A Comprehensive Analysis of Novel Variations Associated with Bile Duct Cancer: Insights into Expression, Methylation, and 3D Protein Structure
Abstract
1. Introduction
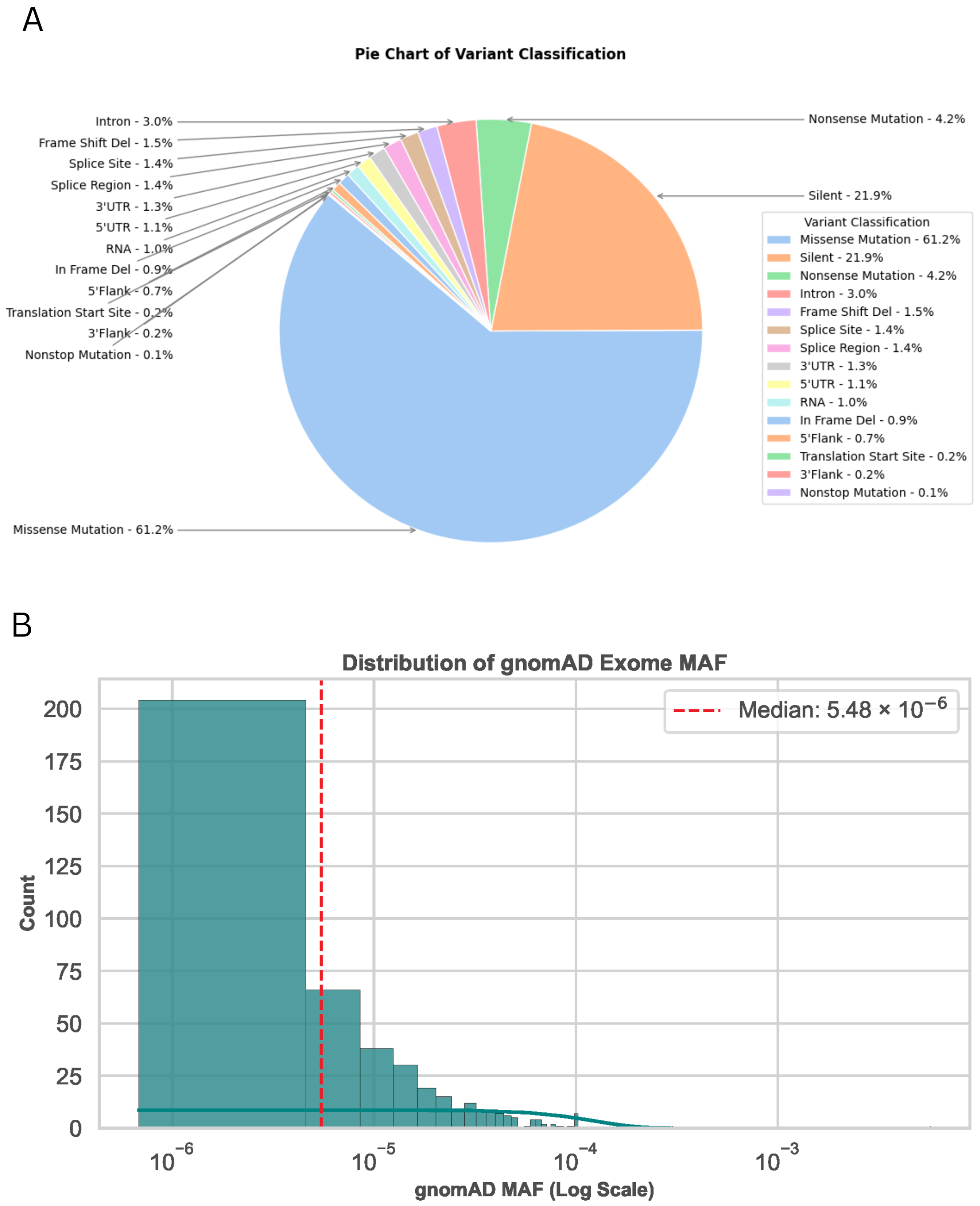
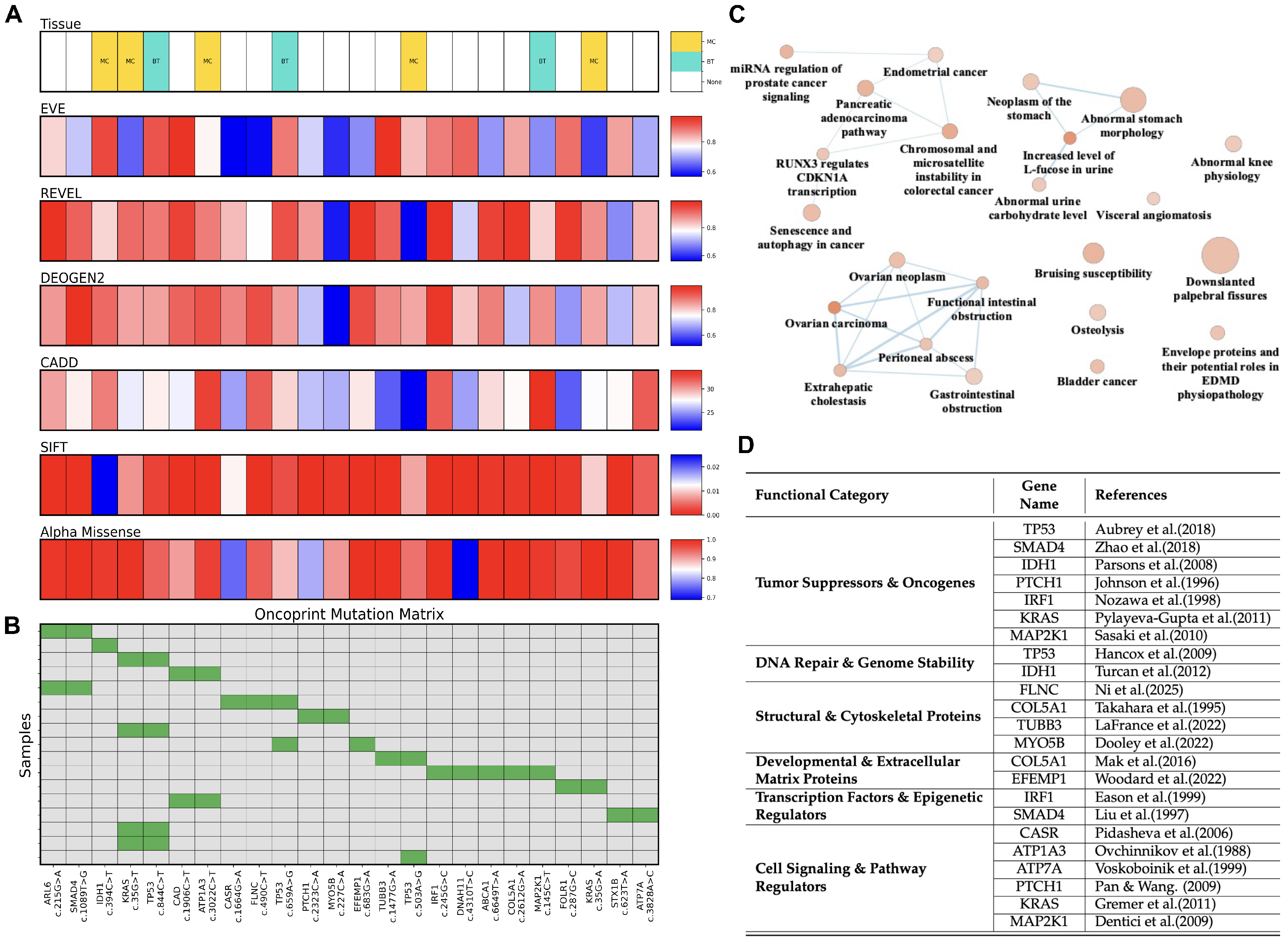
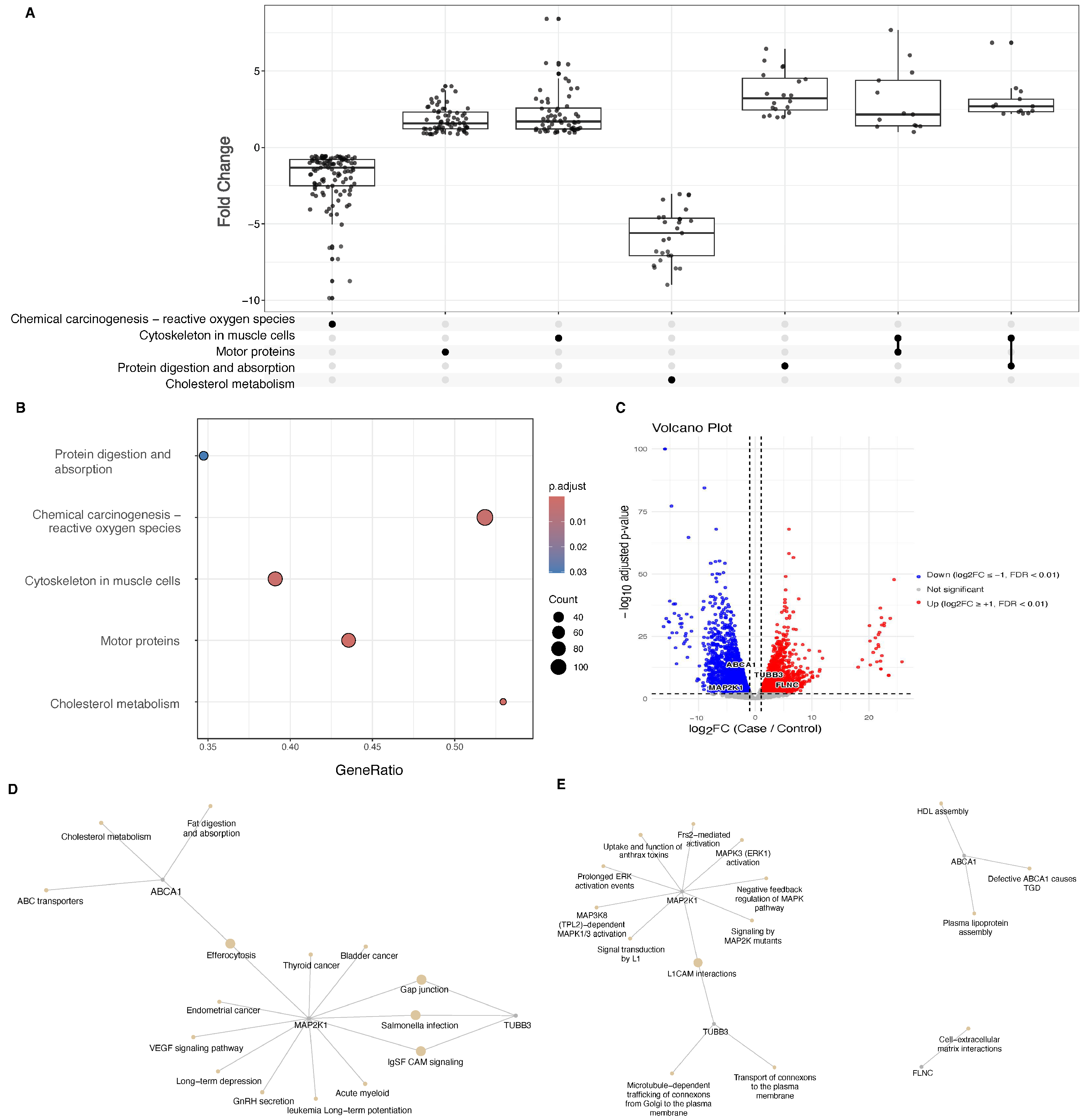
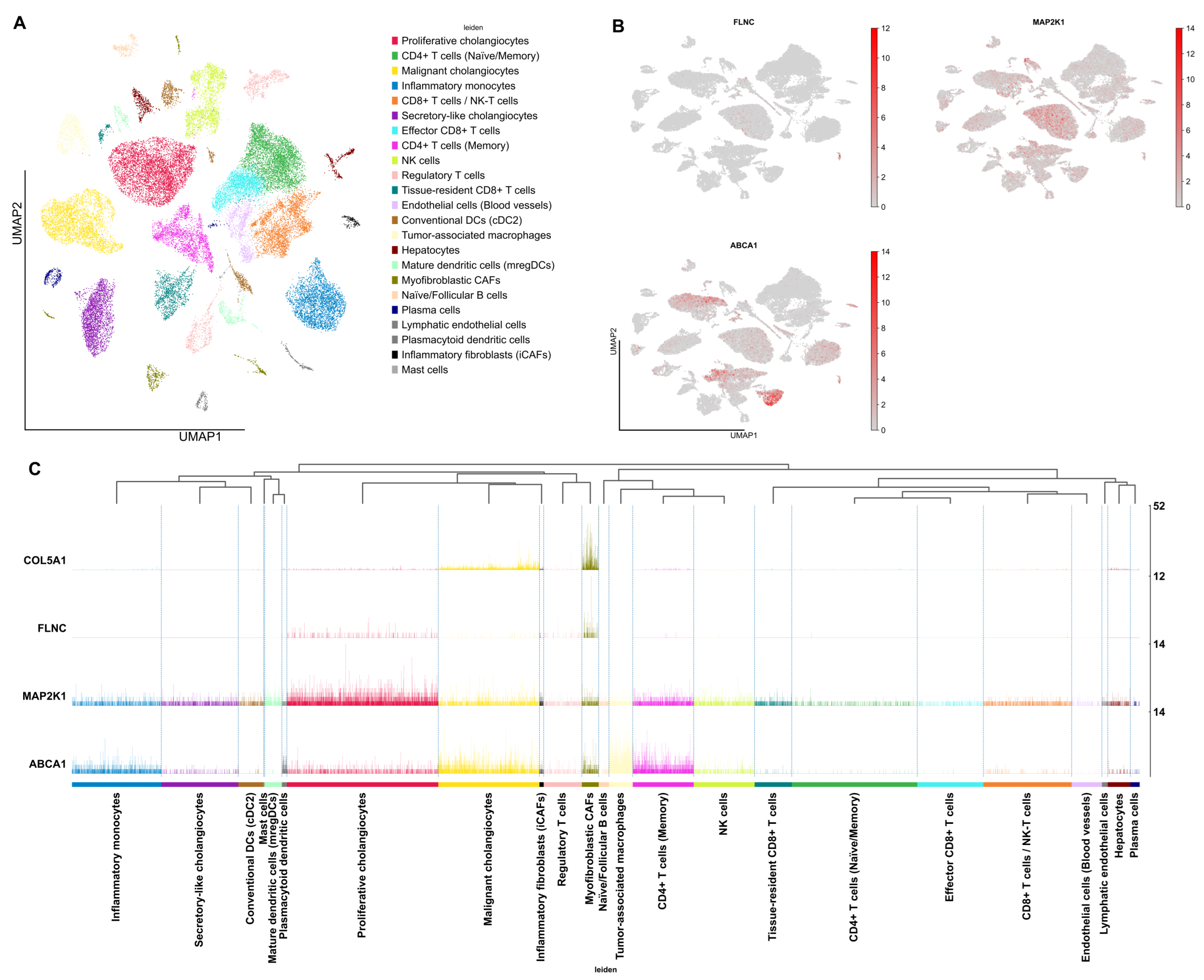
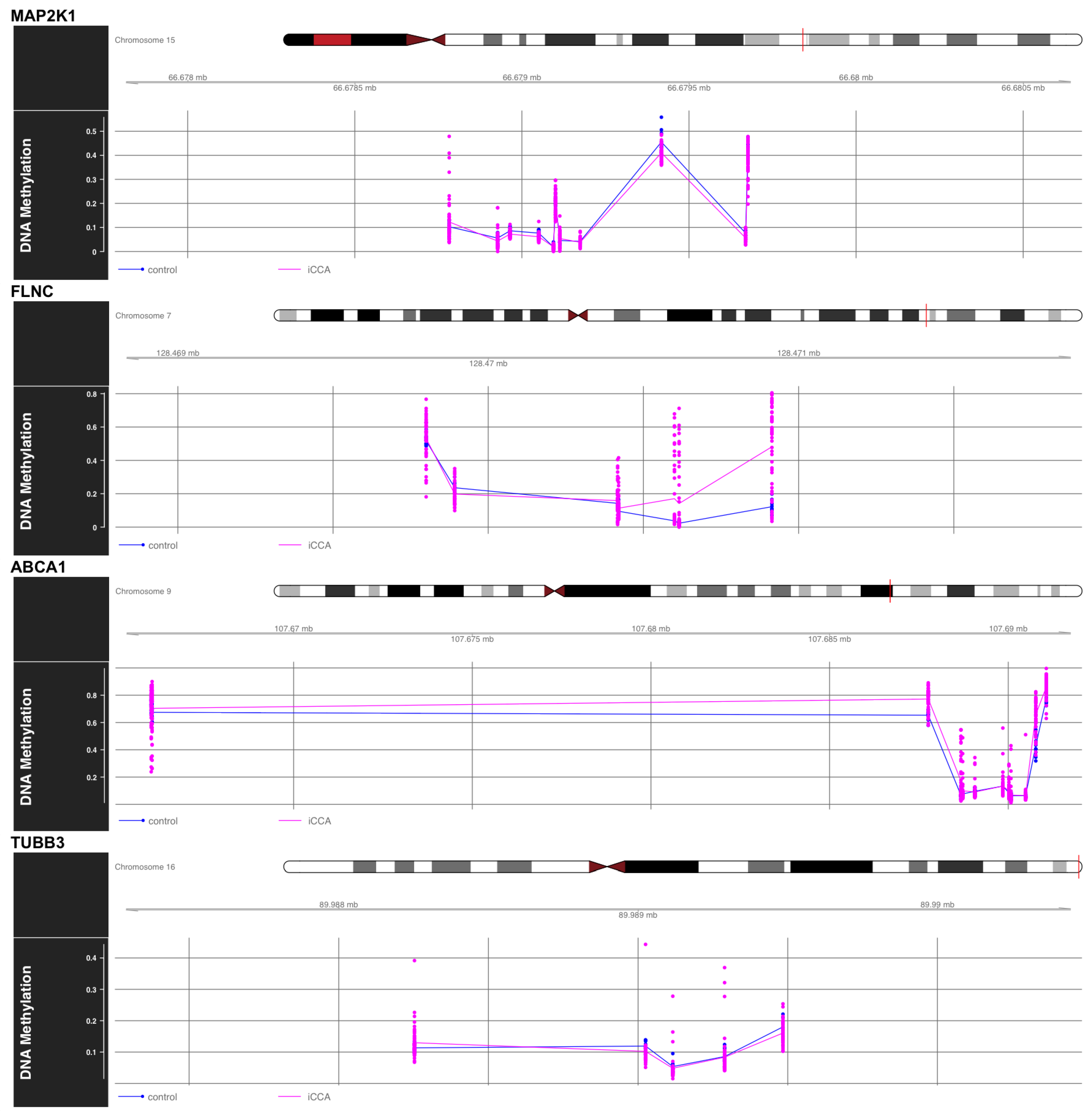
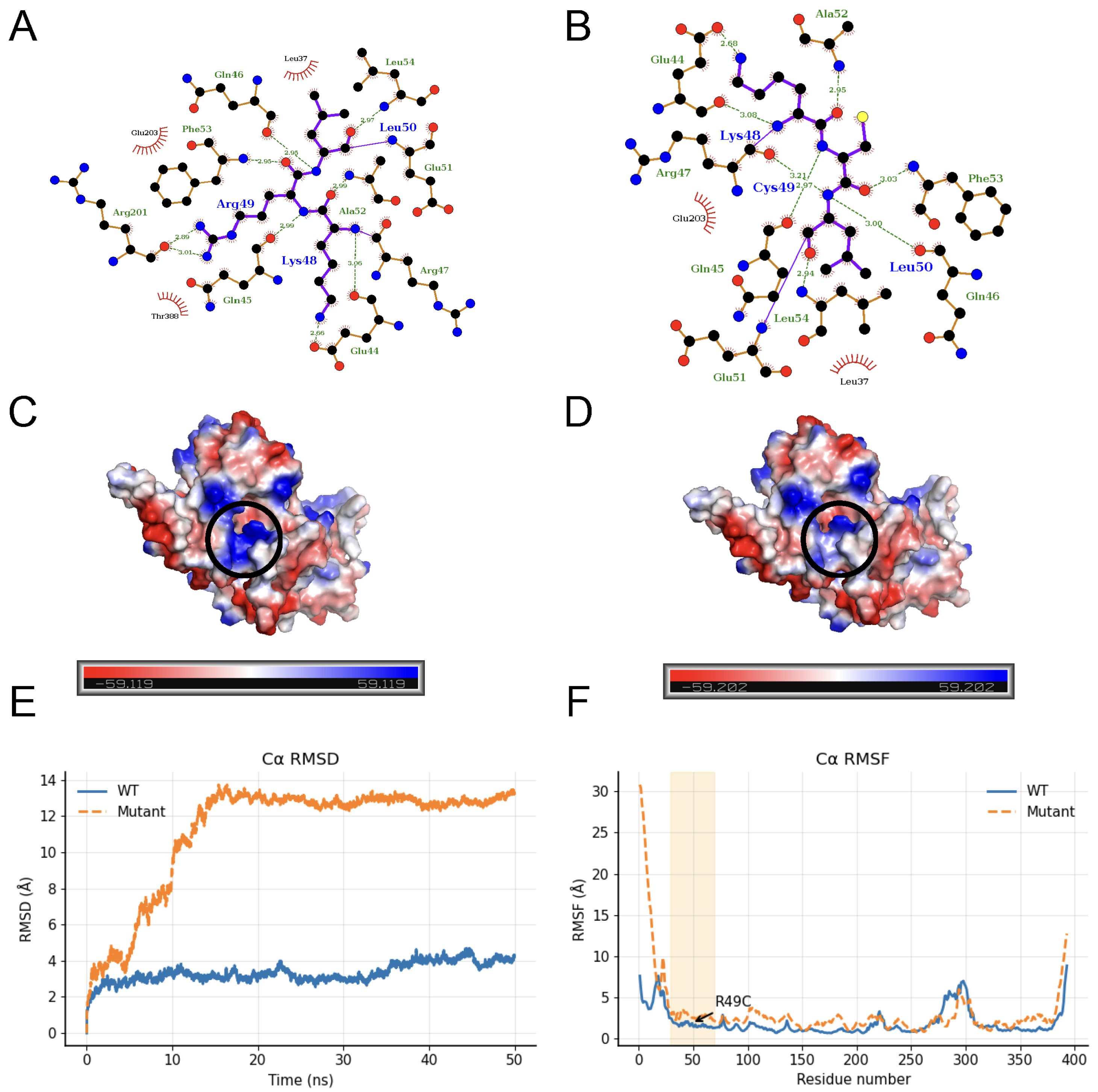
2. Results
3. Discussion
4. Materials and Methods
4.1. Data Collection and Variant Extraction
4.2. Variation Annotation
4.3. Performance Evaluation of Predictive Tools
4.4. Transcriptomic and Pathway Analysis of Differentially Expressed Genes in Bile Duct Tumors
4.5. Single-Cell RNA-Seq Analysis of iCCA
4.6. Differential DNA Methylation Analysis
4.7. Structural Modeling and Molecular Dynamics Simulations
5. Conclusions
5.1. Limitation
5.2. Future Aspect
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CCA | Cholangiocarcinoma |
| iCCA | Intrahepatic cholangiocarcinoma |
| TCGA | The Cancer Genome Atlas |
| VUS | Variants of uncertain significance |
| DEGs | Differentially expressed genes |
| DMPs | Differentially methylated positions |
| DMRs | Differentially methylated regions |
| CAF | Cancer-associated fibroblasts |
| HDL | High-density lipoprotein |
| AUC | Area under the curve |
| UMAP | Uniform Manifold Approximation and Projection |
| PCA | Principal Component Analysis |
| GSEA | Gene set enrichment analysis |
| scRNA-seq | Single-cell RNA sequencing |
| RNA-seq | RNA sequencing |
| HVGs | Highly variable genes |
| RMSD | Root Mean Square Deviation |
| RMSF | Root Mean Square Fluctuation |
| MD | Molecular dynamics |
| FDR | False discovery rate |
| MAF | Minor allele frequency |
| SNPs | Single nucleotide polymorphisms |
| NES | Normalized enrichment score |
| MAPK | Mitogen-activated protein kinase |
| GDC | Genomic Data Commons |
| GEO | Gene Expression Omnibus |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Brindley, P.J.; Bachini, M.; Ilyas, S.I.; Khan, S.A.; Loukas, A.; Sirica, A.E.; Teh, B.T.; Wongkham, S.; Gores, G.J. Cholangiocarcinoma. Nat. Rev. Dis. Prim. 2021, 7, 65. [Google Scholar] [CrossRef]
- Chan-On, W.; Nairismägi, M.L.; Ong, C.K.; Lim, W.K.; Dima, S.; Pairojkul, C.; Lim, K.H.; McPherson, J.R.; Cutcutache, I.; Heng, H.L.; et al. Exome sequencing identifies distinct mutational patterns in liver fluke–related and non-infection-related bile duct cancers. Nat. Genet. 2013, 45, 1474–1478. [Google Scholar] [CrossRef] [PubMed]
- Statistics About Bile Duct Cancer|Cholangiocarcinoma Stats. Available online: https://www.cancer.org/cancer/types/bile-duct-cancer/about/key-statistics.html (accessed on 2 July 2025).
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Everhart, J.E.; Ruhl, C.E. Burden of digestive diseases in the United States Part III: Liver, biliary tract, and pancreas. Gastroenterology 2009, 136, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Carotenuto, M.; Sacco, A.; Forgione, L.; Normanno, N. Genomic alterations in cholangiocarcinoma: Clinical significance and relevance to therapy. Explor. Target. Antitumor Ther. 2022, 3, 200. [Google Scholar] [CrossRef]
- Chen, S.; Francioli, L.C.; Goodrich, J.K.; Collins, R.L.; Kanai, M.; Wang, Q.; Karczewski, K.J.; Farjoun, Y.; Banks, E.; Donnelly, S.; et al. A Genomic Mutational Constraint Map Using Variation in 76,156 Human Genomes. Nature 2024, 625, 92–100, Erratum in Nature 2024, 626, E1. [Google Scholar] [CrossRef]
- Frazer, J.; Notin, P.; Dias, M.; Gomez, A.; Min, J.K.; Brock, K.; Gal, Y.; Marks, D.S. Disease variant prediction with deep generative models of evolutionary data. Nature 2021, 599, 91–95, Erratum in Nature 2022, 601, E7. [Google Scholar] [CrossRef]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, D.; Tanyalcin, I.; Ferté, J.; Gazzo, A.; Orlando, G.; Lenaerts, T.; Rooman, M.; Vranken, W. DEOGEN2: Prediction and interactive visualization of single amino acid variant deleteriousness in human proteins. Nucleic Acids Res. 2017, 45, W201–W206. [Google Scholar] [CrossRef]
- Ioannidis, N.M.; Rothstein, J.H.; Pejaver, V.; Middha, S.; McDonnell, S.K.; Baheti, S.; Musolf, A.; Li, Q.; Holzinger, E.; Karyadi, D.; et al. REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 2016, 99, 877–885. [Google Scholar] [CrossRef]
- Tordai, H.; Torres, O.; Csepi, M.; Padányi, R.; Lukács, G.L.; Hegedűs, T. Analysis of AlphaMissense data in different protein groups and structural context. Sci. Data 2024, 11, 495. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Kelly, G.L.; Janic, A.; Herold, M.J.; Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018, 25, 104–113. [Google Scholar] [CrossRef]
- Zhao, M.; Mishra, L.; Deng, C.-X. The role of TGF-β/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018, 14, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.L.; Rothman, A.L.; Xie, J.; Goodrich, L.V.; Bare, J.W.; Bonifas, J.M.; Quinn, A.G.; Myers, R.M.; Cox, D.R.; Epstein, E.H., Jr.; et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 1996, 272, 1668–1671. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, H.; Oda, E.; Ueda, S.; Tamura, G.; Maesawa, C.; Muto, T.; Taniguchi, T.; Tanaka, N. Functionally inactivating point mutation in the tumor-suppressor IRF-1 gene identified in human gastric cancer. Int. J. Cancer 1998, 77, 522–527. [Google Scholar] [CrossRef]
- Pylayeva-Gupta, Y.; Grabocka, E.; Bar-Sagi, D. RAS oncogenes: Weaving a tumorigenic web. Nat. Rev. Cancer 2011, 11, 761–774. [Google Scholar] [CrossRef]
- Sasaki, H.; Hikosaka, Y.; Kawano, O.; Moriyama, S.; Yano, M.; Fujii, Y. MEK1 and AKT2 mutations in Japanese lung cancer. J. Thorac. Oncol. 2010, 5, 597–600. [Google Scholar] [CrossRef]
- Hancox, R.J.; Poulton, R.; Welch, D.; Olova, N.; McLachlan, C.R.; Greene, J.M.; Sears, M.R.; Caspi, A.; Moffitt, T.E.; Robertson, S.P.; et al. Accelerated, decline, in, lung, function, in, cigarette, smokers, is, assoc, iated with TP53/MDM2 polymorphisms. Hum. Genet. 2009, 126, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Turcan, S.; Rohle, D.; Goenka, A.; Walsh, L.A.; Fang, F.; Yilmaz, E.; Campos, C.; Fabius, A.W.; Lu, C.; Ward, P.S.; et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012, 483, 479–483. [Google Scholar] [CrossRef]
- Ni, P.; Li, L.; Zhu, Y.; Du, K.; Nov, P.; Wang, D.; Wang, C.; Kou, Q.; Li, Y.; Zhang, Y.; et al. Unveiling the multifaceted role of the FLNC gene: Implications for cancer diagnosis and prognosis. Eur. J. Med. Res. 2025, 30, 608. [Google Scholar] [CrossRef]
- Takahara, K.; Hoffman, G.G.; Greenspan, D.S. Complete structural organization of the human alpha 1(V) collagen gene (COL5A1): Divergence from the conserved organization of other characterized fibrillar collagen genes. Genomics 1995, 29, 588–597. [Google Scholar] [CrossRef]
- LaFrance, B.J.; Roostalu, J.; Henkin, G.; Greber, B.J.; Zhang, R.; Normanno, D.; McCollum, C.O.; Surrey, T.; Nogales, E. Structural transitions in the GTP cap visualized by cryo-electron microscopy of catalytically inactive microtubules. Proc. Natl. Acad. Sci. USA 2022, 119, e2114994119. [Google Scholar] [CrossRef]
- Dooley, S.A.; Engevik, K.A.; Digrazia, J.; Stubler, R.; Kaji, I.; Krystofiak, E.; Engevik, A.C. Myosin 5b is required for proper localization of the intermicrovillar adhesion complex in the intestinal brush border. Am. J. Physiol.–Gastrointest. Liver Physiol. 2022, 323, G501–G510. [Google Scholar] [CrossRef]
- Mak, K.M.; Png, C.Y.; Lee, D.J. Type V collagen in health, disease, and fibrosis. Anat. Rec. 2016, 299, 613–629. [Google Scholar] [CrossRef]
- Woodard, D.R.; Daniel, S.; Nakahara, E.; Abbas, A.; DiCesare, S.M.; Collier, G.E.; Hulleman, J.D. A loss-of-function cysteine mutant in fibulin-3 (EFEMP1) forms aberrant extracellular disulfide-linked homodimers and alters extracellular matrix composition. Hum. Mutat. 2022, 43, 1945–1955. [Google Scholar] [CrossRef]
- Eason, D.D.; Shepherd, A.T.; Blanck, G. Interferon regulatory factor 1 tryptophan 11 to arginine point mutation abolishes DNA binding. Biochim. Biophys. Acta 1999, 1446, 140–144. [Google Scholar] [CrossRef]
- Liu, F.; Pouponnot, C.; Massague, J. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional complexes. Genes Dev. 1997, 11, 3157–3167. [Google Scholar] [CrossRef] [PubMed]
- Pidasheva, S.; Grant, M.; Canaff, L.; Ercan, O.; Kumar, U.; Hendy, G.N. Calcium-sensing receptor dimerizes in the endoplasmic reticulum: Biochemical and biophysical characterization of CASR mutants retained intracellularly. Hum. Mol. Genet. 2006, 15, 2200–2209. [Google Scholar] [CrossRef]
- Ovchinnikov, Y.A.; Monastyrskaya, G.S.; Broude, N.E.; Ushkaryov, Y.A.; Melkov, A.M.; Smirnov, Y.V.; Malyshev, I.V.; Allikmets, R.L.; Kostina, M.B.; Dulubova, I.E.; et al. Family of human Na+,K+-ATPase genes. Structure of the gene for the catalytic subunit (alpha III-form) and its relationship with structural features of the protein. FEBS Lett. 1988, 233, 87–94. [Google Scholar] [CrossRef]
- Voskoboinik, I.; Strausak, D.; Greenough, M.; Brooks, H.; Petris, M.; Smith, S.; Mercer, J.F.; Camakaris, J. Functional analysis of the N-terminal CXXC metal-binding motifs in the human Menkes copper-transporting P-type ATPase expressed in cultured mammalian cells. J. Biol. Chem. 1999, 274, 22008–22012, Erratum in J. Biol. Chem. 1999, 274, 36030. [Google Scholar] [CrossRef]
- Pan, S.; Li, T.-J. PTCH1 mutations in odontogenic keratocysts: Are they related to epithelial cell proliferation? Oral Oncol. 2009, 45, 861–865. [Google Scholar] [CrossRef]
- Gremer, L.; Merbitz-Zahradnik, T.; Dvorsky, R.; Cirstea, I.C.; Kratz, C.P.; Zenker, M.; Wittinghofer, A.; Ahmadian, M.R. Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum. Mutat. 2011, 32, 33–43. [Google Scholar] [CrossRef]
- Dentici, M.L.; Sarkozy, A.; Pantaleoni, F.; Carta, C.; Lepri, F.; Ferese, R.; Cordeddu, V.; Martinelli, S.; Briuglia, S.; Digilio, M.C.; et al. Spectrum of MEK1 and MEK2 gene mutations in cardio-facio-cutaneous syndrome and genotype–phenotype correlations. Eur. J. Hum. Genet. 2009, 17, 733–740. [Google Scholar] [CrossRef]
- Yuan, J.; Ng, W.H.; Tian, Z.; Yap, J.; Baccarini, M.; Chen, Z.; Hu, J. Activating mutations in MEK1 enhance homodimerization and promote tumorigenesis. Sci. Signal. 2018, 11, eaar6795. [Google Scholar] [CrossRef]
- Zen, Y.; Britton, D.; Mitra, V.; Pike, I.; Sarker, D.; Itoh, T.; Heaton, N.; Quaglia, A. Tubulin β-III: A novel immunohistochemical marker for intrahepatic peripheral cholangiocarcinoma. Histopathology 2014, 65, 784–792. [Google Scholar] [CrossRef]
- Shroff, R.T.; King, G.; Colby, S.; Scott, A.J.; Borad, M.; Goff, L.; Matin, K.; Mahipal, A.; Kalyan, A.; Javle, M.M.; et al. SWOG S1815: A Phase III Randomized Trial of Gemcitabine, Cisplatin, and Nab-Paclitaxel Versus Gemcitabine and Cisplatin in Newly Diagnosed, Advanced Biliary Tract Cancers. J. Clin. Oncol. 2025, 43, 536–544. [Google Scholar] [CrossRef]
- Choi, J.H.; Thung, S.N. Recent advances in pathology of intrahepatic cholangiocarcinoma. Cancers 2024, 16, 1537. [Google Scholar] [CrossRef]
- Sirica, A.E.; Gores, G.J. Desmoplastic stroma and cholangiocarcinoma: Clinical implications and therapeutic targeting. Hepatology 2014, 59, 2397–2402. [Google Scholar] [CrossRef]
- Zhu, G.; Wang, Y.; Wang, Y.; Huang, H.; Li, B.; Chen, P.; Chen, C.; Zhang, H.; Li, Y.; Liu, H.; et al. Myofibroblasts derived type V collagen promoting tissue mechanical stress and facilitating metastasis and therapy resistance of lung adenocarcinoma cells. Cell Death Dis. 2024, 15, 493. [Google Scholar] [CrossRef]
- Subrungruang, I.; Thawornkuno, C.; Chawalitchewinkoon-Petmitr, P.; Pairojkul, C.; Wongkham, S.; Petmitr, S. Gene expression profiling of intrahepatic cholangiocarcinoma. Asian Pac. J. Cancer Prev. 2013, 14, 557–563. [Google Scholar] [CrossRef]
- Vitali, E.; Franceschini, B.; Milana, F.; Soldani, C.; Polidoro, M.A.; Carriero, R.; Kunderfranco, P.; Trivellin, G.; Costa, G.; Milardi, G.; et al. Filamin A is involved in human intrahepatic cholangiocarcinoma aggressiveness and progression. Liver Int. 2024, 44, 518–531. [Google Scholar] [CrossRef]
- Qiao, J.; Cui, S.J.; Xu, L.L.; Chen, S.J.; Yao, J.; Jiang, Y.H.; Peng, G.; Fang, C.Y.; Yang, P.Y.; Liu, F. Filamin C, a dysregulated protein in cancer revealed by label-free quantitative proteomic analyses of human gastric cancer cells. Oncotarget 2014, 6, 1171. [Google Scholar] [CrossRef]
- Qi, Y.; Xu, F.; Chen, L.; Li, Y.; Xu, Z.; Zhang, Y.; Wei, W.; Su, N.; Zhang, T.; Fan, F.; et al. Quantitative proteomics reveals FLNC as a potential progression marker for the development of hepatocellular carcinoma. Oncotarget 2016, 7, 68242. [Google Scholar] [CrossRef]
- Seeree, P.; Janvilisri, T.; Kangsamaksin, T.; Tohtong, R.; Kumkate, S. Downregulation of ABCA1 and ABCG1 transporters by simvastatin in cholangiocarcinoma cells. Oncol. Lett. 2019, 18, 5173–5184. [Google Scholar] [CrossRef]
- Zhu, A.X.; Borger, D.R.; Kim, Y.; Cosgrove, D.; Ejaz, A.; Alexandrescu, S.; Groeschl, R.T.; Deshpande, V.; Lindberg, J.M.; Ferrone, C.; et al. Genomic profiling of intrahepatic cholangiocarcinoma: Refining prognosis and identifying therapeutic targets. Ann. Surg. Oncol. 2014, 21, 3827–3834. [Google Scholar] [CrossRef]
- Quinn, L.M.; Haldenby, S.; Antzcak, P.; Fowler, A.; Bullock, K.; Kenny, J.; Gilbert, T.; Andrews, T.; Diaz-Nieto, R.; Fenwick, S.; et al. Genomic profiling of idiopathic peri-hilar cholangiocarcinoma reveals new targets and mutational pathways. Sci. Rep. 2023, 13, 6681. [Google Scholar] [CrossRef]
- Doherty, M.K.; Tam, V.C.; McNamara, M.G.; Jang, R.; Hedley, D.; Chen, E.; Dhani, N.; Tang, P.; Sim, H.-W.; O’Kane, G.M.; et al. Randomised, Phase II study of selumetinib, an oral inhibitor of MEK, in combination with cisplatin and gemcitabine chemotherapy for patients with advanced biliary tract cancer. Br. J. Cancer 2022, 127, 1473–1478. [Google Scholar] [CrossRef]
- Yarchoan, M.; Cope, L.; Ruggieri, A.N.; Anders, R.A.; Noonan, A.M.; Goff, L.W.; Goyal, L.; Lacy, J.; Li, D.; Patel, A.K.; et al. Multicenter randomized phase II trial of atezolizumab with or without cobimetinib in biliary tract cancers. J. Clin. Investig. 2021, 131, e152670. [Google Scholar] [CrossRef]
- Heumann, T.R.; Yarchoan, M.; Murray, J.; Lu, J.; Li, D.; Kunk, P.R.; Azad, N.S.; Kalyan, A.; Wang, H.; Sharon, E.; et al. A randomized phase 2 study of combination atezolizumab and varlilumab (CDX-1127) with or without addition of cobimetinib in previously treated unresectable biliary tract cancer (ETCTN 10476). J. Clin. Oncol. 2024, 42 (Suppl. 16), 4017. [Google Scholar] [CrossRef]
- Goyal, L.; Meric-Bernstam, F.; Hollebecque, A.; Valle, J.W.; Morizane, C.; Karasic, T.B.; Bridgewater, J.A.; Wacheck, V.; He, Y.; Liu, M.; et al. Futibatinib for FGFR2-rearranged intrahepatic cholangiocarcinoma. N. Engl. J. Med. 2023, 388, 228–239. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Jagadeesh, K.A.; Wenger, A.M.; Berger, M.J.; Guturu, H.; Stenson, P.D.; Cooper, D.N.; Bernstein, J.A.; Bejerano, G. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016, 48, 1581–1586. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhi, D.; Wang, K.; Liu, X. MetaRNN: Differentiating rare pathogenic and rare benign missense SNVs and InDels using deep learning. Genome Med. 2022, 14, 115. [Google Scholar] [CrossRef]
- Carter, H.; Douville, C.; Stenson, P.D.; Cooper, D.N.; Karchin, R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genom. 2013, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Schubach, M.; Maass, T.; Nazaretyan, L.; Röner, S.; Kircher, M. CADD v1. 7: Using protein language models, regulatory CNNs and other nucleotide-level scores to improve genome-wide variant predictions. Nucleic Acids Res. 2024, 52, D1143–D1154. [Google Scholar] [CrossRef]
- Kramer, O. Machine Learning for Evolution Strategies; Springer: Berlin, Germany, 2016; Volume 20. [Google Scholar]
- Liu, X.; Li, C.; Mou, C.; Dong, Y.; Tu, Y. dbNSFP v4: A comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020, 12, 103. [Google Scholar] [CrossRef]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g: Profiler—A web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef]
- Kanehisa, M. The KEGG database. In Proceedings of the ‘In Silico’ Simulation of Biological Processes: Novartis Foundation Symposium 247, London, UK, 15 November 2002; pp. 91–103. [Google Scholar]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Köhler, S.; Gargano, M.; Matentzoglu, N.; Carmody, L.C.; Lewis-Smith, D.; Vasilevsky, N.A.; Danis, D.; Balagura, G.; Baynam, G.; Brower, A.M.; et al. The human phenotype ontology in 2021. Nucleic Acids Res. 2021, 49, D1207–D1217. [Google Scholar] [CrossRef] [PubMed]
- Martens, M.; Ammar, A.; Riutta, A.; Waagmeester, A.; Slenter, D.N.; Hanspers, K.; Miller, R.A.; Digles, D.; Lopes, E.N.; Ehrhart, F.; et al. WikiPathways: Connecting communities. Nucleic Acids Res. 2021, 49, D613–D621. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Fertig, E.J.; Jaffe, A.E.; Storey, J.D.; Zhang, Y.; Torres, L.C. sva: Surrogate variable analysis. R Package Version 2019, 3, 882–883. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Sia, D.; Losic, B.; Moeini, A.; Cabellos, L.; Hao, K.; Revill, K.; Bonal, D.; Miltiadous, O.; Zhang, Z.Y.; Hoshida, Y.; et al. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat. Commun. 2015, 6, 6087. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Yao, W.; Liu, X.; He, Y.; Tian, M.; Lu, S.; Wang, Q.; Zheng, Y.; Lv, Z.; Hao, C.; Xue, D.; et al. ScRNA-seq and bulk RNA-seq reveal the characteristics of ferroptosis and establish a risk signature in cholangiocarcinoma. Mol. Ther. Oncolytics 2022, 27, 48–60. [Google Scholar] [CrossRef]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018, 19, 1–5. [Google Scholar] [CrossRef]
- Mackiewicz, A.; Ratajczak, W. Principal components analysis (PCA). Comput. Geosci. 1993, 19, 303–342. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Traag, V.A.; Waltman, L.; Van Eck, N.J. From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Goeppert, B.; Toth, R.; Singer, S.; Albrecht, T.; Lipka, D.B.; Lutsik, P.; Brocks, D.; Baehr, M.; Muecke, O.; Assenov, Y.; et al. Integrative analysis defines distinct prognostic subgroups of intrahepatic cholangiocarcinoma. Hepatology 2019, 69, 2091–2106. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, X.; Wang, Y. A framework for analyzing DNA methylation data from Illumina Infinium HumanMethylation450 BeadChip. BMC Bioinform. 2018, 19, 15–22. [Google Scholar] [CrossRef]
- Fortin, J.P.; Triche Jr, T.J.; Hansen, K.D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 2017, 33, 558–560. [Google Scholar] [CrossRef]
- Hansen, K.D.; Aryee, M.; Timp, W. minfiData: Example Data for the Illumina Methylation 450 k Array. R Package Version 0.54.0. Available online: https://bioconductor.org/packages/minfiData (accessed on 6 June 2025).
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Peters, T.J.; Buckley, M.J.; Chen, Y.; Smyth, G.K.; Goodnow, C.C.; Clark, S.J. Calling differentially methylated regions from whole genome bisulphite sequencing with DMRcate. Nucleic Acids Res. 2021, 49, e109. [Google Scholar] [CrossRef]
- Martorell-Marugán, J.; González-Rumayor, V.; Carmona-Sáez, P. mCSEA: Detecting subtle differentially methylated regions. Bioinformatics 2019, 35, 3257–3262. [Google Scholar] [CrossRef]
- Hahne, F.; Ivanek, R. Visualizing genomic data using Gviz and bioconductor. In Statistical Genomics: Methods and Protocols; Springer: New York, NY, USA, 2016; pp. 335–351. [Google Scholar]
- Wickham, H.; Wickham, M.H. Package ‘Stringr’. 2019. Available online: https://github.com/tidyverse/stringr (accessed on 30 July 2025).
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Campo, M.G. Structural and dynamic properties of SPC/E water. Pap. Phys. 2010, 2, 020001. [Google Scholar] [CrossRef]
- Wall, M.E.; Calabró, G.; Bayly, C.I.; Mobley, D.L.; Warren, G.L. Biomolecular Solvation Structure Revealed by Molecular Dynamics Simulations. J. Am. Chem. Soc. 2019, 141, 4711–4720. [Google Scholar] [CrossRef]
- Aier, I.; Varadwaj, P.K.; Raj, U. Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci. Rep. 2016, 6, 34984. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Carugo, O. How root-mean-square distance (rmsd) values depend on the resolution of protein structures that are compared. J. Appl. Crystallogr. 2003, 36, 125–128. [Google Scholar] [CrossRef]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chan, H.C.S.; Hu, Z. Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Buß, O.; Rudat, J.; Ochsenreither, K. FoldX as protein engineering tool: Better than random based approaches? Comput. Struct. Biotechnol. J. 2018, 16, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Flyvbjerg, H.; Petersen, H.G. Error estimates on averages of correlated data. J. Chem. Phys. 1989, 91, 461. [Google Scholar] [CrossRef]
| Model | AUC | Accuracy | Precision | Recall | F1 |
|---|---|---|---|---|---|
| M-CAP | 0.914286 | 0.421053 | 1.000000 | 0.214286 | 0.352941 |
| MetaRNN | 0.871429 | 0.894737 | 1.000000 | 0.857143 | 0.923077 |
| VEST4 | 0.866667 | 0.869565 | 1.000000 | 0.833333 | 0.909091 |
| CADD | 0.644444 | 0.782609 | 0.842105 | 0.888889 | 0.864865 |
| REVEL | 0.950000 | 0.684211 | 1.000000 | 0.571429 | 0.727273 |
| AlphaMissense | 0.857143 | 0.842105 | 1.000000 | 0.785714 | 0.880000 |
| SIFT | 0.928571 | 0.894737 | 1.000000 | 0.857143 | 0.923077 |
| Polyphen | 0.857143 | 0.789474 | 1.000000 | 0.714286 | 0.833333 |
| EVE | 1.000000 | 0.846154 | 1.000000 | 0.818182 | 0.900000 |
| DEOGEN2 | 0.907692 | 0.888889 | 1.000000 | 0.846154 | 0.916667 |
| MetaLR | 0.892857 | 0.684211 | 0.900000 | 0.642857 | 0.750000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bülbül, A.; Gerdan, G.; Portakal, C.; Bajrami, S.; Boylu Akyerli, C. A Comprehensive Analysis of Novel Variations Associated with Bile Duct Cancer: Insights into Expression, Methylation, and 3D Protein Structure. Int. J. Mol. Sci. 2025, 26, 11244. https://doi.org/10.3390/ijms262311244
Bülbül A, Gerdan G, Portakal C, Bajrami S, Boylu Akyerli C. A Comprehensive Analysis of Novel Variations Associated with Bile Duct Cancer: Insights into Expression, Methylation, and 3D Protein Structure. International Journal of Molecular Sciences. 2025; 26(23):11244. https://doi.org/10.3390/ijms262311244
Chicago/Turabian StyleBülbül, Alper, Gizel Gerdan, Cansu Portakal, Sudenaz Bajrami, and Cemaliye Boylu Akyerli. 2025. "A Comprehensive Analysis of Novel Variations Associated with Bile Duct Cancer: Insights into Expression, Methylation, and 3D Protein Structure" International Journal of Molecular Sciences 26, no. 23: 11244. https://doi.org/10.3390/ijms262311244
APA StyleBülbül, A., Gerdan, G., Portakal, C., Bajrami, S., & Boylu Akyerli, C. (2025). A Comprehensive Analysis of Novel Variations Associated with Bile Duct Cancer: Insights into Expression, Methylation, and 3D Protein Structure. International Journal of Molecular Sciences, 26(23), 11244. https://doi.org/10.3390/ijms262311244






