Feature-Guided Machine Learning for Studying Passive Blood–Brain Barrier Permeability to Aid Drug Discovery
Abstract
1. Introduction
2. Results and Discussions
2.1. Base Model Comparison
2.2. Impact of Resampling Techniques on Model Performance
2.3. Feature Ranking and Interpretation of Key Molecular Features
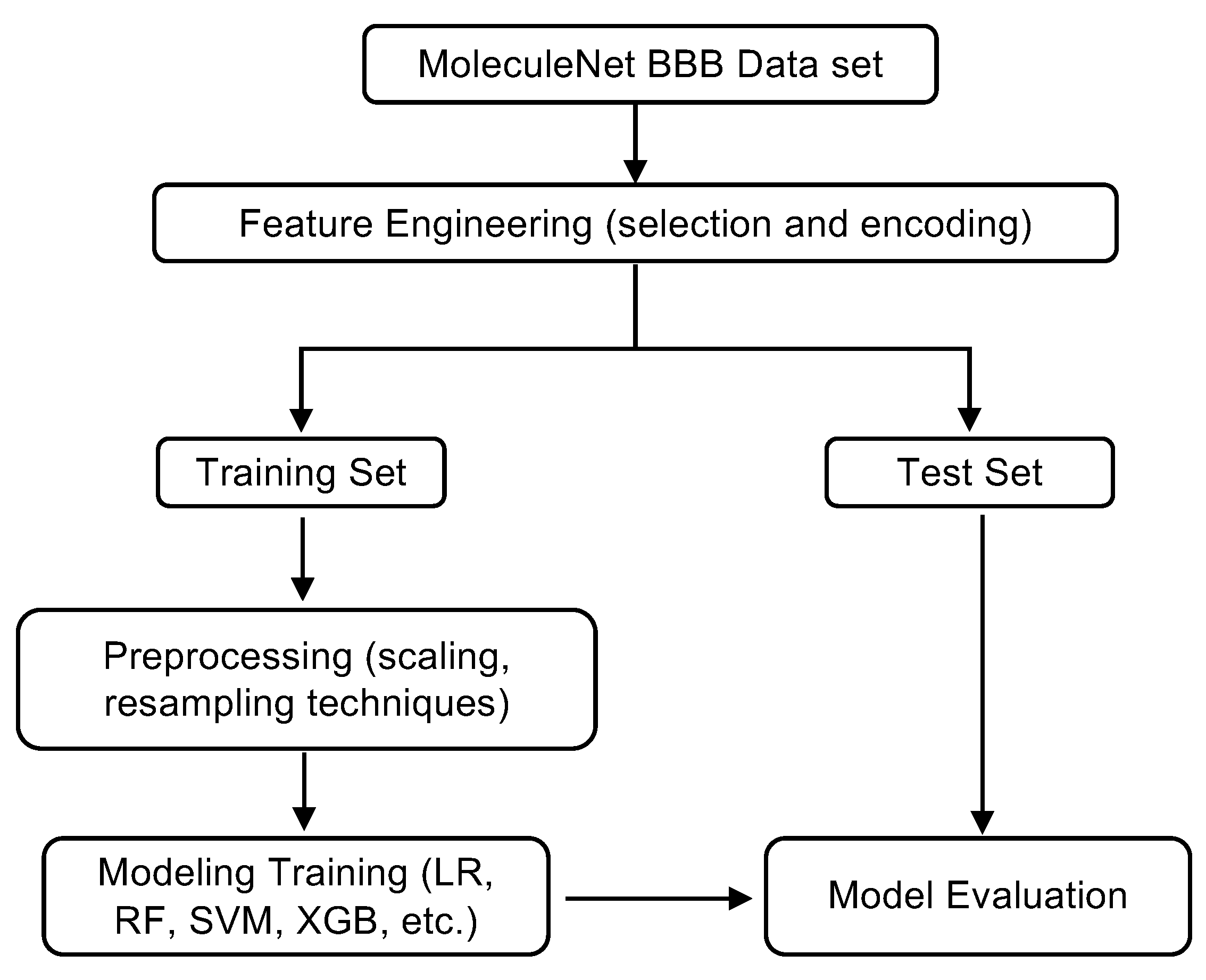
3. Methods
3.1. Dataset Description and Molecular Representations
3.2. Feature Engineering, Preprocessing, Model Training and Evaluation
3.3. Model Evaluation and Interpretation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | Area Under the Curve |
| BBB | Blood-Brain Barrier |
| BBBP | Blood-Brain Barrier Penetration |
| CNS | Central Neural System |
| GaussianNB | Gaussian Naive Bayes |
| k-NN | k-Nearest Neighbors |
| LightGBM | Light Gradient Boosting Machine |
| logP | Lipophilicity |
| MACCS | Molecular ACCess System |
| MD | Molecular Dynamics |
| MFP | Morgan FingerPrint |
| MLP | Multi-Layer Perceptron |
| RBF | Radial Basis Function |
| ROC | Receiver Operating Characteristic |
| SMILES | Simplified Molecular Input Line Entry System |
| SMOTE | Synthetic Minority Oversampling Technique |
| SVM | Support Vector Machines |
| TPSA | Topological Polar Surface Area |
| XGBoost | eXtreme Gradient Boosting |
References
- Jeffrey, P.; Summerfield, S. Assessment of the blood–brain barrier in CNS drug discovery. Neurobiol. Dis. 2010, 37, 33–37. [Google Scholar] [CrossRef]
- Alavijeh, M.S.; Chishty, M.; Qaiser, M.Z.; Palmer, A.M. Drug metabolism and pharmacokinetics, the blood–brain barrier, and central nervous system drug discovery. NeuroRx 2005, 2, 554–571. [Google Scholar] [CrossRef]
- Pardridge, W.M. CNS drug design based on principles of blood–brain barrier transport. J. Neurochem. 1998, 70, 1781–1792. [Google Scholar] [CrossRef]
- Pardridge, W.M. Blood–brain barrier endogenous transporters as therapeutic targets: A new model for small molecule CNS drug discovery. Expert Opin. Ther. Targets 2015, 19, 1059–1072. [Google Scholar] [CrossRef]
- Pardridge, W.M. The blood–brain barrier: Bottleneck in brain drug development. NeuroRx 2005, 2, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Cecchelli, R.; Berezowski, V.; Lundquist, S.; Culot, M.; Renftel, M.; Dehouck, M.P.; Fenart, L. Modelling of the blood–brain barrier in drug discovery and development. Nat. Rev. Drug Discov. 2007, 6, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Pardridge, W.M. Alzheimer’s disease drug development and the problem of the blood–brain barrier. Alzheimer’s Dement. 2009, 5, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Markou, A.; Chiamulera, C.; Geyer, M.A.; Tricklebank, M.; Steckler, T. Removing obstacles in neuroscience drug discovery: The future path for animal models. Neuropsychopharmacology 2009, 34, 74–89. [Google Scholar] [CrossRef]
- Spencer, B.J.; Verma, I.M. Targeted delivery of proteins across the blood–brain barrier. Proc. Natl. Acad. Sci. USA 2007, 104, 7594–7599. [Google Scholar] [CrossRef]
- Lim, S.; Kim, W.J.; Kim, Y.H.; Lee, S.; Koo, J.H.; Lee, J.A.; Yoon, H.; Kim, D.H.; Park, H.J.; Kim, H.M.; et al. dNP2 is a blood–brain barrier-permeable peptide enabling ctCTLA-4 protein delivery to ameliorate experimental autoimmune encephalomyelitis. Nat. Commun. 2015, 6, 8244. [Google Scholar] [CrossRef]
- Pardridge, W.M. Blood-brain barrier drug targeting: The future of brain drug development. Mol. Interv. 2003, 3, 90. [Google Scholar] [CrossRef] [PubMed]
- Banks, W.A. Characteristics of compounds that cross the blood–brain barrier. BMC Neurol. 2009, 9, S3. [Google Scholar] [CrossRef]
- Ohtsuki, S.; Terasaki, T. Contribution of carrier-mediated transport systems to the blood–brain barrier as a supporting and protecting interface for the brain; importance for CNS drug discovery and development. Pharm. Res. 2007, 24, 1745–1758. [Google Scholar] [CrossRef] [PubMed]
- Kaisar, M.A.; Sajja, R.K.; Prasad, S.; Abhyankar, V.V.; Liles, T.; Cucullo, L. New experimental models of the blood–brain barrier for CNS drug discovery. Expert Opin. Drug Discov. 2017, 12, 89–103. [Google Scholar] [CrossRef]
- Garberg, P.; Ball, M.; Borg, N.; Cecchelli, R.; Fenart, L.; Hurst, R.; Lindmark, T.; Mabondzo, A.; Nilsson, J.; Raub, T.; et al. In vitro models for the blood–brain barrier. Toxicol. Vitr. 2005, 19, 299–334. [Google Scholar] [CrossRef]
- Wilhelm, I.; Krizbai, I.A. In vitro models of the blood–brain barrier for the study of drug delivery to the brain. Mol. Pharm. 2014, 11, 1949–1963. [Google Scholar] [CrossRef]
- Dehouck, M.P.; Jolliet-Riant, P.; Brée, F.; Fruchart, J.C.; Cecchelli, R.; Tillement, J.P. Drug transfer across the blood–brain barrier: Correlation between in vitro and in vivo models. J. Neurochem. 1992, 58, 1790–1797. [Google Scholar] [CrossRef]
- Kafa, H.; Wang, J.T.W.; Rubio, N.; Venner, K.; Anderson, G.; Pach, E.; Ballesteros, B.; Preston, J.E.; Abbott, N.J.; Al-Jamal, K.T. The interaction of carbon nanotubes with an in vitro blood–brain barrier model and mouse brain in vivo. Biomaterials 2015, 53, 437–452. [Google Scholar] [CrossRef]
- Ruck, T.; Bittner, S.; Meuth, S.G. Blood-brain barrier modeling: Challenges and perspectives. Neural Regen. Res. 2015, 10, 889–891. [Google Scholar] [CrossRef]
- Gidwani, M.; Singh, A.V. Nanoparticle enabled drug delivery across the blood brain barrier: In vivo and in vitro models, opportunities and challenges. Curr. Pharm. Biotechnol. 2013, 14, 1201–1212. [Google Scholar] [CrossRef] [PubMed]
- Shah, B.; Dong, X. Current status of in vitro models of the blood–brain barrier. Curr. Drug Deliv. 2022, 19, 1034–1046. [Google Scholar]
- Bujak, R.; Struck-Lewicka, W.; Kaliszan, M.; Kaliszan, R.; Markuszewski, M.J. Blood–brain barrier permeability mechanisms in view of quantitative structure–activity relationships (QSAR). J. Pharm. Biomed. Anal. 2015, 108, 29–37. [Google Scholar] [CrossRef]
- Vucicevic, J.; Nikolic, K.; Dobričić, V.; Agbaba, D. Prediction of blood–brain barrier permeation of α-adrenergic and imidazoline receptor ligands using PAMPA technique and quantitative-structure permeability relationship analysis. Eur. J. Pharm. Sci. 2015, 68, 94–105. [Google Scholar] [CrossRef]
- Liu, R.; Sun, H.; So, S.S. Development of quantitative structure- property relationship models for early ADME evaluation in drug discovery. 2. Blood-brain barrier penetration. J. Chem. Inf. Comput. Sci. 2001, 41, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Golmohammadi, H.; Dashtbozorgi, Z.; Acree, W.E., Jr. Quantitative structure–activity relationship prediction of blood-to-brain partitioning behavior using support vector machine. Eur. J. Pharm. Sci. 2012, 47, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wu, M.B.; Lin, J.P.; Yang, L.R. Quantitative structure–activity relationship: Promising advances in drug discovery platforms. Expert Opin. Drug Discov. 2015, 10, 1283–1300. [Google Scholar] [CrossRef]
- Kortagere, S.; Chekmarev, D.; Welsh, W.J.; Ekins, S. New predictive models for blood–brain barrier permeability of drug-like molecules. Pharm. Res. 2008, 25, 1836–1845. [Google Scholar] [CrossRef]
- Vilar, S.; Chakrabarti, M.; Costanzi, S. Prediction of passive blood–brain partitioning: Straightforward and effective classification models based on in silico derived physicochemical descriptors. J. Mol. Graph. Model. 2010, 28, 899–903. [Google Scholar] [CrossRef]
- Mensch, J.; Jaroskova, L.; Sanderson, W.; Melis, A.; Mackie, C.; Verreck, G.; Brewster, M.E.; Augustijns, P. Application of PAMPA-models to predict BBB permeability including efflux ratio, plasma protein binding and physicochemical parameters. Int. J. Pharm. 2010, 395, 182–197. [Google Scholar] [CrossRef]
- Shityakov, S.; Neuhaus, W.; Dandekar, T.; Förster, C. Analysing molecular polar surface descriptors to predict blood–brain barrier permeation. Int. J. Comput. Biol. Drug Des. 2013, 6, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Bickel, U. How to measure drug transport across the blood–brain barrier. NeuroRx 2005, 2, 15–26. [Google Scholar] [CrossRef]
- Jackson, S.; Meeks, C.; Vezina, A.; Robey, R.W.; Tanner, K.; Gottesman, M.M. Model systems for studying the blood–brain barrier: Applications and challenges. Biomaterials 2019, 214, 119217. [Google Scholar] [CrossRef] [PubMed]
- Abbott, N.J. Blood–brain barrier structure and function and the challenges for CNS drug delivery. J. Inherit. Metab. Dis. 2013, 36, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Hajal, C.; Le Roi, B.; Kamm, R.D.; Maoz, B.M. Biology and models of the blood–brain barrier. Annu. Rev. Biomed. Eng. 2021, 23, 359–384. [Google Scholar] [CrossRef]
- Carpenter, T.S.; Kirshner, D.A.; Lau, E.Y.; Wong, S.E.; Nilmeier, J.P.; Lightstone, F.C. A method to predict blood–brain barrier permeability of drug-like compounds using molecular dynamics simulations. Biophys. J. 2014, 107, 630–641. [Google Scholar] [CrossRef]
- Shamloo, A.; Pedram, M.Z.; Heidari, H.; Alasty, A. Computing the blood brain barrier (BBB) diffusion coefficient: A molecular dynamics approach. J. Magn. Magn. Mater. 2016, 410, 187–197. [Google Scholar] [CrossRef]
- Goliaei, A.; Adhikari, U.; Berkowitz, M.L. Opening of the blood–brain barrier tight junction due to shock wave induced bubble collapse: A molecular dynamics simulation study. ACS Chem. Neurosci. 2015, 6, 1296–1301. [Google Scholar] [CrossRef]
- Man, V.H.; Li, M.S.; Derreumaux, P.; Wang, J.; Nguyen, T.T.; Nangia, S.; Nguyen, P.H. Molecular mechanism of ultrasound interaction with a blood brain barrier model. J. Chem. Phys. 2020, 153, 045104. [Google Scholar] [CrossRef]
- Rajagopal, N.; Irudayanathan, F.J.; Nangia, S. Computational nanoscopy of tight junctions at the blood–brain barrier interface. Int. J. Mol. Sci. 2019, 20, 5583. [Google Scholar] [CrossRef] [PubMed]
- Salo-Ahen, O.M.; Alanko, I.; Bhadane, R.; Bonvin, A.M.; Honorato, R.V.; Hossain, S.; Juffer, A.H.; Kabedev, A.; Lahtela-Kakkonen, M.; Larsen, A.S.; et al. Molecular dynamics simulations in drug discovery and pharmaceutical development. Processes 2020, 9, 71. [Google Scholar] [CrossRef]
- Borhani, D.W.; Shaw, D.E. The future of molecular dynamics simulations in drug discovery. J. Comput.-Aided Mol. Des. 2012, 26, 15–26. [Google Scholar] [CrossRef]
- Durrant, J.D.; McCammon, J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011, 9, 71. [Google Scholar] [CrossRef]
- Saikia, S.; Bordoloi, M. Molecular docking: Challenges, advances and its use in drug discovery perspective. Curr. Drug Targets 2019, 20, 501–521. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Xia, L.Y.; Chen, H.H.; Huang, H.H.; Liang, Y. Improved classification of blood–brain-barrier drugs using deep learning. Sci. Rep. 2019, 9, 8802. [Google Scholar] [CrossRef] [PubMed]
- Ansari, M.Y.; Chandrasekar, V.; Singh, A.V.; Dakua, S.P. Re-routing drugs to blood brain barrier: A comprehensive analysis of machine learning approaches with fingerprint amalgamation and data balancing. IEEE Access 2022, 11, 9890–9906. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, H.; Wu, Z.; Wang, T.; Li, W.; Tang, Y.; Liu, G. In silico prediction of blood–brain barrier permeability of compounds by machine learning and resampling methods. ChemMedChem 2018, 13, 2189–2201. [Google Scholar] [CrossRef]
- Yang, Q.; Fan, L.; Hao, E.; Hou, X.; Deng, J.; Xia, Z.; Du, Z. Machine Learning Exploration of the Relationship Between Drugs and the Blood–Brain Barrier: Guiding Molecular Modification. Pharm. Res. 2024, 41, 863–875. [Google Scholar] [CrossRef]
- Mazumdar, B.; Sarma, P.K.D.; Mahanta, H.J.; Sastry, G.N. Machine learning based dynamic consensus model for predicting blood–brain barrier permeability. Comput. Biol. Med. 2023, 160, 106984. [Google Scholar] [CrossRef]
- Wu, Z.; Ramsundar, B.; Feinberg, E.N.; Gomes, J.; Geniesse, C.; Pappu, A.S.; Leswing, K.; Pande, V. MoleculeNet: A benchmark for molecular machine learning. Chem. Sci. 2018, 9, 513–530. [Google Scholar] [CrossRef] [PubMed]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Han, H.; Wang, W.Y.; Mao, B.H. Borderline-SMOTE: A new over-sampling method in imbalanced data sets learning. In Proceedings of the International Conference on Intelligent Computing, Hefei, China, 23–26 August 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 878–887. [Google Scholar]
- Johnson, J.M.; Khoshgoftaar, T.M. Survey on deep learning with class imbalance. J. Big Data 2019, 6, 27. [Google Scholar] [CrossRef]
- Xue, J.; Ma, J. Extreme Sample Imbalance Classification Model Based on Sample Skewness Self-Adaptation. Symmetry 2023, 15, 1082. [Google Scholar] [CrossRef]
- Poduslo, J.F.; Curran, G.L. Polyamine modification increases the permeability of proteins at the blood-nerve and blood–brain barriers. J. Neurochem. 1996, 66, 1599–1609. [Google Scholar] [CrossRef]
- Fong, C.W. Permeability of the blood–brain barrier: Molecular mechanism of transport of drugs and physiologically important compounds. J. Membr. Biol. 2015, 248, 651–669. [Google Scholar] [CrossRef]
- Pardridge, W.M.; Sakiyama, R.; Fierer, G. Transport of propranolol and lidocaine through the rat blood–brain barrier. Primary role of globulin-bound drug. J. Clin. Investig. 1983, 71, 900–908. [Google Scholar] [CrossRef]
- Olesen, J.; Hougård, K.; Hertz, M. Isoproterenol and propranolol: Ability to cross the blood–brain barrier and effects on cerebral circulation in man. Stroke 1978, 9, 344–349. [Google Scholar] [CrossRef]
- Medeiros, A.; O’Brien, T. Ampicillin-resistant Haemophilus influenzae type B possessing a TEM-type β-lactamase but little permeability barrier to ampicillin. Lancet 1975, 305, 716–719. [Google Scholar] [CrossRef] [PubMed]
- Nau, R.; Sorgel, F.; Eiffert, H. Penetration of drugs through the blood-cerebrospinal fluid/blood–brain barrier for treatment of central nervous system infections. Clin. Microbiol. Rev. 2010, 23, 858–883. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.C.; Wang, G.P.; Shan, H.L.; Liang, W.Q.; Gao, J.Q. Predicting blood–brain barrier penetration from molecular weight and number of polar atoms. Eur. J. Pharm. Biopharm. 2008, 70, 462–466. [Google Scholar] [CrossRef]
- Dichiara, M.; Amata, B.; Turnaturi, R.; Marrazzo, A.; Amata, E. Tuning properties for blood–brain barrier permeation: A statistics-based analysis. ACS Chem. Neurosci. 2019, 11, 34–44. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Berman, S.M.; Kuczenski, R.; McCracken, J.T.; London, E.D. Potential adverse effects of amphetamine treatment on brain and behavior: A review. Mol. Psychiatry 2009, 14, 123–142. [Google Scholar] [CrossRef] [PubMed]
- Spigelman, M.K.; Zappulla, R.A.; Johnson, J.; Goldsmith, S.J.; Malis, L.I.; Holland, J.F. Etoposide-induced blood–brain barrier disruption: Effect of drug compared with that of solvents. J. Neurosurg. 1984, 61, 674–678. [Google Scholar] [CrossRef] [PubMed]
- Landrum, G. Rdkit documentation. Release 2013, 1, 4. [Google Scholar]
- Charman, W.N.; Porter, C.J.; Mithani, S.; Dressman, J.B. Physicochemical and physiological mechanisms for the effects of food on drug absorption: The role of lipids and pH. J. Pharm. Sci. 1997, 86, 269–282. [Google Scholar] [CrossRef]
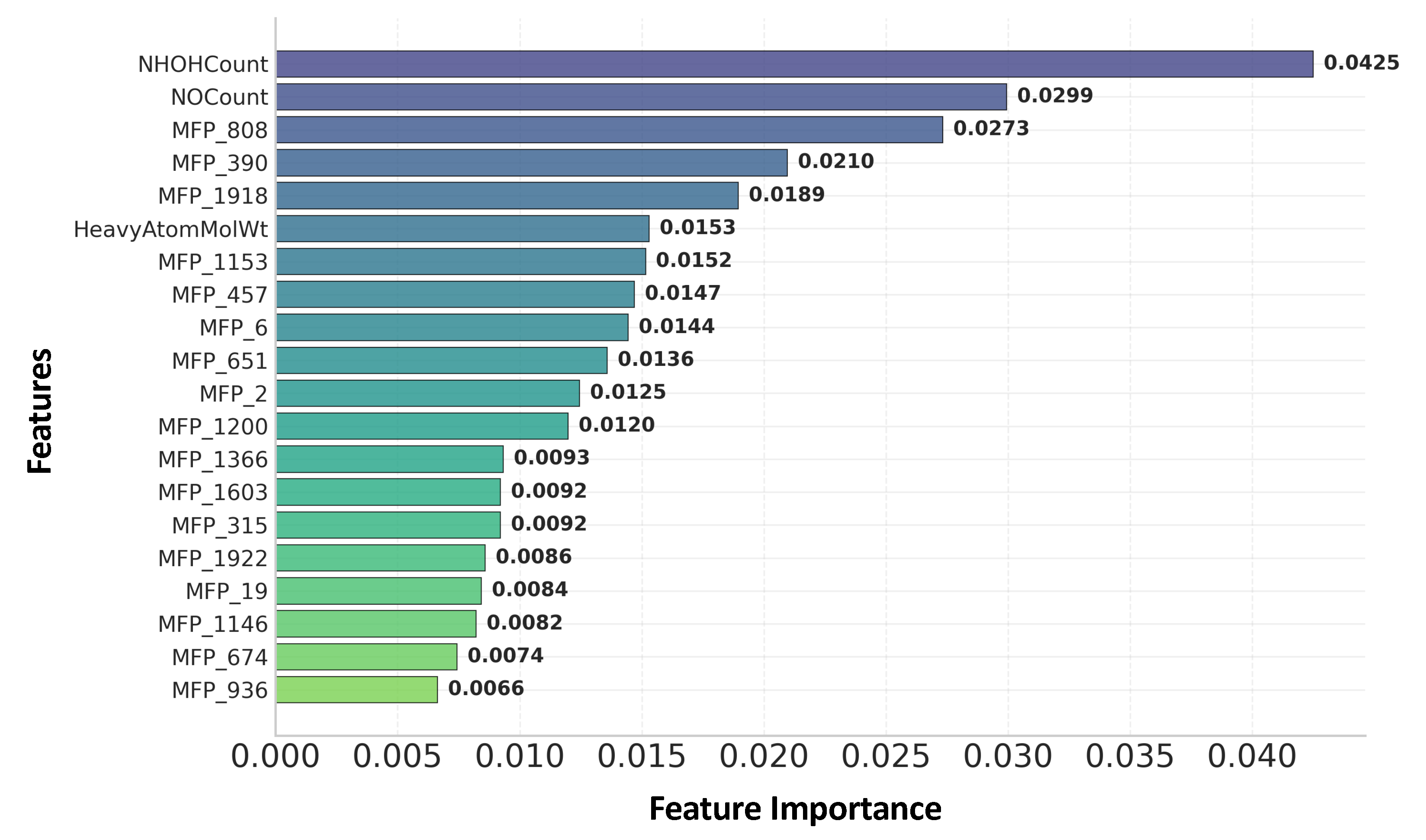
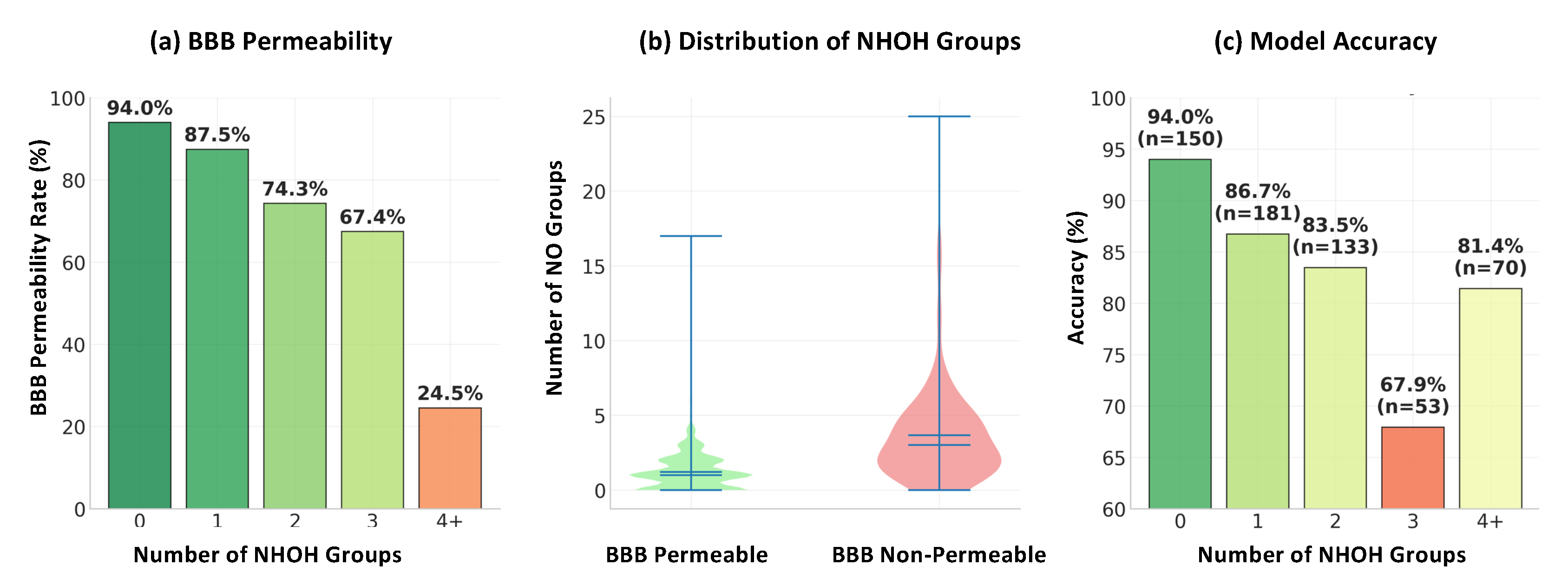
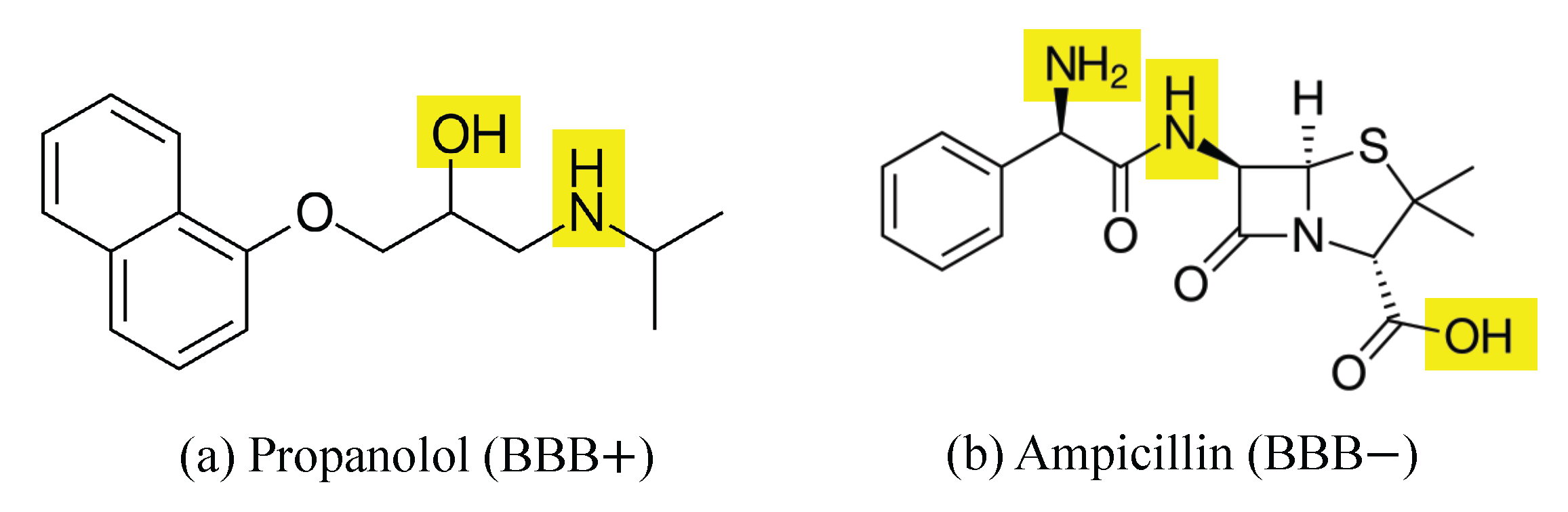

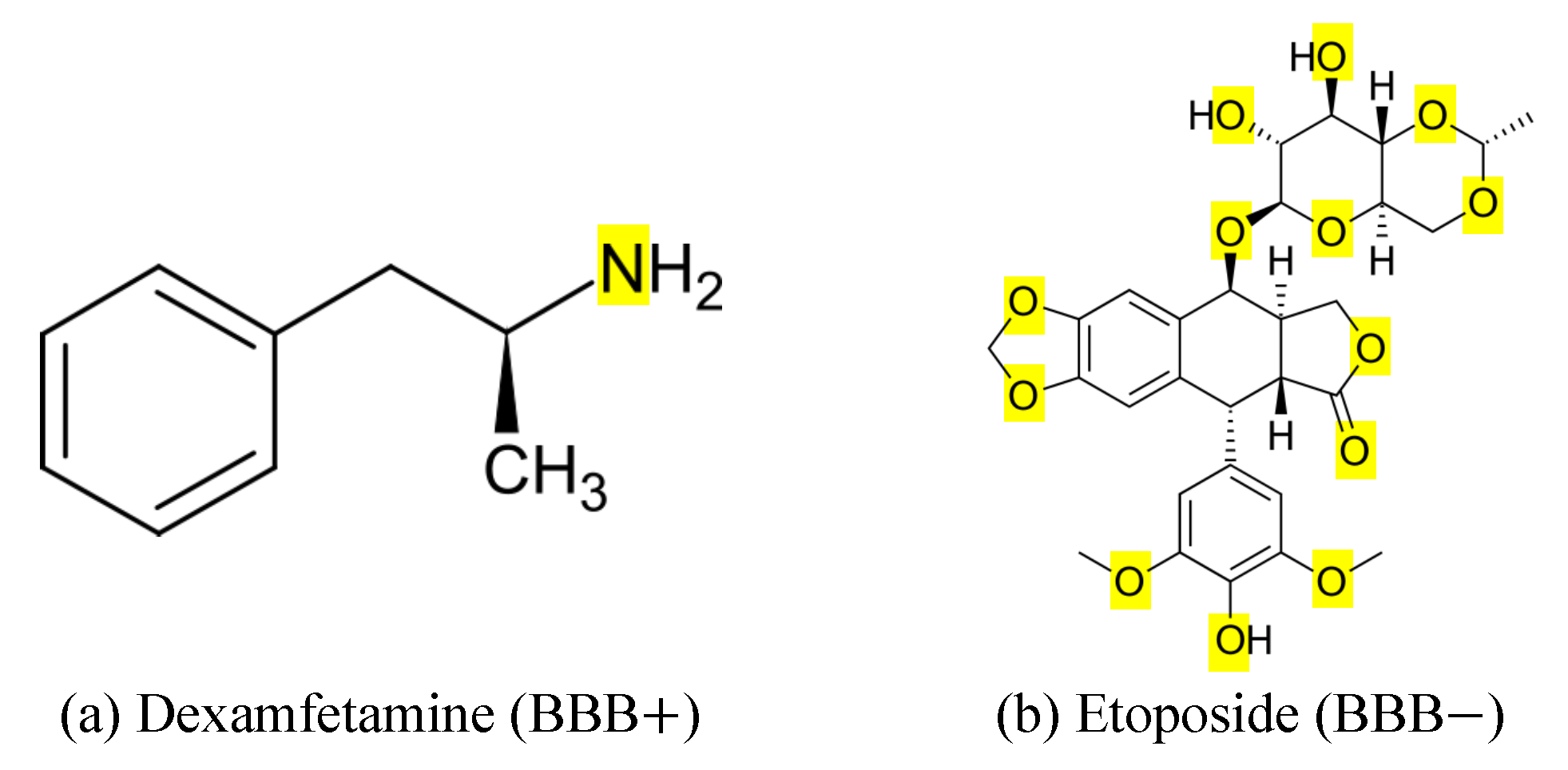
| Model | Run Time (s) | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|
| LogisticRegression | 0.32 | 0.881 | 0.891 | 0.962 | 0.925 |
| RandomForestClassifier | 0.04 | 0.877 | 0.876 | 0.978 | 0.924 |
| XGBClassifier | 0.06 | 0.876 | 0.888 | 0.958 | 0.922 |
| GradientBoostingClassifier | 0.13 | 0.876 | 0.882 | 0.966 | 0.922 |
| LGBMClassifier | 0.05 | 0.872 | 0.891 | 0.949 | 0.919 |
| MLPClassifier | 0.37 | 0.860 | 0.882 | 0.944 | 0.912 |
| AdaBoostClassifier | 0.04 | 0.855 | 0.863 | 0.964 | 0.910 |
| DecisionTreeClassifier | 0.01 | 0.823 | 0.887 | 0.882 | 0.884 |
| KNeighborsClassifier | 0.00 | 0.814 | 0.844 | 0.928 | 0.884 |
| DummyClassifier_most_frequent | 0.00 | 0.763 | 0.763 | 1.000 | 0.866 |
| GaussianNB | 0.00 | 0.658 | 0.824 | 0.701 | 0.758 |
| Metric | Without SMOTE | SMOTE | Borderline SMOTE | Undersampling |
|---|---|---|---|---|
| True Positives (TP) | 420 | 409 (−11) | 403 (−17) | 389 (−31) |
| True Negatives (TN) | 82 | 93 (+11) | 92 (+10) | 96 (+14) |
| False Positives (FP) | 57 | 46 (−11) | 47 (−10) | 43 (−14) |
| False Negatives (FN) | 28 | 39 (+11) | 45 (+17) | 59 (+31) |
| Correct Predictions | 502 | 502 (0) | 495 (−7) | 485 (−17) |
| Incorrect Predictions | 85 | 85 (0) | 92 (+7) | 102 (+17) |
| Accuracy | 0.855 | 0.855 (+0.000) | 0.843 (−0.012) | 0.826 (−0.029) |
| Precision | 0.881 | 0.899 (+0.018) | 0.896 (+0.015) | 0.900 (+0.020) |
| Recall | 0.938 | 0.913 (−0.025) | 0.900 (−0.038) | 0.868 (−0.070) |
| F1 Score | 0.908 | 0.906 (−0.002) | 0.898 (−0.011) | 0.884 (−0.024) |
| ROC AUC | 0.764 | 0.791 (+0.027) | 0.781 (+0.017) | 0.779 (+0.016) |
| Average Precision | 0.873 | 0.887 (+0.014) | 0.882 (+0.009) | 0.882 (+0.009) |
| Metric | Without SMOTE | SMOTE | Borderline SMOTE | Undersampling |
|---|---|---|---|---|
| True Positives (TP) | 443 | 442 (−1) | 440 (−3) | 428 (−15) |
| True Negatives (TN) | 87 | 90 (+3) | 88 (+1) | 97 (+10) |
| False Positives (FP) | 52 | 49 (−3) | 51 (−1) | 42 (−10) |
| False Negatives (FN) | 5 | 6 (+1) | 8 (+3) | 20 (+15) |
| Correct Predictions | 530 | 532 (+2) | 528 (−2) | 525 (−5) |
| Incorrect Predictions | 57 | 55 (−2) | 59 (+2) | 62 (+5) |
| Accuracy | 0.903 | 0.906 (+0.003) | 0.900 (−0.004) | 0.894 (−0.009) |
| Precision | 0.895 | 0.900 (+0.005) | 0.896 (+0.001) | 0.911 (+0.016) |
| Recall | 0.989 | 0.987 (−0.002) | 0.982 (−0.007) | 0.955 (−0.034) |
| F1 Score | 0.940 | 0.941 (+0.001) | 0.937 (−0.003) | 0.933 (−0.008) |
| ROC AUC | 0.807 | 0.817 (+0.010) | 0.808 (+0.001) | 0.827 (+0.020) |
| Average Precision | 0.893 | 0.898 (+0.005) | 0.894 (+0.001) | 0.904 (+0.011) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, B.; Liu, S. Feature-Guided Machine Learning for Studying Passive Blood–Brain Barrier Permeability to Aid Drug Discovery. Int. J. Mol. Sci. 2025, 26, 11228. https://doi.org/10.3390/ijms262211228
Zhu B, Liu S. Feature-Guided Machine Learning for Studying Passive Blood–Brain Barrier Permeability to Aid Drug Discovery. International Journal of Molecular Sciences. 2025; 26(22):11228. https://doi.org/10.3390/ijms262211228
Chicago/Turabian StyleZhu, Baining, and Suwei Liu. 2025. "Feature-Guided Machine Learning for Studying Passive Blood–Brain Barrier Permeability to Aid Drug Discovery" International Journal of Molecular Sciences 26, no. 22: 11228. https://doi.org/10.3390/ijms262211228
APA StyleZhu, B., & Liu, S. (2025). Feature-Guided Machine Learning for Studying Passive Blood–Brain Barrier Permeability to Aid Drug Discovery. International Journal of Molecular Sciences, 26(22), 11228. https://doi.org/10.3390/ijms262211228





