Special Issue “Melanoma: From Molecular Pathology to Therapeutic Approaches”
Conflicts of Interest
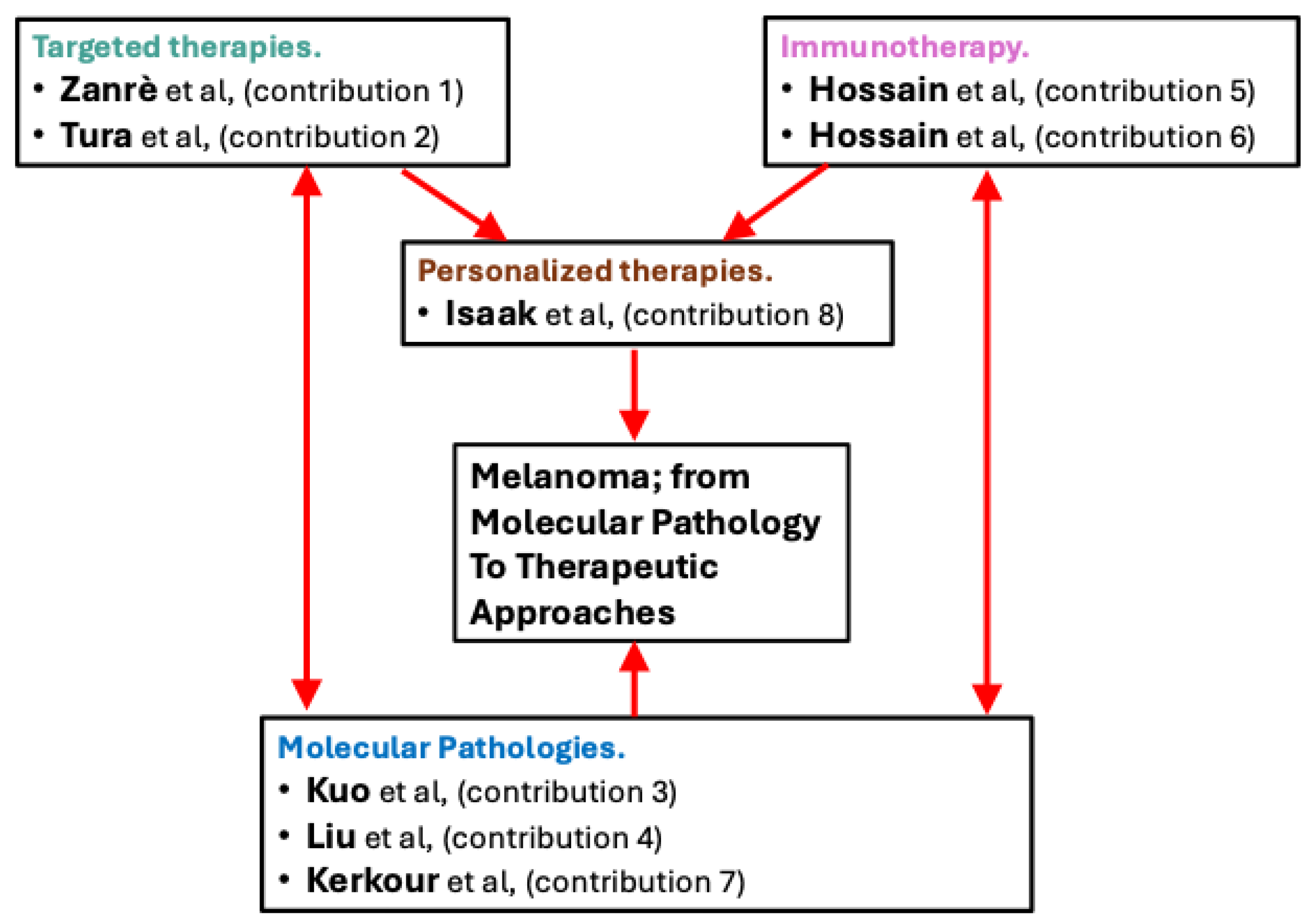
List of Contributions
- Zanrè, V.; Bellinato, F.; Cardile, A.; Passarini, C.; Di Bella, S.; Menegazzi, M. BRAF-Mutated Melanoma Cell Lines Develop Distinct Molecular Signatures After Prolonged Exposure to AZ628 or Dabrafenib: Potential Benefits of the Antiretroviral Treatments Cabotegravir or Doravirine on BRAF-Inhibitor-Resistant Cells. Int. J. Mol. Sci. 2024, 25, 11939. https://doi.org/10.3390/ijms252211939.
- Tura, A.; Herfs, V.; Maaßen, T.; Zuo, H.; Vardanyan, S.; Prasuhn, M.; Ranjbar, M.; Kakkassery, V.; Grisanti, S. Quercetin Impairs the Growth of Uveal Melanoma Cells by Interfering with Glucose Uptake and Metabolism. Int. J. Mol. Sci. 2024, 25, 4292. https://doi.org/10.3390/ijms25084292.
- Kuo, F.-C.; Tsai, H.-Y.; Cheng, B.-L.; Tsai, K.-J.; Chen, P.-C.; Huang, Y.-B.; Liu, C.-J.; Wu, D.-C.; Wu, M.-C.; Huang, B.; et al. Endothelial Mitochondria Transfer to Melanoma Induces M2-Type Macrophage Polarization and Promotes Tumor Growth by the Nrf2/HO-1-Mediated Pathway. Int. J. Mol. Sci. 2024, 25, 1857. https://doi.org/10.3390/ijms25031857.
- Liu, Z.; Krstic, A.; Neve, A.; Casalou, C.; Rauch, N.; Wynne, K.; Cassidy, H.; McCann, A.; Kavanagh, E.; McCann, B.; et al. Kinase Suppressor of RAS 1 (KSR1) Maintains the Transformed Phenotype of BRAFV600E Mutant Human Melanoma Cells. Int. J. Mol. Sci. 2023, 24, 11821. https://doi.org/10.3390/ijms241411821.
- Hossain, S.M.; Ly, K.; Sung, Y.J.; Braithwaite, A.; Li, K. Immune Checkpoint Inhibitor Therapy for Metastatic Melanoma: What Should We Focus on to Improve the Clinical Outcomes? Int. J. Mol. Sci. 2024, 25, 10120. https://doi.org/10.3390/ijms251810120.
- Hossain, S.M.; Carpenter, C.; Eccles, M.R. Genomic and Epigenomic Biomarkers of Immune Checkpoint Immunotherapy Response in Melanoma: Current and Future Perspectives. Int. J. Mol. Sci. 2024, 25, 7252. https://doi.org/10.3390/ijms25137252.
- Kerkour, T.; Zhou, C.; Hollestein, L.; Mooyaart, A. Genetic Concordance in Primary Cutaneous Melanoma and Matched Metastasis: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2023, 24, 16281. https://doi.org/10.3390/ijms242216281.
- Isaak, A.J.; Clements, G.R.; Buenaventura, R.G.M.; Merlino, G.; Yu, Y. Development of Personalized Strategies for Precisely Battling Malignant Melanoma. Int. J. Mol. Sci. 2024, 25, 5023. https://doi.org/10.3390/ijms25095023.
References
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; de Vries, E.; Whiteman, D.C.; Bray, F. Global Burden of Cutaneous Melanoma in 2020 and Projections to 2040. JAMA Dermatol. 2022, 158, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Kaštelan, S.; Pavičić, A.D.; Pašalić, D.; Nikuševa-Martić, T.; Čanović, S.; Kovačević, P.; Konjevoda, S. Biological characteristics and clinical management of uveal and conjunctival melanoma. Oncol Res. 2024, 32, 1265–1285. [Google Scholar] [CrossRef]
- Qin, Z.; Zheng, M. Advances in targeted therapy and immunotherapy for melanoma. Exp. Ther. Med. 2023, 26, 416. [Google Scholar] [CrossRef] [PubMed]
- Holder, A.M.; Dedeilia, A.; Sierra-Davidson, K.; Cohen, S.; Liu, D.; Parikh, A.; Boland, G.M. Defining clinically useful biomarkers of immune checkpoint inhibitors in solid tumours. Nat. Rev. Cancer 2024, 24, 498–512. [Google Scholar] [CrossRef] [PubMed]
- Mourah, S.; Louveau, B.; Dumaz, N. Mechanisms of resistance and predictive biomarkers of response to targeted therapies and immunotherapies in metastatic melanoma. Curr. Opin. Oncol. 2020, 32, 91–97. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eccles, M.R. Special Issue “Melanoma: From Molecular Pathology to Therapeutic Approaches”. Int. J. Mol. Sci. 2025, 26, 11199. https://doi.org/10.3390/ijms262211199
Eccles MR. Special Issue “Melanoma: From Molecular Pathology to Therapeutic Approaches”. International Journal of Molecular Sciences. 2025; 26(22):11199. https://doi.org/10.3390/ijms262211199
Chicago/Turabian StyleEccles, Michael R. 2025. "Special Issue “Melanoma: From Molecular Pathology to Therapeutic Approaches”" International Journal of Molecular Sciences 26, no. 22: 11199. https://doi.org/10.3390/ijms262211199
APA StyleEccles, M. R. (2025). Special Issue “Melanoma: From Molecular Pathology to Therapeutic Approaches”. International Journal of Molecular Sciences, 26(22), 11199. https://doi.org/10.3390/ijms262211199




