Integrative Epigenomic and Targeted Protein Analysis in MRONJ: Correlating DNA Methylation with Bone Biomarkers
Abstract
1. Introduction
2. Results
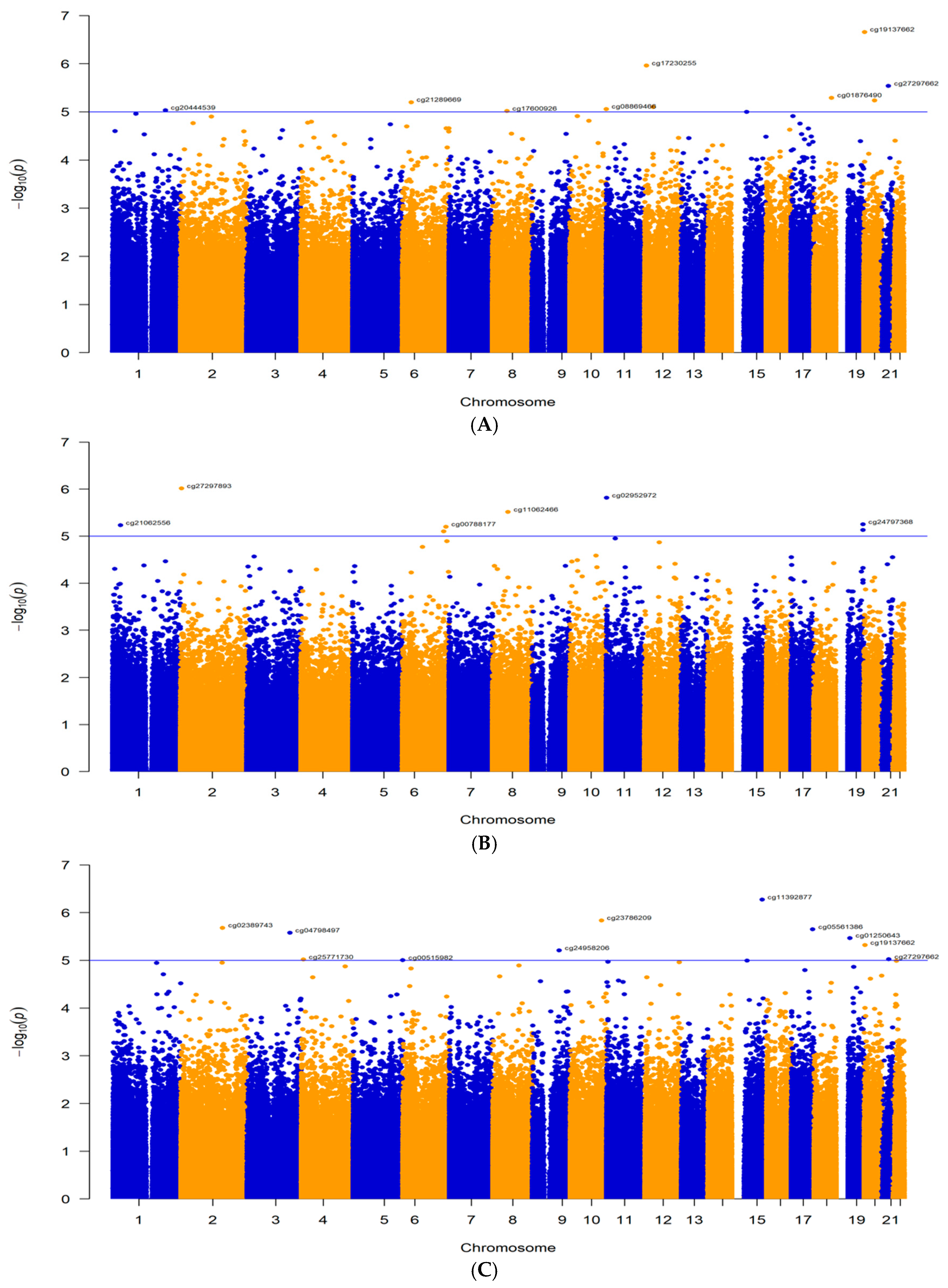
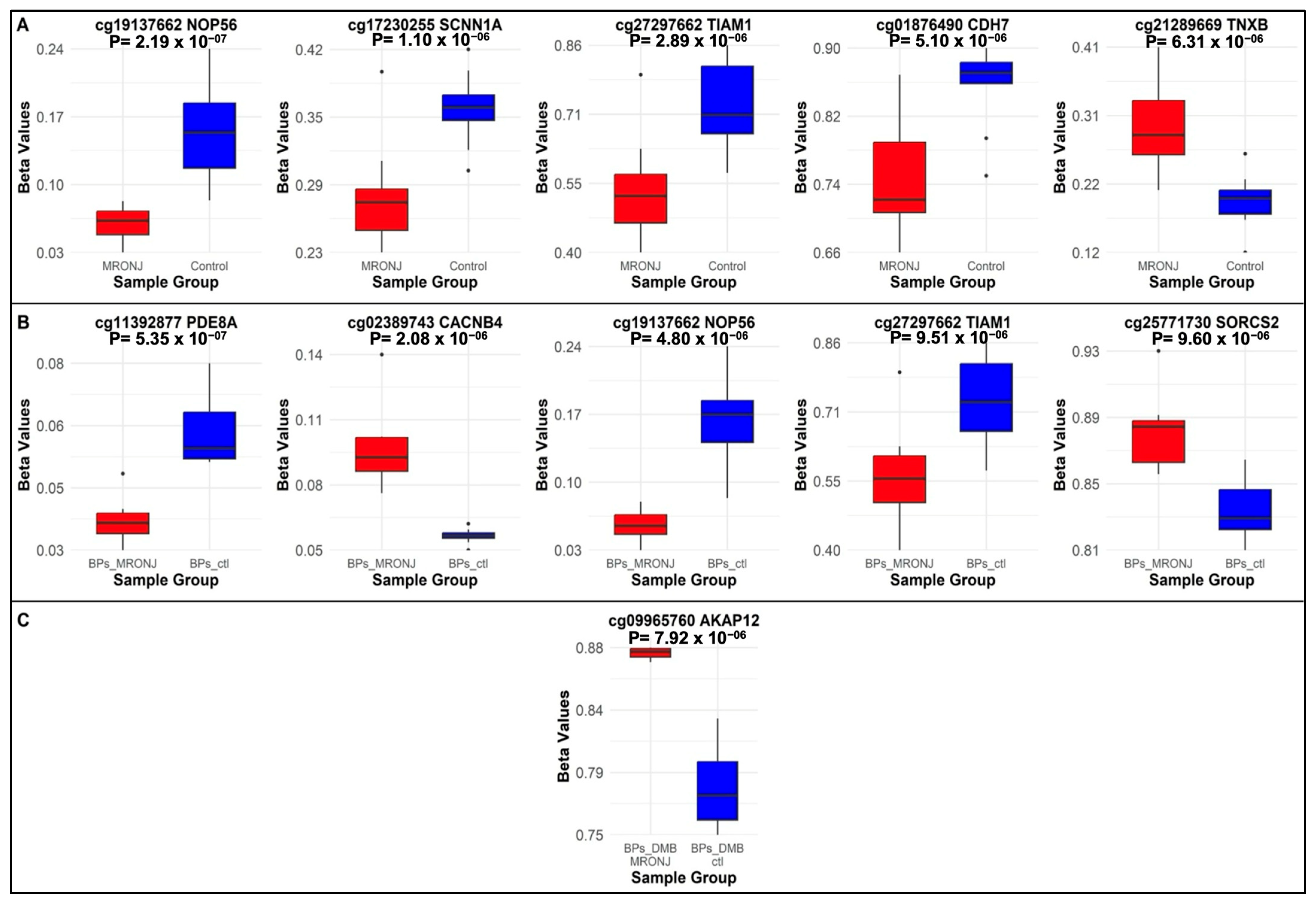
2.1. Differentially Methylated Probes
2.1.1. Main Analysis: MRONJ vs. Controls
2.1.2. First Subgroup Analysis: BPs-MRONJ vs. BPs-Controls
2.1.3. Second Subgroup Analysis: BPs/DMB MRONJ vs. BPs/DMB
2.2. DMRs
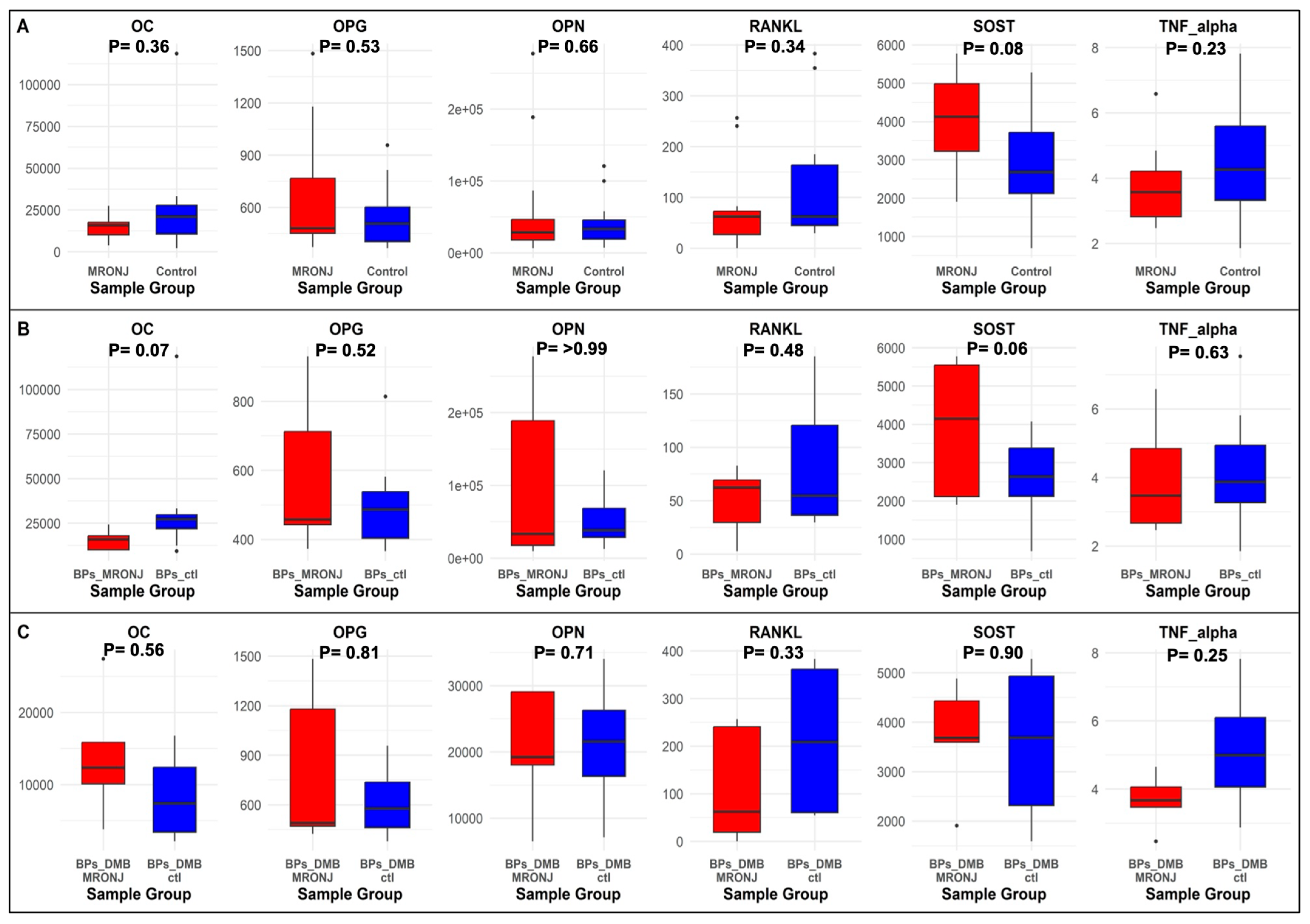
2.3. Bone Protein Biomarkers
2.4. Correlation Analysis
3. Discussion
4. Materials and Methods
4.1. DNA Methylation Profiling and Processing
4.1.1. Data Processing and Quality Controls (QC)
4.1.2. Assessment of Samples Quality and Normalization
4.1.3. Gender Confirmation
4.1.4. Removing Poor-Quality Probes
4.1.5. Removing Probes at Sex Chromosomes and SNPs
4.1.6. Removing Flagged Probes
4.2. Assessment of Cytokine and Bone Protein Levels
4.3. Statistical Analysis
4.3.1. Descriptive Statistics
4.3.2. Estimating Cell Type Proportions
4.3.3. DMPs Analysis
4.3.4. DMRs Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruggiero, S.L.; Dodson, T.B.; Aghaloo, T.; Carlson, E.R.; Ward, B.B.; Kademani, D. American Association of Oral and Maxillofacial Surgeons’ Position Paper on Medication-Related Osteonecrosis of the Jaws-2022 Update. J. Oral Maxillofac. Surg. 2022, 80, 920–943. [Google Scholar] [CrossRef]
- Wajda, B.G.; Ferrie, L.E.; Abbott, A.G.; Elmi Assadzadeh, G.; Monument, M.J.; Kendal, J.K. Denosumab vs. Zoledronic Acid for Metastatic Bone Disease: A Comprehensive Systematic Review and Meta-Analysis of Randomized Controlled Trials. Cancers 2025, 17, 388. [Google Scholar] [CrossRef] [PubMed]
- Sarasquete, M.E.; García-Sanz, R.; Marín, L.; Alcoceba, M.; Chillón, M.C.; Balanzategui, A.; Santamaria, C.; Rosiñol, L.; de la Rubia, J.; Hernandez, M.T.; et al. Bisphosphonate-related osteonecrosis of the jaw is associated with polymorphisms of the cytochrome P450 CYP2C8 in multiple myeloma: A genome-wide single nucleotide polymorphism analysis. Blood 2008, 112, 2709–2712. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, P.; Cartsos, V.M.; Palaska, P.K.; Shen, Y.; Floratos, A.; Zavras, A.I. Genomewide pharmacogenetics of bisphosphonate-induced osteonecrosis of the jaw: The role of RBMS3. Oncologist 2012, 17, 279–287. [Google Scholar] [CrossRef]
- Yang, G.; Singh, S.; McDonough, C.W.; Lamba, J.K.; Hamadeh, I.; Holliday, L.S.; Wang, D.; Katz, J.; Lakatos, P.A.; Balla, B.; et al. Genome-wide Association Study Identified Chromosome 8 Locus Associated with Medication-Related Osteonecrosis of the Jaw. Clin. Pharmacol. Ther. 2021, 110, 1558–1569. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- Delgado-Calle, J.; Sañudo, C.; Fernández, A.F.; García-Renedo, R.; Fraga, M.F.; Riancho, J.A. Role of DNA methylation in the regulation of the RANKL-OPG system in human bone. Epigenetics 2012, 7, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Li, Y. Modern epigenetics methods in biological research. Methods 2021, 187, 104–113. [Google Scholar] [CrossRef]
- Drake, M.T.; Clarke, B.L.; Khosla, S. Bisphosphonates: Mechanism of action and role in clinical practice. Mayo Clin. Proc. 2008, 83, 1032–1045. [Google Scholar] [CrossRef]
- Nagy, V.; Penninger, J.M. The RANKL-RANK Story. Gerontology 2015, 61, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.E.; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A.; Kaul, R.; et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, D.; Liu, S.; Huang, J. The roles of NOP56 in cancer and SCA36. Pathol. Oncol. Res. 2023, 29, 1610884. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.Y.; Jin, X.D.; Zhang, H.; Chen, J.; Yang, G.Z.; Wang, X.D.; Tang, L.; Lu, X.Y.; Lu, L.; Li, Q.N. Role of epithelial sodium channel in rat osteoclast differentiation and bone resorption. J. South. Med. Univ 2016, 36, 1148–1152. [Google Scholar]
- Perin, M.; Chinigo, G.; Genova, T.; Mussano, F.; Munaron, L. The Impact of Plasma Membrane Ion Channels on Bone Remodeling in Response to Mechanical Stress, Oxidative Imbalance, and Acidosis. Antioxidants 2023, 12, 689. [Google Scholar] [CrossRef]
- Shao, W.; Orlando, R.C.; Awayda, M.S. Bisphosphonates stimulate an endogenous nonselective cation channel in Xenopus oocytes: Potential mechanism of action. Am. J. Physiol. Cell Physiol. 2005, 289, C248–C256. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.Y.; Putney, J.W., Jr. Calcium signaling in osteoclasts. Biochim. Biophys. Acta 2011, 1813, 979–983. [Google Scholar] [CrossRef]
- Wang, X.; Qu, Z.; Zhao, S.; Luo, L.; Yan, L. Wnt/beta-catenin signaling pathway: Proteins’ roles in osteoporosis and cancer diseases and the regulatory effects of natural compounds on osteoporosis. Mol. Med. 2024, 30, 193. [Google Scholar] [CrossRef]
- McClung, M.R. Sclerostin antibodies in osteoporosis: Latest evidence and therapeutic potential. Ther. Adv. Musculoskelet. Dis. 2017, 9, 263–270. [Google Scholar] [CrossRef]
- Han, G.; Zuo, J.; Holliday, L.S. Specialized Roles for Actin in Osteoclasts: Unanswered Questions and Therapeutic Opportunities. Biomolecules 2019, 9, 17. [Google Scholar] [CrossRef]
- Boissier, P.; Huynh-Do, U. The guanine nucleotide exchange factor Tiam1: A Janus-faced molecule in cellular signaling. Cell Signal 2014, 26, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Huang, Q.; Jia, W.; Feng, S.; Liu, L.; Li, X.; Tao, D.; Xie, D. YAP1 induces invadopodia formation by transcriptionally activating TIAM1 through enhancer in breast cancer. Oncogene 2022, 41, 3830–3845. [Google Scholar] [CrossRef]
- Wang, B.; Zheng, B.; Cao, L.; Liao, K.; Huang, D.; Zhang, Y.; Jiang, Y.; Zheng, S. T-lymphoma invasion and metastasis 1 promotes invadopodia formation and is regulated by the PI3K/Akt signaling pathway in hepatocellular carcinoma. Exp. Cell Res. 2021, 407, 112806. [Google Scholar] [CrossRef]
- Luxenburg, C.; Parsons, J.T.; Addadi, L.; Geiger, B. Involvement of the Src-cortactin pathway in podosome formation and turnover during polarization of cultured osteoclasts. J. Cell Sci. 2006, 119, 4878–4888. [Google Scholar] [CrossRef]
- Matsubara, T.; Myoui, A.; Ikeda, F.; Hata, K.; Yoshikawa, H.; Nishimura, R.; Yoneda, T. Critical role of cortactin in actin ring formation and osteoclastic bone resorption. J. Bone Miner. Metab. 2006, 24, 368–372. [Google Scholar] [CrossRef]
- Tehrani, S.; Faccio, R.; Chandrasekar, I.; Ross, F.P.; Cooper, J.A. Cortactin has an essential and specific role in osteoclast actin assembly. Mol. Biol. Cell 2006, 17, 2882–2895. [Google Scholar] [CrossRef]
- Kajitani, N.; Yamada, T.; Kawakami, K.; Matsumoto, K.I. TNX deficiency results in bone loss due to an increase in multinucleated osteoclasts. Biochem. Biophys. Res. Commun. 2019, 512, 659–664. [Google Scholar] [CrossRef]
- Rodan, G.A.; Bourret, L.A.; Harvey, A.; Mensi, T. Cyclic AMP and cyclic GMP: Mediators of the mechanical effects on bone remodeling. Science 1975, 189, 467–469. [Google Scholar] [CrossRef]
- Chabre, O.; Conklin, B.R.; Lin, H.Y.; Lodish, H.F.; Wilson, E.; Ives, H.E.; Catanzariti, L.; Hemmings, B.A.; Bourne, H.R. A recombinant calcitonin receptor independently stimulates 3′,5′-cyclic adenosine monophosphate and Ca2+/inositol phosphate signaling pathways. Mol. Endocrinol. 1992, 6, 551–556. [Google Scholar] [CrossRef]
- Martin, T.J. PTH1R Actions on Bone Using the cAMP/Protein Kinase A Pathway. Front. Endocrinol. 2021, 12, 833221. [Google Scholar] [CrossRef]
- Pekkinen, M.; Ahlstrom, M.E.B.; Riehle, U.; Huttunen, M.M.; Lamberg-Allardt, C.J.E. Effects of phosphodiesterase 7 inhibition by RNA interference on the gene expression and differentiation of human mesenchymal stem cell-derived osteoblasts. Bone 2008, 43, 84–91. [Google Scholar] [CrossRef]
- Qasim, H.; McConnell, B.K. AKAP12 Signaling Complex: Impacts of Compartmentalizing cAMP-Dependent Signaling Pathways in the Heart and Various Signaling Systems. J. Am. Heart Assoc. 2020, 9, e016615. [Google Scholar] [CrossRef]
- Chen, Y.J.; Chang, W.A.; Hsu, Y.L.; Chen, C.H.; Kuo, P.L. Deduction of Novel Genes Potentially Involved in Osteoblasts of Rheumatoid Arthritis Using Next-Generation Sequencing and Bioinformatic Approaches. Int. J. Mol. Sci. 2017, 18, 2396. [Google Scholar] [CrossRef]
- Johnson, A.G.; Lapointe, C.P.; Wang, J.; Corsepius, N.C.; Choi, J.; Fuchs, G.; Puglisi, J.D. RACK1 on and off the ribosome. RNA 2019, 25, 881–895. [Google Scholar] [CrossRef]
- Kershner, L.; Welshhans, K. RACK1 is necessary for the formation of point contacts and regulates axon growth. Dev. Neurobiol. 2017, 77, 1038–1056. [Google Scholar] [CrossRef]
- Park, J.H.; Jeong, E.; Lin, J.; Ko, R.; Kim, J.H.; Yi, S.; Choi, Y.; Kang, I.C.; Lee, D.; Lee, S.Y. RACK1 interaction with c-Src is essential for osteoclast function. Exp. Mol. Med. 2019, 51, 1–9. [Google Scholar] [CrossRef]
- Yang, C.; Jin, X.; Liu, X.; Wu, G.; Yang, W.; Pang, B.; Jiang, J.; Liao, D.; Zhang, Y. TRIM15 forms a regulatory loop with the AKT/FOXO1 axis and LASP1 to modulate the sensitivity of HCC cells to TKIs. Cell Death Dis. 2023, 14, 47. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, W.; Wei, X.Y.; Hu, J.Z.; Hu, X.; Liu, H.; Lu, J.; Shen, S.; Ji, M.L. TRIM15 drives chondrocyte senescence and osteoarthritis progression. Sci. Transl. Med. 2025, 17, eadq1735. [Google Scholar] [CrossRef]
- Peraldi, R.; Kmita, M. 40 years of the homeobox: Mechanisms of Hox spatial-temporal collinearity in vertebrates. Development 2024, 151, dev202508. [Google Scholar] [CrossRef]
- Myers, C.; Charboneau, A.; Boudreau, N. Homeobox B3 promotes capillary morphogenesis and angiogenesis. J. Cell Biol. 2000, 148, 343–351. [Google Scholar] [CrossRef]
- Kachgal, S.; Mace, K.A.; Boudreau, N.J. The dual roles of homeobox genes in vascularization and wound healing. Cell Adh Migr. 2012, 6, 457–470. [Google Scholar] [CrossRef]
- Chung, N.; Jee, B.K.; Chae, S.W.; Jeon, Y.W.; Lee, K.H.; Rha, H.K. HOX gene analysis of endothelial cell differentiation in human bone marrow-derived mesenchymal stem cells. Mol. Biol. Rep. 2009, 36, 227–235. [Google Scholar] [CrossRef]
- Gatti, D.; Viapiana, O.; Adami, S.; Idolazzi, L.; Fracassi, E.; Rossini, M. Bisphosphonate treatment of postmenopausal osteoporosis is associated with a dose dependent increase in serum sclerostin. Bone 2012, 50, 739–742. [Google Scholar] [CrossRef] [PubMed]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Zhang, X.; Huang, C.C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef]
- EPIC Array Power Calculations. Available online: https://epigenetics.essex.ac.uk/shiny/EPICDNAmPowerCalcs/ (accessed on 11 November 2025).
- Peters, T.J.; Buckley, M.J.; Statham, A.L.; Pidsley, R.; Samaras, K.; Lord, R.V.; Clark, S.J.; Molloy, P.L. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 2015, 8, 6. [Google Scholar] [CrossRef]

| Variable | All Participants (n = 24) | ||
|---|---|---|---|
| Cases (n = 12) | Control (n = 12) | p Value | |
| * Continuous | |||
| Age (years) | 68.5 (59.25–71.5) | 67.5 (57.0–72.0) | 0.73 |
| BMI (kg/m2) | 28.4 (24.5–32.1) | 27.25 (25.4–28.8) | 0.71 |
| * Categorical | |||
| Sex at birth | >0.99 | ||
| Female | 8 (66.7.%) | 9 (75%) | |
| Male | 4 (33.3%) | 3 (25%) | |
| Race Groups | 0.32 | ||
| White | 8 (66.7%) | 11 (91.7%) | |
| Black | 4 (33.3%) | 1 (8.3%) | |
| Smoking status | >0.99 | ||
| Current smoker | 0 (0.0%) | 1 (8.3%) | |
| Previous smoker | 7 (58.3%) | 7 (58.3%) | |
| Never smoke | 5 (41.7%) | 4 (33.3%) | |
| AAOMS Staging of MRONJ | |||
| Stage 1 | 7 (58.3%) | - | |
| Stage2 | 3 (25%) | - | |
| Stage3 | 0 (0.0%) | - | |
| Stage4 | 0 (0.0%) | - | |
| NA | 2 (16.7%) | ||
| Lesion location | |||
| Maxilla | 2 (16.7%) | - | |
| Mandible | 7 (58.3%) | - | |
| Anterior–posterior | 1 (8.3%) | - | |
| Lingual/buccal | 0 (0.0%) | - | |
| NA | 2 (16.7%) | - | |
| Medications | |||
| Bisphosphonates | 7 (58.3%) | 8 (66.7%) | >0.99 |
| Sequential therapy (BPs/DMB) | 5 (41.7%) | 4 (33.3%) | |
| No | Ilumina ID | CHR | MAP INFO | Gene Name | Feature | Main Analysis | First Subgroup Analysis | Second Subgroup Analysis | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| logFC | p | logFC | p | logFC | p | ||||||
| 1 | cg19137662 * | chr20 | 2652716 | NOP56 | Promoter | −1.38 | 2.19 × 10−7 | −1.60 | 4.80 × 10−6 | −1.01 | 0.01 |
| 2 | cg17230255 * | chr12 | 6364225 | SCNN1A | Promoter | −0.60 | 1.10 × 10−6 | −0.68 | 2.27 × 10−5 | −0.45 | 0.01 |
| 3 | cg27297662 | chr21 | 31136999 | TIAM1 | Intron | −1.30 | 2.89 × 10−6 | −1.53 | 9.51 × 10−6 | −0.85 | 0.02 |
| 4 | cg01876490 | chr18 | 65766883 | CDH7 | Intron | −1.10 | 5.10 × 10−6 | −1.17 | 0.0002 | −0.97 | 0.01 |
| 5 | cg21289669 | chr6 | 32096987 | TNXB | Exon | 0.84 | 6.31 × 10−6 | 0.88 | 0.0003 | 0.82 | 0.005 |
| 6 | cg11392877 | chr15 | 84981545 | PDE8A | Promoter | −0.55 | 0.0005 | −0.98 | 5.35 × 10−7 | 0.13 | 0.48 |
| 7 | cg02389743 * | chr2 | 152099098 | CACNB4 | Promoter | 0.52 | 0.0006 | 0.92 | 2.08 × 10−6 | −0.07 | 0.71 |
| 8 | cg25771730 | chr4 | 7425468 | SORCS2 | Intron | 0.42 | 0.004 | 0.82 | 9.60 × 10−6 | −0.17 | 0.38 |
| 9 | cg00515982 | chr5 | 181243930 | RACK1 | Promoter | −0.51 | 0.0004 | −0.81 | 9.94 × 10−6 | −0.02 | 0.91 |
| 10 | cg09965760 | chr6 | 151300350 | AKAP12 | Intron | 0.44 | 0.0003 | 0.15 | 0.22 | 0.89 | 7.92 × 10−6 |
| No | CHR | Gene Name | Main Analysis | First Subgroup Analysis | Second Subgroup Analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. CpG | Mean Diff | p | No. CpG | Mean Diff | p | No. CpG | Mean Diff | p | |||
| 1 | chr6 | TNXB | 20 | 0.07 | 3.30 × 10−10 | 10 | 0.09 | 2.76 × 10−7 | ND | ND | ND |
| 2 | chr17 | HOXB-AS3, HOXB3, HOXB4 | 15 | −0.06 | 4.27 × 10−8 | ND | ND | ND | ND | ND | ND |
| 3 | chr4 | SPON2 | 13 | −0.03 | 2.76 × 10−7 | 12 | −0.04 | 4.27 × 10−8 | ND | ND | ND |
| 4 | chr6 | TRIM15 | 9 | 0.07 | 1.43 × 10−5 | 12 | 0.1 | 3.30 × 10−10 | ND | ND | ND |
| No. | DMP | Methylation Status | Region | Main Analysis (n = 24) | First Subgroup Analysis (n = 15) | Second Subgroup Analysis (n = 9) | Biomarker | |||
|---|---|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | |||||
| 1 | cg19137662 (NOP56) | Hypomethylation | Promoter | 0.48 | 0.02 | 0.57 | 0.03 | −0.40 | 0.28 | OC |
| 0.32 | 0.13 | 0.38 | 0.16 | 0.73 | 0.025 | RANKL | ||||
| 2 | cg17230255 (SCNN1A) | Hypomethylation | Promoter | 0.56 | 0.004 | 0.66 | 0.01 | 0.44 | 0.24 | TNF- alpha |
| 3 | cg11392877 (PDE8A) | Hypomethylation | Promoter | −0.28 | 0.19 | −0.51 | 0.05 | 0.63 | 0.07 | SOST |
| 0.24 | 0.25 | 0.09 | 0.75 | 0.82 | 0.01 | OPG | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshammari, R.A.; Tantawy, M.; Wang, D.; Castro-Hall, E.; Abreu, M.; Villa, A.; Katz, J.; Holliday, L.S.; Gong, Y. Integrative Epigenomic and Targeted Protein Analysis in MRONJ: Correlating DNA Methylation with Bone Biomarkers. Int. J. Mol. Sci. 2025, 26, 11208. https://doi.org/10.3390/ijms262211208
Alshammari RA, Tantawy M, Wang D, Castro-Hall E, Abreu M, Villa A, Katz J, Holliday LS, Gong Y. Integrative Epigenomic and Targeted Protein Analysis in MRONJ: Correlating DNA Methylation with Bone Biomarkers. International Journal of Molecular Sciences. 2025; 26(22):11208. https://doi.org/10.3390/ijms262211208
Chicago/Turabian StyleAlshammari, Raed Awadh, Marwa Tantawy, Danxin Wang, Elysse Castro-Hall, Maria Abreu, Alessandro Villa, Joseph Katz, Lexie Shannon Holliday, and Yan Gong. 2025. "Integrative Epigenomic and Targeted Protein Analysis in MRONJ: Correlating DNA Methylation with Bone Biomarkers" International Journal of Molecular Sciences 26, no. 22: 11208. https://doi.org/10.3390/ijms262211208
APA StyleAlshammari, R. A., Tantawy, M., Wang, D., Castro-Hall, E., Abreu, M., Villa, A., Katz, J., Holliday, L. S., & Gong, Y. (2025). Integrative Epigenomic and Targeted Protein Analysis in MRONJ: Correlating DNA Methylation with Bone Biomarkers. International Journal of Molecular Sciences, 26(22), 11208. https://doi.org/10.3390/ijms262211208






