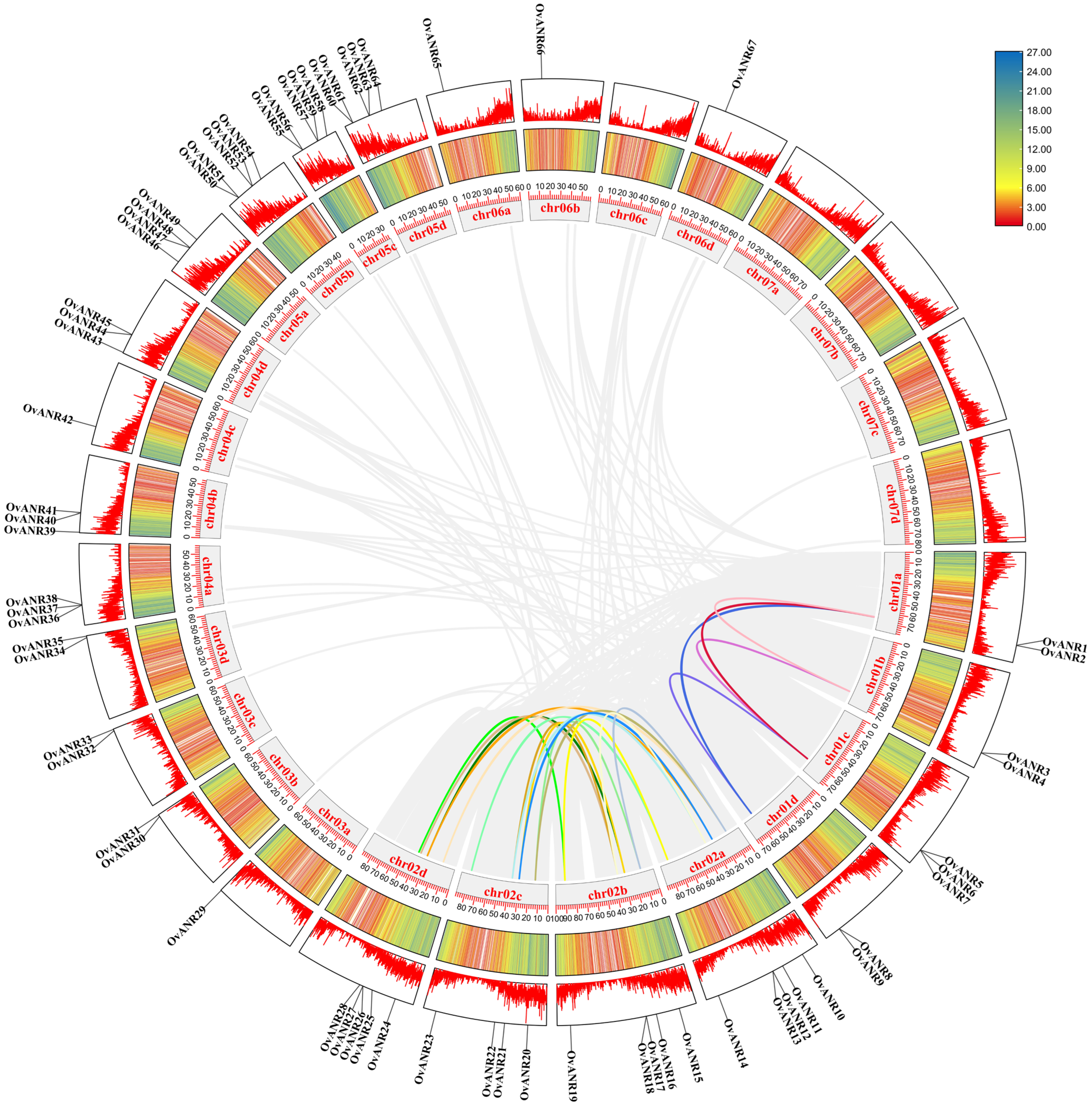
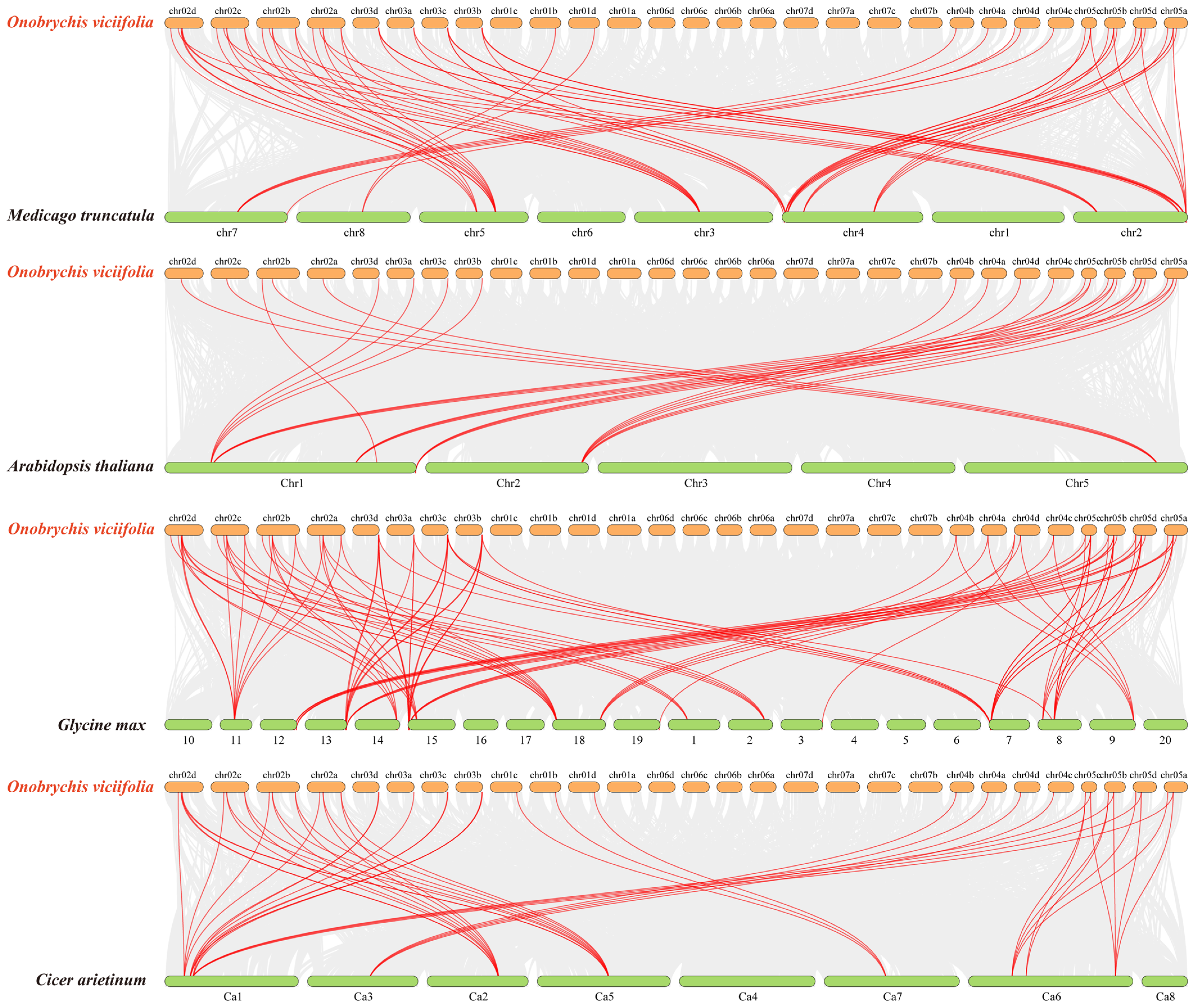
2.6. Chromosomal Localization and Collinearity Analysis of OvANR Genes
2.7. The Expression Pattern Analysis of OvANR Genes in Sainfoin
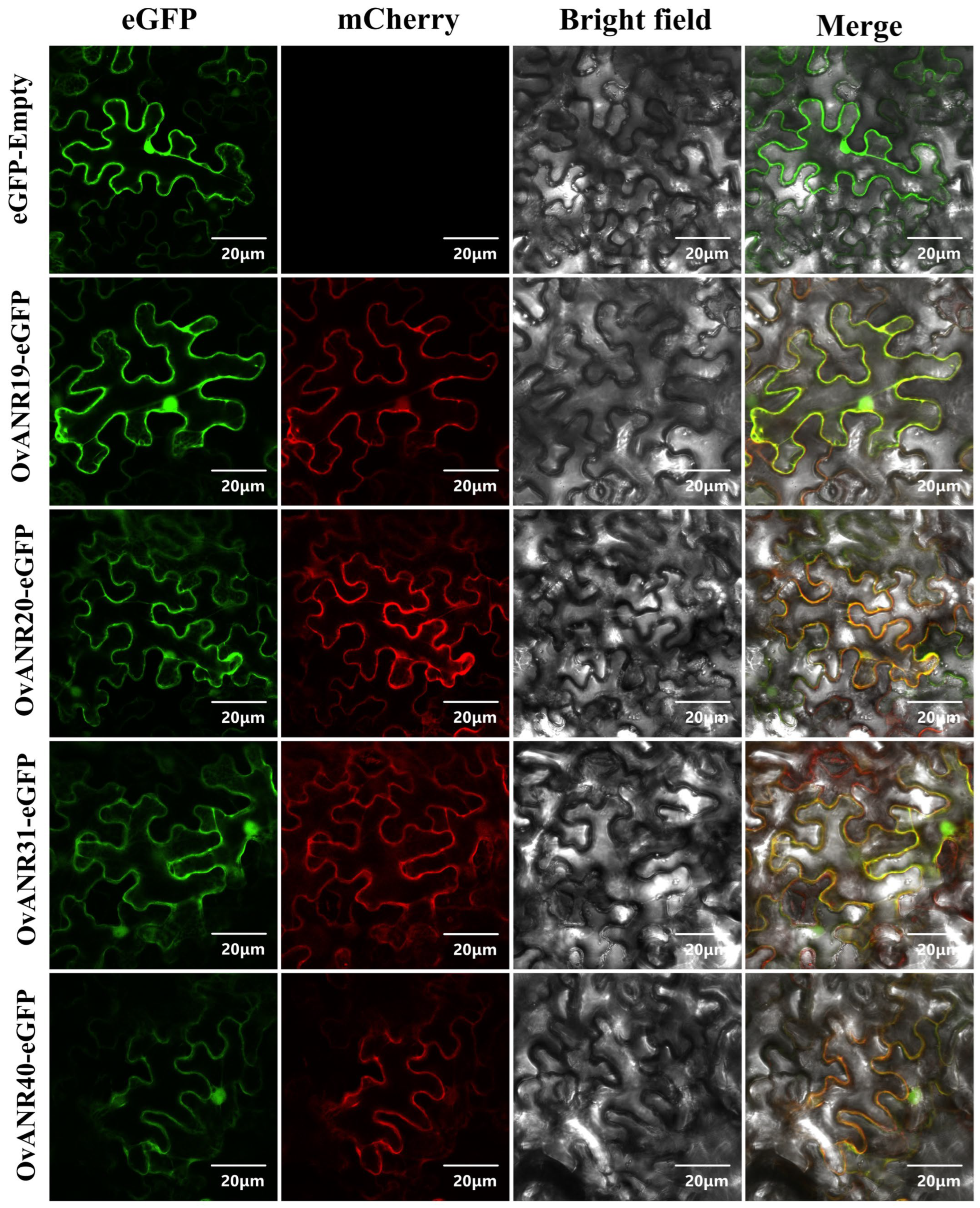
2.8. Subcellular Localization of OvANRs
3. Discussion
4. Materials and Methods
4.1. Identification of the OvANR Gene Family and Analysis of Protein Physicochemical Properties in Sainfoin
4.2. Evolutionary, Gene Structure, Conserved Domain, and Motif Analysis
4.3. Prediction of Secondary and Tertiary Structures
4.4. Cis-Acting Elements Analysis
4.5. Phylogenetic Analysis
4.6. Chromosomal Location and Synteny Analysis
4.7. Cold and Drought Transcriptome Data
4.8. Plant Materials and Treatments
4.9. RT-qPCR Analysis
4.10. Subcellular Localization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Irani, S.; Majidi, M.M.; Mirlohi, A.; Zargar, M.; Karami, M. Assessment of drought tolerance in sainfoin: Physiological and drought tolerance indices. Agron. J. 2015, 107, 1771–1781. [Google Scholar] [CrossRef]
- Re, G.A.; Piluzza, G.; Sulas, L.; Franca, A.; Porqueddu, C.; Sanna, F.; Bullitta, S. Condensed tannin accumulation and nitrogen fixation potential of Onobrychis viciifolia Scop. Grown in a Mediterranean environment. J. Sci. Food Agric. 2014, 94, 639–645. [Google Scholar] [CrossRef]
- Aufrère, J.; Theodoridou, K.; Baumont, R. Agronomic and nutritional value of Sainfoin. Fourrages 2013, 213, 63–75. [Google Scholar]
- Sakhraoui, A.; Ltaeif, H.B.; Sakhraoui, A.; Villalba, J.J.; Castillo, J.M.; Rouz, S. Sainfoin (Onobrychis viciifolia): A legume with great ecological and agronomical potential under climate change. J. Agric. Sci. 2024, 162, 307–331. [Google Scholar] [CrossRef]
- Hayot Carbonero, C.; Mueller-Harvey, I.; Brown, T.A.; Smith, L. Sainfoin (Onobrychis viciifolia): A beneficial forage legume. Plant Genet. Resour. 2011, 9, 70–85. [Google Scholar] [CrossRef]
- Bhattarai, S.; Coulman, B.; Beattie, A.D.; Biligetu, B. Assessment of sainfoin (Onobrychis viciifolia Scop.) germplasm for agro-morphological traits and nutritive value. Grass Forage Sci. 2018, 73, 958–966. [Google Scholar] [CrossRef]
- Kozlova, Z.V.; Matais, L.N.; Glushkova, O.A. Influence of sainfoin on soil fertility and agro-economic indicators of fodder crop rotations under conditions of East Siberia. Multifunct. Adapt. Feed. Prod. 2020, 23, 67–72. [Google Scholar]
- McMahon, L.R.; Majak, W.; McAllister, T.A.; Hall, J.W.; Jones, G.A.; Popp, J.D.; Cheng, K.J. Effect of sainfoin on in vitro digestion of fresh alfalfa and bloat in steers. Can. J. Anim. Sci. 1999, 79, 203–212. [Google Scholar] [CrossRef]
- Huyen, N.T.; Desrues, O.; Alferink, S.J.J.; Zandstra, T.; Verstegen, M.W.A.; Hendriks, W.H.; Pellikaan, W.F. Inclusion of sainfoin (Onobrychis viciifolia) silage in dairy cow rations affects nutrient digestibility, nitrogen utilization, energy balance, and methane emissions. J. Dairy. Sci. 2016, 99, 3566–3577. [Google Scholar] [CrossRef] [PubMed]
- Min, B.R.; Barry, T.N.; Attwood, G.T.; and McNabb, W.C. The Effect of Condensed Tannins on the Nutrition and Health of Ruminants Fed Fresh Temperate Forages: A Review. Anim. Feed Sci. Technol. 2003, 106, 3–19. [Google Scholar] [CrossRef]
- Shimizu-Inatsugi, R.; Terada, A.; Hirose, K.; Kudoh, H.; Sese, J.; Shimizu, K.K. Plant adaptive radiation mediated by polyploid plasticity in transcriptomes. Mol. Ecol. 2017, 26, 193–207. [Google Scholar] [CrossRef]
- He, J.Y.; Tian, D.Y.; Li, X.; Wang, X.M.; Wang, T.T.; Wang, Z.Y.; Zang, H.; He, X.F.; Zhang, T.J.; Yun, Q.Z.; et al. A chromosome-level genome assembly for Onobrychis viciifolia reveals gene copy number gain underlying enhanced proanthocyanidin biosynthesis. Commun. Biol. 2024, 7, 19. [Google Scholar] [CrossRef]
- Dave, K.; Kumar, A.; Dave, N.; Jain, M.; Dhanda, P.S.; Yadav, A.; Kaushik, P. Climate change impacts on legume physiology and ecosystem dynamics: A multifaceted perspective. Sustainability 2024, 16, 6026. [Google Scholar] [CrossRef]
- Ergon, Å.; Amdahl, H. Winter survival in red clover: Experimental evidence for interactions among stresses. BMC Plant Biol. 2024, 24, 467. [Google Scholar] [CrossRef]
- Lu, J.; Li, N.; Li, G.; Tian, Z.; Shi, L.; Wang, Y.; Cai, Y.; Zhang, K.; Sun, W.; Wang, D.; et al. N-glycosylation of SnRK2s affects NADPH maintenance in peroxisomes during prolonged ABA signaling. Nat. Commun. 2024, 15, 6630. [Google Scholar] [CrossRef]
- Chen, J.; Li, F.F.; Li, Y.X.; Wang, Y.S.; Wang, C.Z.; Yuan, D.B.; Jiang, Y.M. Exogenous procyanidin treatment delays senescence of harvested banana fruit by enhancing antioxidant responses and in vivo procyanidin content. Postharvest Biol. Technol. 2019, 158, 110999. [Google Scholar] [CrossRef]
- Huang, W.Y.; Wu, H.; Li, D.J.; Song, J.F.; Xiao, Y.D.; Liu, C.Q.; Zhou, J.Z.; Sui, Z.Q. Protective effects of blueberry anthocyanins against H2O2-induced oxidative injuries in human retinal pigment epithelial cells. J. Agric. Food Chem. 2018, 66, 1638–1648. [Google Scholar] [CrossRef] [PubMed]
- Visser, J.; van Staden, P.J.; Soma, P.; Buys, A.V.; Pretorius, E. The stabilizing effect of an oligomeric proanthocyanidin on red blood cell membrane structure of poorly controlled Type II diabetes. Nutr. Diabetes 2017, 7, e275. [Google Scholar] [CrossRef]
- Yu, K.J.; Song, Y.S.; Lin, J.X.; Dixon, R.A. The complexities of proanthocyanidin biosynthesis and its regulation in plants. Plant Commun. 2023, 4, 100498. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Xie, D.-Y.; Sharma, S.B. Proanthocyanidins—A final frontier in flavonoid research? New Phytol. 2005, 165, 9–28. [Google Scholar] [CrossRef] [PubMed]
- Lu, N.; Rao, X.; Li, Y.; Jun, J.H.; Dixon, R.A. Dissecting the transcriptional regulation of proanthocyanidin and anthocyanin biosynthesis in soybean (Glycine max). Plant Biotechnol. J. 2021, 19, 1429–1442. [Google Scholar] [CrossRef]
- Bao, C.; Niu, M.; Liu, Z.; Wu, Y.; Cao, B.; Zhou, M.; Yuan, X.; Jia, L.; Cui, J.; Shen, Z.; et al. VrMYB90 negatively regulates proanthocyanidin biosynthesis by repressing VrANR in mung bean (Vigna radiata L.). Planta 2025, 262, 70. [Google Scholar] [CrossRef]
- Wang, P.; Liu, Y.; Zhang, L.; Wang, W.; Hou, H.; Zhao, Y.; Jiang, X.; Yu, J.; Tan, H.; Wang, Y.; et al. Functional demonstration of plant flavonoid carbocations proposed to be involved in the biosynthesis of proanthocyanidins. Plant J. 2020, 101, 18–36. [Google Scholar] [CrossRef]
- Liang, B.; Ye, X.; Li, H.; Li, F.; Wang, S.; Jiang, C.; Wang, J.; Wang, P. Genome-wide identification and analysis of anthocyanidin reductase gene family in lychee (Litchi chinensis Sonn.). Genes 2024, 15, 757. [Google Scholar] [CrossRef]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Korban, S.S. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 2012, 63, 2437–2447. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.D.; Tan, Z.W.; Li, L.; Yu, Y.L.; Xu, L.J.; Yang, H.Q.; Dong, W.; Liang, H.Z. Cloning and expression analysis of anthocyanidin reductase gene ANR in Carthamus tinctorius L. J. Nucl. Agric. Sci. 2022, 36, 517–526. [Google Scholar]
- Trainin, T.; Harel-Beja, R.; Bar-Ya’akov, I.; Ben-Simhon, Z.; Yahalomi, R.; Borochov-Neori, H.; Ophir, R.; Sherman, A.; Doron-Faigenboim, A.; Holland, D. Fine mapping of the “black” peel color in pomegranate (Punica granatum L.) strongly suggests that a mutation in the anthocyanidin reductase (ANR) gene is responsible for the trait. Front. Plant Sci. 2021, 12, 642019. [Google Scholar] [CrossRef]
- Zhu, Q.; Yu, S.; Zeng, D.; Liu, H.; Wang, H.; Yang, Z.; Xie, X.; Shen, R.; Tan, J.; Li, H.; et al. Development of “Purple Endosperm Rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol. Plant 2017, 10, 918–929. [Google Scholar] [CrossRef]
- Xin, Y.; Meng, S.; Ma, B.; He, W.; He, N. Mulberry genes MnANR and MnLAR confer transgenic plants with resistance to Botrytis cinerea. Plant Sci. 2020, 296, 110473. [Google Scholar] [CrossRef] [PubMed]
- Bargunam, S.; Roy, R.; Shetty, D.; Amisha, S.H.; Shukla, V.S.; Babu, V.S. Melatonin-governed growth and metabolome divergence: Circadian and stress responses in key plant species. Plant Physiol. Biochem. 2025, 221, 109635. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, M.; Bao, L.; Long, J.; Cui, X.; Zheng, Z.; Zhao, X.; Huang, Y.; Jiao, F.; Su, C.; et al. Metabolomic and transcriptomic analysis of flavonoids biosynthesis mechanisms in mulberry fruit (Hongguo 2) under exogenous hormone treatments. Plant Physiol. Biochem. 2024, 212, 108773. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.; Shi, C.; Peng, Y.; Tan, H.; Xin, P.; Yang, Y.; Wang, F.; Li, X.; Chu, J.; Huang, J.; et al. Brassinosteroid-activated BRI1-EMS-SUPPRESSOR 1 inhibits flavonoid biosynthesis and coordinates growth and UV-B stress responses in plants. Plant Cell 2020, 32, 3224–3239. [Google Scholar] [CrossRef]
- Rodrigues, A.; Adamo, M.; Crozet, P.; Margalha, L.; Confraria, A.; Martinho, C.; Elias, A.; Rabissi, A.; Lumbreras, V.; González-Guzmán, M.; et al. ABI1 and PP2CA Phosphatases Are Negative Regulators of Snf1-Related Protein Kinase1 Signaling in Arabidopsis. Plant Cell 2013, 25, 3871–3884. [Google Scholar] [CrossRef]
- Jia, Y.; Meng, W.; Chen, G.; Fan, X.; Zhang, Y.; Ding, A.; Xu, M.; Hu, G.; Tan, M.; Xiang, Z. The regulation mechanism of MYC on MeJA-induced flavonoid synthesis in Dendrobium officinale. J. Plant Growth Regul. 2025, 44, 217–232. [Google Scholar] [CrossRef]
- Wang, Y.; Hou, Y.; Qiu, J.; Wang, H.; Wang, S.; Tang, L.; Tong, X.; Zhang, J. Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72-AOC’ pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol. 2020, 228, 1336–1353. [Google Scholar] [CrossRef]
- Pan, J.; Hu, Y.; Wang, H.; Guo, Q.; Chen, Y.; Howe, G.A.; Yu, D. Molecular mechanism underlying the synergetic effect of jasmonate on abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 2020, 32, 3846–3865. [Google Scholar] [CrossRef]
- Li, S.; Dong, Y.; Li, D.; Shi, S.; Zhao, N.; Liao, J.; Liu, Y.; Chen, H. Eggplant transcription factor SmMYB5 integrates jasmonate and light signaling during anthocyanin biosynthesis. Plant Physiol. 2024, 194, 1139–1165. [Google Scholar] [CrossRef]
- Zhu, C.; Sang, T.; Zhang, Z.; Wang, Y.; Lin, Z.; Wang, W.; Lang, Z.; Zhu, J.K.; Wang, P. Maintaining basal B-RAF kinase activity for abscisic acid signaling via reciprocal phosphoregulation of a single serine residue. J. Integr. Plant Biol. 2025, 67, 2848–2862. [Google Scholar] [CrossRef] [PubMed]
- Mascheretti, I.; Alfieri, M.; Lauria, M.; Locatelli, F.; Consonni, R.; Cusano, E.; Dougué Kentsop, R.A.; Laura, M.; Ottolina, G.; Faoro, F.; et al. New insight into justicidin B pathway and production in Linum austriacum. Int. J. Mol. Sci. 2021, 22, 2507. [Google Scholar] [CrossRef]
- Andrews, G.; Fan, K.; Pratt, H.E.; Phalke, N.; Karlsson, E.K.; Lindblad-Toh, K.; Gazal, S.; Moore, J.E.; Weng, Z.; Andrews, G.; et al. Mammalian evolution of human cis-regulatory elements and transcription factor binding sites. Science 2023, 380, eabn7930. [Google Scholar] [CrossRef] [PubMed]
- Gourlay, G.; Hawkins, B.J.; Albert, A.; Schnitzler, J.P.; Peter Constabel, C. Condensed tannins as antioxidants that protect poplar against oxidative stress from drought and UV-B. Plant Cell Environ. 2022, 45, 362–377. [Google Scholar] [CrossRef] [PubMed]
- Bogs, J.; Downey, M.O.; Harvey, J.S.; Ashton, A.R.; Tanner, G.J.; Robinson, S.P. Proanthocyanidin Synthesis and Expression of Genes Encoding Leucoanthocyanidin Reductase and Anthocyanidin Reductase in Developing Grape Berries and Grapevine Leaves. Plant Physiol. 2005, 139, 652–663. [Google Scholar] [CrossRef]
- Jonker, A.; Yu, P. The occurrence, biosynthesis, and molecular structure of proanthocyanidins and their effects on legume forage protein precipitation, digestion, and absorption in the ruminant digestive tract. Int. J. Mol. Sci. 2017, 18, 1105. [Google Scholar] [CrossRef]
- Shao, Z.H.; Vanden Hoek, T.L.; Li, C.Q.; Schumacker, P.T.; Becker, L.B.; Chan, K.C.; Qin, Y.; Yin, J.J.; Yuan, C.S. Synergistic effect of Scutellaria baicalensis and grape seed proanthocyanidins on scavenging reactive oxygen species in vitro. Am. J. Chin. Med. 2004, 32, 89–95. [Google Scholar] [CrossRef]
- AMukherjee, D.; Saha, D.; Acharya, D.; Mukherjee, A.; Ghosh, T.C. Interplay between gene expression and gene architecture as a consequence of gene and genome duplications: Evidence from metabolic genes of Arabidopsis thaliana. Physiol. Mol. Biol. Plants 2022, 28, 1091–1108. [Google Scholar] [CrossRef]
- Cui, C.; Yang, L.J.; Liu, Z.W.; Shu, X.; Zhang, W.W.; Gao, Y.; Wang, Y.X.; Wang, T.; Chen, C.C.; Guo, R.T.; et al. Substrate specificity of a branch of aromatic dioxygenases determined by three distinct motifs. Nat. Commun. 2024, 15, 7682. [Google Scholar] [CrossRef] [PubMed]
- Baulin, E.F.; Bohdan, D.R.; Kowalski, D.; Serwatka, M.; Świerczyńska, J.; Żyra, Z.; Bujnicki, J.M. ARTEM: A method for RNA and DNA tertiary motif identification with backbone permutations. Genome Biol. 2025, 26, 226. [Google Scholar] [CrossRef] [PubMed]
- Feenstra, K.A.; Pirovano, W.; Krab, K.; Heringa, J. Sequence harmony: Detecting functional specificity from alignments. Nucleic Acids Res. 2007, 35, W495–W498. [Google Scholar] [CrossRef]
- Vergara, F.; Rymen, B.; Kuwahara, A.; Sawada, Y.; Sato, M.; Hirai, M.Y. Autopolyploidization, geographic origin, and metabolome evolution in Arabidopsis thaliana. Am. J. Bot. 2017, 104, 905–914. [Google Scholar] [CrossRef]
- de Oliveira, P.M.C.; de Oliveira, S.M. The zipper effect: Why different positions along the chromosome suffer different selection pressures. Phys. A 2011, 390, 492–498. [Google Scholar] [CrossRef]
- Tek, A.L.; Nagaki, K.; Yıldız Akkamış, H.; Tanaka, K.; Kobayashi, H. Chromosome-specific barcode system with centromeric repeat in cultivated soybean and wild progenitor. Life Sci. Alliance 2024, 7, e202402802. [Google Scholar] [CrossRef] [PubMed]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Schnable, J.C.; Freeling, M.; Lyons, E. Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biol. Evol. 2012, 4, 265–277. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, R.; Jiang, K.-W.; Qi, J.; Hu, Y.; Guo, J.; Zhu, R.; Zhang, T.; Egan, A.N.; Yi, T.-S.; et al. Nuclear phylotranscriptomics and phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen-fixing symbiosis in Fabaceae. Mol. Plant 2021, 14, 748–773. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183, Erratum in Nature 2010, 465, 120. [Google Scholar] [CrossRef]
- Rawal, H.C.; Singh, N.K.; Sharma, T.R. Conservation, Divergence, and Genome-Wide Distribution of PAL and POX A Gene Families in Plants. Int. J. Genom. 2013, 2013, 678969. [Google Scholar]
- Li, W.; Wen, Y.; Quan, J.; Gao, M.; Shang, C.; Liu, X.; Liu, G.; Hu, X.; Li, J. Regulation of jasmonic acid signalling in tomato cold stress response: Insights into the MYB15-LOXD and MYB15-MYC2-LOXD regulatory modules. Plant Biotechnol. J. 2025, 23, 4246–4260. [Google Scholar] [CrossRef]
- Liu, N.; Zhao, H.; Hou, L.; Zhang, C.; Bo, W.; Pang, X.; Li, Y. HPLC-MS/MS-based and transcriptome analysis reveal the effects of ABA and MeJA on jujube (Ziziphus jujuba Mill.) cracking. Food Chem. 2023, 421, 136155. [Google Scholar] [CrossRef]
- Du, C.X.; Si, Y.Y.; Wang, Z.; Guo, Y.T.; Li, Y.P.; Liu, C.; Fan, H.F. Overexpression of CsPP2-A1 in cucumber enhanced salt tolerance by participating ABA-JA signaling pathway and antioxidant system. Environ. Exp. Bot. 2022, 204, 105095. [Google Scholar] [CrossRef]
- Luo, P.; Shen, Y.; Jin, S.; Huang, S.; Cheng, X.; Wang, Z.; Li, P.; Zhao, J.; Bao, M.; Ning, G. Overexpression of Rosa rugosa anthocyanidin reductase enhances tobacco tolerance to abiotic stress through increased ROS scavenging and modulation of ABA signaling. Plant Sci. 2016, 245, 35–49. [Google Scholar] [CrossRef] [PubMed]
- An, X.H.; Tian, Y.; Chen, K.Q.; Liu, X.J.; Liu, D.D.; Xie, X.B.; Cheng, C.G.; Cong, P.H.; Hao, Y.J. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015, 56, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kidokoro, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulatory networks in plant responses to drought and cold stress. Plant Physiol. 2024, 195, 170–189. [Google Scholar] [CrossRef]
- Benner, C.; Konovalov, S.; Mackintosh, C.; Hutt, K.R.; Stunnenberg, R.; Garcia-Bassets, I. Decoding a signature-based model of transcription cofactor recruitment dictated by cardinal cis-regulatory elements in proximal promoter regions. PLoS Genet. 2013, 9, e1003906. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019, 223, 246–260. [Google Scholar] [CrossRef]
- Zhu, Y.; Peng, Q.Z.; Li, K.G.; Xie, D.Y. Molecular cloning and functional characterization of the anthocyanidin reductase gene from Vitis bellula. Planta 2014, 240, 381–398. [Google Scholar] [CrossRef]
- Nesi, N.; Lucas, M.-O.; Auger, B.; Baron, C.; Lécureuil, A.; Guerche, P.; Kronenberger, J.; Lepiniec, L.; Debeaujon, I.; Renard, M. Promoter of the Arabidopsis thaliana BAN gene is active in proanthocyanidin-accumulating cells of the Brassica napus seed coat. Plant Cell Rep. 2009, 28, 601–617. [Google Scholar] [CrossRef]
- Xie, D.; Sharma, S.B.; Paiva, N.L.; Ferreira, D.; Dixon, R.A. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 2003, 299, 396–399. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.M.; Mordaunt, M.; Lopez, P.; Uppala, A.; Rosati, D.; Rodrigues, N.P.; Grabitz, P.; Rife, S.C. Scite: A smart citation index that displays the context of citations and classifies their intent using deep learning. bioRxiv 2021, 2, 882–898. [Google Scholar] [CrossRef]
- Yang, M.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI’s Conserved Domain Database and Tools for Protein Domain Analysis. Curr. Protoc. Bioinform. 2019, 69, e90. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2022, 51, D384–D388. [Google Scholar] [CrossRef]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPM: A self-optimized method for protein secondary structure prediction. Protein Eng. 1994, 7, 157–164. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.Z.Y.; Jiang, W.B.; Luo, Y.J.; Huang, H.J.; Yi, D.X.; Pang, Y.Z. Analyses on flavonoids and transcriptome reveal key MYB gene for proanthocyanidins regulation in Onobrychis viciifolia. Front. Plant Sci. 2022, 13, 941918. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Jiang, G.; Wang, J.; He, H.; Liu, L.; Du, P.; Li, H.; Wang, F.; Xie, Q. Genome-Wide Identification and Hormone-Induced Expression Analysis of the Anthocyanidin Reductase Gene Family in Sainfoin (Onobrychis viciifolia Scop.). Int. J. Mol. Sci. 2025, 26, 11256. https://doi.org/10.3390/ijms262311256
Hu Y, Jiang G, Wang J, He H, Liu L, Du P, Li H, Wang F, Xie Q. Genome-Wide Identification and Hormone-Induced Expression Analysis of the Anthocyanidin Reductase Gene Family in Sainfoin (Onobrychis viciifolia Scop.). International Journal of Molecular Sciences. 2025; 26(23):11256. https://doi.org/10.3390/ijms262311256
Chicago/Turabian StyleHu, Yuqing, Guangzhi Jiang, Jiayin Wang, Huan He, Lele Liu, Pingping Du, Hongbin Li, Fei Wang, and Quanliang Xie. 2025. "Genome-Wide Identification and Hormone-Induced Expression Analysis of the Anthocyanidin Reductase Gene Family in Sainfoin (Onobrychis viciifolia Scop.)" International Journal of Molecular Sciences 26, no. 23: 11256. https://doi.org/10.3390/ijms262311256
APA StyleHu, Y., Jiang, G., Wang, J., He, H., Liu, L., Du, P., Li, H., Wang, F., & Xie, Q. (2025). Genome-Wide Identification and Hormone-Induced Expression Analysis of the Anthocyanidin Reductase Gene Family in Sainfoin (Onobrychis viciifolia Scop.). International Journal of Molecular Sciences, 26(23), 11256. https://doi.org/10.3390/ijms262311256





