From CMS to iCMS/IMF: Developing Roadmap to Precision Therapy in Colorectal Cancer
Abstract
1. Introduction
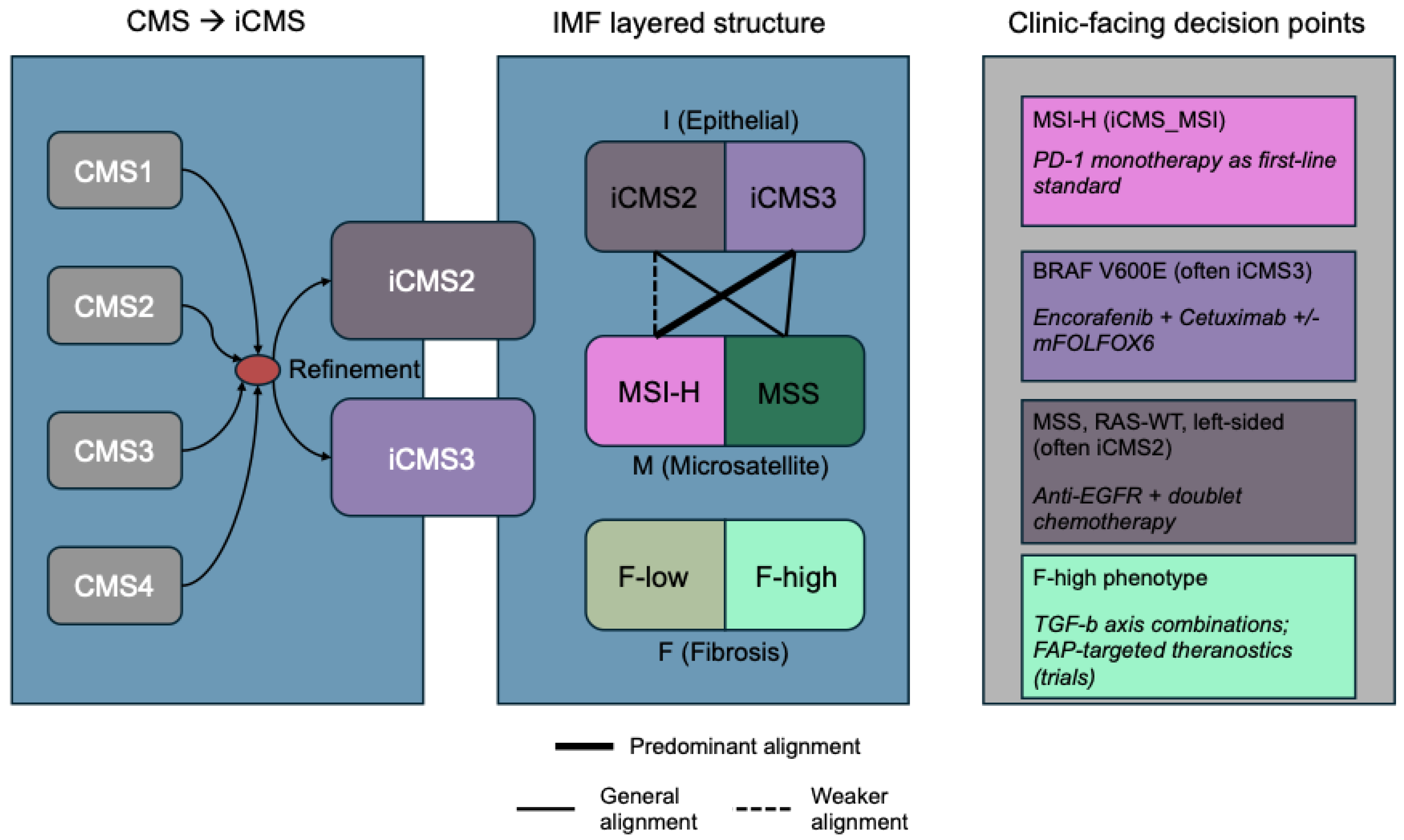
2. Concepts and Definitions: From CMS to iCMS to IMF
3. The Fibrosis Axis and the Contributions of Single-Cell and Spatial Transcriptomics
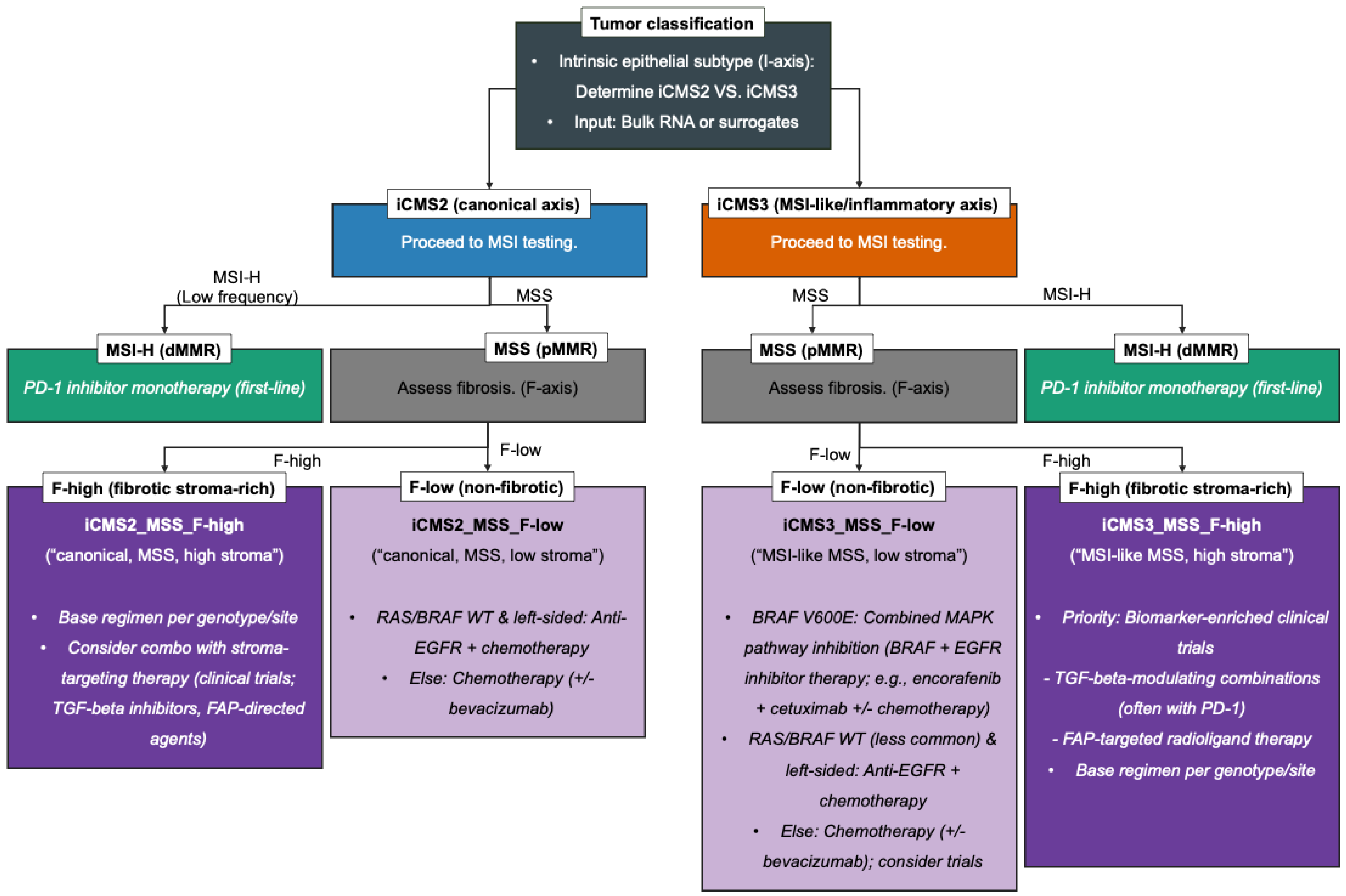
4. Therapeutic Mapping: Decision Points by iCMS and IMF
4.1. MSI-H/iCMS3: First-Line PD-1 Blockade and Resistance Considerations
4.2. iCMS3_MSS: MSI-H-like Biology and Combination-Immunotherapy Hypotheses (Trial Priorities, Not Standards)
4.3. MAPK/BRAF V600E (Often iCMS3): Targeted Therapy and First-Line Expansion
4.4. iCMS2 (Canonical Axis): The EGFR-First Principle in RAS-WT, Left-Sided Disease
4.5. F-High Phenotypes: Anti-Fibrosis/TGF-β Strategies and FAP Theranostics
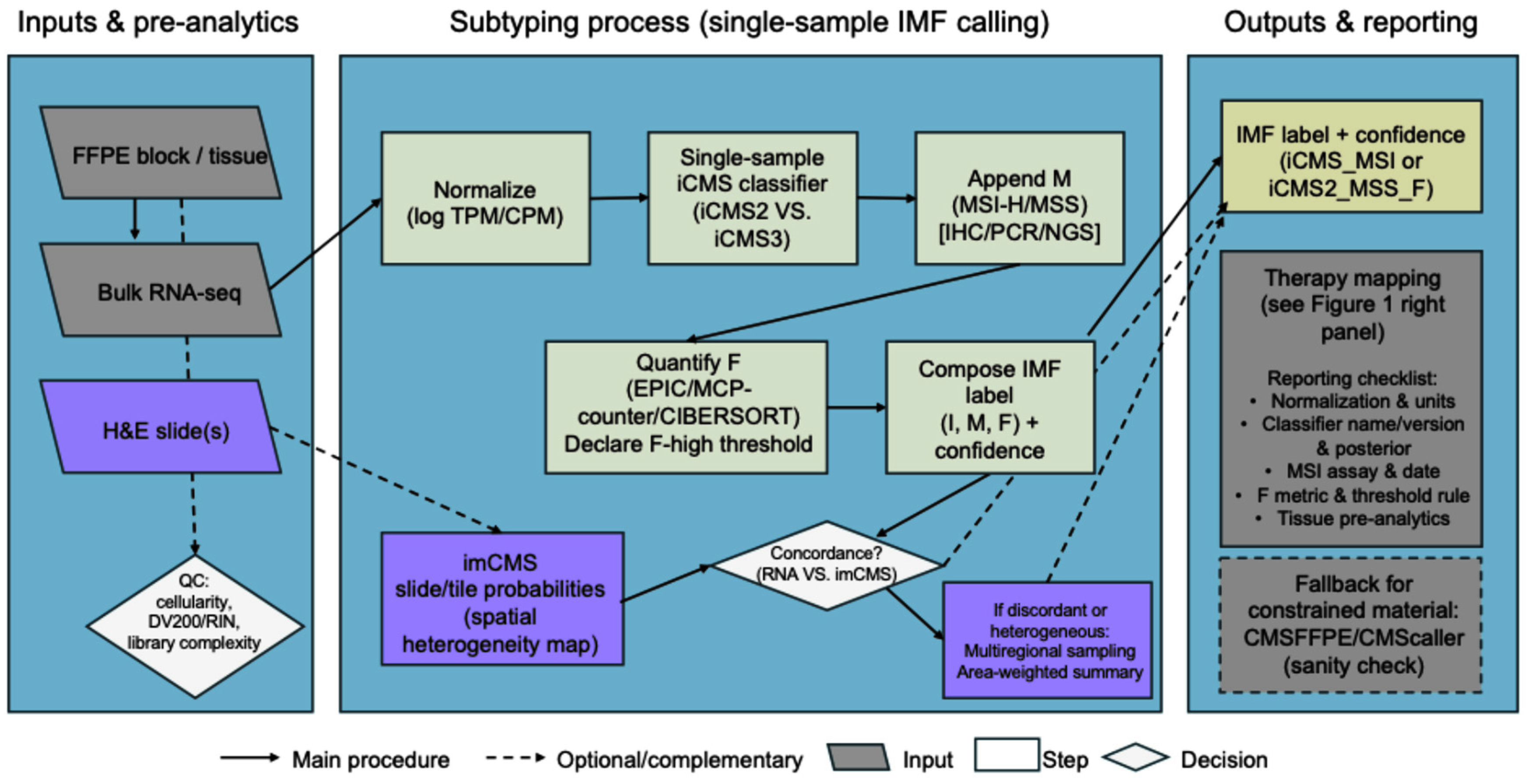
5. Suggested Implementation Guide: Calling iCMS/IMF in Clinical and Research Cohorts
5.1. Quality Control Prerequisites for IMF Classification Pipeline
5.2. Inputs and Step-by-Step Pipeline to Call IMF (I + M + F)
5.3. FFPE/Low-Quality Material: Compatibility and Specialized Classifiers
5.4. Complementary Modalities: Digital Pathology and Multiregional Profiling
6. Subtype Concordance Across Sites, Organ-Specific Context, and Patient-Derived Organoid-Stroma Translation
7. Discussion
8. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AE | Adverse event |
| AUC | Area under curve |
| CAF | Cancer-associated fibroblast |
| cfDNA | Cell-free DNA |
| CI | Confidence interval |
| CIMP | CpG-island methylator phenotype |
| CMS | Consensus molecular subtypes |
| CMSFFPE | FFPE-curated CMS classifier |
| CRC | Colorectal cancer |
| CRIS | CRC intrinsic subtypes |
| ctDNA | Circulating tumor DNA |
| dMMR | Deficient mismatch repair |
| EC | Encorafenib + cetuximab |
| ECM | Extracellular matrix |
| EMT | Epithelial–mesenchymal transition |
| EPIC | Estimate the proportion of immune and cancer cells |
| FAPI | FAP-inhibitor |
| FAPI-PET | FAP-targeted PET |
| FDA | Food and Drug Administration |
| FFPE | Formalin-fixed, paraffin-embedded |
| GR | Growth rate |
| H&E | Hematoxylin and eosin |
| HR | Hazard ratio |
| ICI | Immune checkpoint inhibitor |
| iCMS | Epithelial–intrinsic consensus molecular subtypes |
| IHC | Immunohistochemistry |
| imCMS | Image-based CMS |
| IMF | Intrinsic subtype-MSI-fibrosis |
| MCP | Microenvironment cell populations |
| mCRC | Metastatic CRC |
| mFOLFOX6 | Modified fluorouracil/leucovorin/oxaliplatin |
| mo | Month |
| MSI | Microsatellite instability |
| MSI-H | MSI-high |
| MSS | Microsatellite-stable |
| NGS | Next-generation sequencing |
| NS | Not significant |
| ORR | Objective response rate |
| OS | Overall survival |
| PCR | Polymerase chain reaction |
| PDO | Patient-derived organoid |
| PDX | Patient-derived xenograft |
| PFS | Progression-free survival |
| pMMR | Proficient mismatch repair |
| QC | Quality control |
| RIN | RNA integrity number |
| SOC | Standard of care |
| SOP | Standard operating procedure |
| TMB | Tumor mutational burden |
| TME | Tumor microenvironment |
| TRT | Targeted-radiogland therapy |
| UMI | Unique molecular identifier |
| WT | Wild-type |
| yr | Year |
References
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Joanito, I.; Wirapati, P.; Zhao, N.; Nawaz, Z.; Yeo, G.; Lee, F.; Eng, C.L.P.; Macalinao, D.C.; Kahraman, M.; Srinivasan, H.; et al. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat. Genet. 2022, 54, 963–975. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.O.; Hong, Y.; Etlioglu, H.E.; Cho, Y.B.; Pomella, V.; Van den Bosch, B.; Vanhecke, J.; Verbandt, S.; Hong, H.; Min, J.W.; et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 2020, 52, 594–603. [Google Scholar] [CrossRef]
- Pelka, K.; Hofree, M.; Chen, J.H.; Sarkizova, S.; Pirl, J.D.; Jorgji, V.; Bejnood, A.; Dionne, D.; Ge, W.H.; Xu, K.H.; et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell 2021, 184, 4734–4752.e20. [Google Scholar] [CrossRef]
- Becht, E.; de Reynies, A.; Giraldo, N.A.; Pilati, C.; Buttard, B.; Lacroix, L.; Selves, J.; Sautes-Fridman, C.; Laurent-Puig, P.; Fridman, W.H. Immune and Stromal Classification of Colorectal Cancer Is Associated with Molecular Subtypes and Relevant for Precision Immunotherapy. Clin. Cancer Res. 2016, 22, 4057–4066. [Google Scholar] [CrossRef]
- Isella, C.; Brundu, F.; Bellomo, S.E.; Galimi, F.; Zanella, E.; Porporato, R.; Petti, C.; Fiori, A.; Orzan, F.; Senetta, R.; et al. Selective analysis of cancer-cell intrinsic transcriptional traits defines novel clinically relevant subtypes of colorectal cancer. Nat. Commun. 2017, 8, 15107. [Google Scholar] [CrossRef]
- Cascianelli, S.; Barbera, C.; Ulla, A.A.; Grassi, E.; Lupo, B.; Pasini, D.; Bertotti, A.; Trusolino, L.; Medico, E.; Isella, C.; et al. Multi-label transcriptional classification of colorectal cancer reflects tumor cell population heterogeneity. Genome Med. 2023, 15, 37. [Google Scholar] [CrossRef]
- Sirinukunwattana, K.; Domingo, E.; Richman, S.D.; Redmond, K.L.; Blake, A.; Verrill, C.; Leedham, S.J.; Chatzipli, A.; Hardy, C.; Whalley, C.M.; et al. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut 2021, 70, 544–554. [Google Scholar] [CrossRef]
- Dunne, P.D.; Arends, M.J. Molecular pathological classification of colorectal cancer-an update. Virchows Arch. 2024, 484, 273–285, Erratum in Virchows Arch. 2024, 484, 287. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Fu, M.; Wang, M.; Wei, Y.; Wei, X. Targeting TGF-beta signal transduction for fibrosis and cancer therapy. Mol. Cancer 2022, 21, 104. [Google Scholar] [CrossRef] [PubMed]
- Andre, T.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef]
- Diaz, L.A., Jr.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): Final analysis of a randomised, open-label, phase 3 study. Lancet Oncol. 2022, 23, 659–670. [Google Scholar] [CrossRef]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef]
- Tabernero, J.; Grothey, A.; Van Cutsem, E.; Yaeger, R.; Wasan, H.; Yoshino, T.; Desai, J.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib Plus Cetuximab as a New Standard of Care for Previously Treated BRAF V600E-Mutant Metastatic Colorectal Cancer: Updated Survival Results and Subgroup Analyses from the BEACON Study. J. Clin. Oncol. 2021, 39, 273–284. [Google Scholar] [CrossRef]
- Yoshino, T.; Hooda, N.; Younan, D.; Muro, K.; Shitara, K.; Heinemann, V.; O’Neil, B.H.; Herrero, F.R.; Peeters, M.; Soeda, J.; et al. A meta-analysis of efficacy and safety data from head-to-head first-line trials of epidermal growth factor receptor inhibitors versus bevacizumab in adult patients with RAS wild-type metastatic colorectal cancer by sidedness. Eur. J. Cancer 2024, 202, 113975. [Google Scholar] [CrossRef]
- Mori, Y.; Dendl, K.; Cardinale, J.; Kratochwil, C.; Giesel, F.L.; Haberkorn, U. FAPI PET: Fibroblast Activation Protein Inhibitor Use in Oncologic and Nononcologic Disease. Radiology 2023, 306, e220749. [Google Scholar] [CrossRef] [PubMed]
- Privé, B.M.; Boussihmad, M.A.; Timmermans, B.; van Gemert, W.A.; Peters, S.M.B.; Derks, Y.H.W.; van Lith, S.A.M.; Mehra, N.; Nagarajah, J.; Heskamp, S.; et al. Fibroblast activation protein-targeted radionuclide therapy: Background, opportunities, and challenges of first (pre)clinical studies. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 1906–1918. [Google Scholar] [CrossRef] [PubMed]
- Racle, J.; Gfeller, D. EPIC: A Tool to Estimate the Proportions of Different Cell Types from Bulk Gene Expression Data. Methods Mol. Biol. 2020, 2120, 233–248. [Google Scholar]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautes-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218, Erratum in Genome Biol. 2016, 17, 249. [Google Scholar] [CrossRef] [PubMed]
- de Back, T.R.; Wu, T.; Schafrat, P.J.; Ten Hoorn, S.; Tan, M.; He, L.; van Hooff, S.R.; Koster, J.; Nijman, L.E.; Vink, G.R.; et al. A consensus molecular subtypes classification strategy for clinical colorectal cancer tissues. Life Sci. Alliance 2024, 7, e202402730, Erratum in Life Sci. Alliance 2024, 7, e202402889. https://doi.org/10.26508/lsa.202402889. [Google Scholar] [CrossRef]
- Langerud, J.; Eilertsen, I.A.; Moosavi, S.H.; Klokkerud, S.M.K.; Reims, H.M.; Backe, I.F.; Hektoen, M.; Sjo, O.H.; Jeanmougin, M.; Tejpar, S.; et al. Multiregional transcriptomics identifies congruent consensus subtypes with prognostic value beyond tumor heterogeneity of colorectal cancer. Nat. Commun. 2024, 15, 4342. [Google Scholar] [CrossRef]
- Koncina, E.; Nurmik, M.; Pozdeev, V.I.; Gilson, C.; Tsenkova, M.; Begaj, R.; Stang, S.; Gaigneaux, A.; Weindorfer, C.; Rodriguez, F.; et al. IL1R1(+) cancer-associated fibroblasts drive tumor development and immunosuppression in colorectal cancer. Nat. Commun. 2023, 14, 4251. [Google Scholar] [CrossRef]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Canellas, A.; Hernando-Momblona, X.; et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Waldner, M.J.; Neurath, M.F. TGFbeta and the Tumor Microenvironment in Colorectal Cancer. Cells 2023, 12, 1139. [Google Scholar] [CrossRef]
- Strating, E.; van de Loo, A.; Elias, S.; Lam, M.; Kranenburg, O. Fibroblast Activation Protein Inhibitor-PET Imaging in Colorectal Cancer. PET Clin. 2023, 18, 325–335. [Google Scholar] [CrossRef]
- Valdeolivas, A.; Amberg, B.; Giroud, N.; Richardson, M.; Galvez, E.J.C.; Badillo, S.; Julien-Laferriere, A.; Turos, D.; Voith von Voithenberg, L.; Wells, I.; et al. Profiling the heterogeneity of colorectal cancer consensus molecular subtypes using spatial transcriptomics. NPJ Precis. Oncol. 2024, 8, 10. [Google Scholar] [CrossRef]
- Xiao, J.; Yu, X.; Meng, F.; Zhang, Y.; Zhou, W.; Ren, Y.; Li, J.; Sun, Y.; Sun, H.; Chen, G.; et al. Integrating spatial and single-cell transcriptomics reveals tumor heterogeneity and intercellular networks in colorectal cancer. Cell Death Dis. 2024, 15, 326. [Google Scholar] [CrossRef]
- Feng, Y.; Ma, W.; Zang, Y.; Guo, Y.; Li, Y.; Zhang, Y.; Dong, X.; Liu, Y.; Zhan, X.; Pan, Z.; et al. Spatially organized tumor-stroma boundary determines the efficacy of immunotherapy in colorectal cancer patients. Nat. Commun. 2024, 15, 10259. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, D.; Li, Y.; Qi, L.; Si, W.; Bo, Y.; Chen, X.; Ye, Z.; Fan, H.; Liu, B.; et al. Spatiotemporal single-cell analysis decodes cellular dynamics underlying different responses to immunotherapy in colorectal cancer. Cancer Cell 2024, 42, 1268–1285.e7. [Google Scholar] [CrossRef] [PubMed]
- Andre, T.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.J.A.; Smith, D.; Garcia-Carbonero, R.; Alcaide-Garcia, J.; Gibbs, P.; et al. Pembrolizumab versus chemotherapy in microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer: 5-year follow-up from the randomized phase III KEYNOTE-177 study. Ann. Oncol. 2025, 36, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Heregger, R.; Huemer, F.; Steiner, M.; Gonzalez-Martinez, A.; Greil, R.; Weiss, L. Unraveling Resistance to Immunotherapy in MSI-High Colorectal Cancer. Cancers 2023, 15, 5090. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, D.; Xiao, B.; Zhou, C.; Jiang, W.; Tang, J.; Li, Y.; Zhang, R.; Han, K.; Hou, Z.; et al. B2M and JAK1/2-mutated MSI-H Colorectal Carcinomas Can Benefit From Anti-PD-1 Therapy. J. Immunother. 2022, 45, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Fakih, M.; Raghav, K.P.S.; Chang, D.Z.; Larson, T.; Cohn, A.L.; Huyck, T.K.; Cosgrove, D.; Fiorillo, J.A.; Tam, R.; D’Adamo, D.; et al. Regorafenib plus nivolumab in patients with mismatch repair-proficient/microsatellite stable metastatic colorectal cancer: A single-arm, open-label, multicentre phase 2 study. eClinicalMedicine 2023, 58, 101917. [Google Scholar] [CrossRef]
- Eng, C.; Kim, T.W.; Bendell, J.; Argiles, G.; Tebbutt, N.C.; Di Bartolomeo, M.; Falcone, A.; Fakih, M.; Kozloff, M.; Segal, N.H.; et al. Atezolizumab with or without cobimetinib versus regorafenib in previously treated metastatic colorectal cancer (IMblaze370): A multicentre, open-label, phase 3, randomised, controlled trial. Lancet Oncol. 2019, 20, 849–861, Erratum in Lancet Oncol. 2019, 20, e293. [Google Scholar] [CrossRef]
- Kawazoe, A.; Xu, R.H.; Garcia-Alfonso, P.; Passhak, M.; Teng, H.W.; Shergill, A.; Gumus, M.; Qvortrup, C.; Stintzing, S.; Towns, K.; et al. Lenvatinib Plus Pembrolizumab Versus Standard of Care for Previously Treated Metastatic Colorectal Cancer: Final Analysis of the Randomized, Open-Label, Phase III LEAP-017 Study. J. Clin. Oncol. 2024, 42, 2918–2927. [Google Scholar] [CrossRef] [PubMed]
- Segal, N.H.; Melero, I.; Moreno, V.; Steeghs, N.; Marabelle, A.; Rohrberg, K.; Rodriguez-Ruiz, M.E.; Eder, J.P.; Eng, C.; Manji, G.A.; et al. CEA-CD3 bispecific antibody cibisatamab with or without atezolizumab in patients with CEA-positive solid tumours: Results of two multi-institutional Phase 1 trials. Nat. Commun. 2024, 15, 4091. [Google Scholar] [CrossRef]
- Ambrosini, M.; Rousseau, B.; Manca, P.; Artz, O.; Marabelle, A.; Andre, T.; Maddalena, G.; Mazzoli, G.; Intini, R.; Cohen, R.; et al. Immune checkpoint inhibitors for POLE or POLD1 proofreading-deficient metastatic colorectal cancer. Ann. Oncol. 2024, 35, 643–655. [Google Scholar] [CrossRef]
- Fabrizio, D.A.; George, T.J., Jr.; Dunne, R.F.; Frampton, G.; Sun, J.; Gowen, K.; Kennedy, M.; Greenbowe, J.; Schrock, A.B.; Hezel, A.F.; et al. Beyond microsatellite testing: Assessment of tumor mutational burden identifies subsets of colorectal cancer who may respond to immune checkpoint inhibition. J. Gastrointest. Oncol. 2018, 9, 610–617. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Andre, T.; Atreya, C.E.; Schellens, J.H.M.; Yoshino, T.; Bendell, J.C.; Hollebecque, A.; McRee, A.J.; Siena, S.; Middleton, G.; et al. Combined BRAF, EGFR, and MEK Inhibition in Patients with BRAF(V600E)-Mutant Colorectal Cancer. Cancer Discov. 2018, 8, 428–443. [Google Scholar] [CrossRef]
- Elez, E.; Yoshino, T.; Shen, L.; Lonardi, S.; Van Cutsem, E.; Eng, C.; Kim, T.W.; Wasan, H.S.; Desai, J.; Ciardiello, F.; et al. Encorafenib, Cetuximab, and mFOLFOX6 in BRAF-Mutated Colorectal Cancer. N. Engl. J. Med. 2025, 392, 2425–2437. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. FDA Grants Accelerated Approval to Encorafenib with Cetuximab and mFOLFOX6 for Metastatic Colorectal Cancer with a BRAF V600E Mutation; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2024.
- Watanabe, J.; Muro, K.; Shitara, K.; Yamazaki, K.; Shiozawa, M.; Ohori, H.; Takashima, A.; Yokota, M.; Makiyama, A.; Akazawa, N.; et al. Panitumumab vs Bevacizumab Added to Standard First-line Chemotherapy and Overall Survival Among Patients With RAS Wild-type, Left-Sided Metastatic Colorectal Cancer: A Randomized Clinical Trial. JAMA 2023, 329, 1271–1282. [Google Scholar] [CrossRef]
- Airoldi, M.; Bartolini, M.; Fazio, R.; Farinatti, S.; Dapra, V.; Santoro, A.; Puccini, A. First-Line Therapy in Metastatic, RAS Wild-Type, Left-Sided Colorectal Cancer: Should Everyone Receive Anti-EGFR Therapy? Curr. Oncol. Rep. 2024, 26, 1489–1501. [Google Scholar] [CrossRef]
- Morris, V.K.; Kennedy, E.B.; Baxter, N.N.; Benson, A.B., 3rd; Cercek, A.; Cho, M.; Ciombor, K.K.; Cremolini, C.; Davis, A.; Deming, D.A.; et al. Treatment of Metastatic Colorectal Cancer: ASCO Guideline. J. Clin. Oncol. 2023, 41, 678–700. [Google Scholar] [CrossRef]
- Cheng, Z.; Wang, S.; Xu, S.; Du, B.; Li, X.; Li, Y. FAPI PET/CT in Diagnostic and Treatment Management of Colorectal Cancer: Review of Current Research Status. J. Clin. Med. 2023, 12, 577. [Google Scholar] [CrossRef] [PubMed]
- Baum, R.P.; Schuchardt, C.; Singh, A.; Chantadisai, M.; Robiller, F.C.; Zhang, J.; Mueller, D.; Eismant, A.; Almaguel, F.; Zboralski, D.; et al. Feasibility, Biodistribution, and Preliminary Dosimetry in Peptide-Targeted Radionuclide Therapy of Diverse Adenocarcinomas Using (177)Lu-FAP-2286: First-in-Humans Results. J. Nucl. Med. 2022, 63, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Bentestuen, M.; Al-Obaydi, N.; Zacho, H.D. FAPI-avid nonmalignant PET/CT findings: An expedited systematic review. Semin. Nucl. Med. 2023, 53, 694–705. [Google Scholar] [CrossRef]
- Galbiati, A.; Bocci, M.; Ravazza, D.; Mock, J.; Gilardoni, E.; Neri, D.; Cazzamalli, S. Preclinical Evaluation of (177)Lu-OncoFAP-23, a Multivalent FAP-Targeted Radiopharmaceutical Therapeutic for Solid Tumors. J. Nucl. Med. 2024, 65, 1604–1610. [Google Scholar] [CrossRef] [PubMed]
- McConathy, J.; Menda, Y.; Rodon, J.; Goenka, A.H.; Moy, R.H.; Morse, S.; Demange, A.; Aimone, P.; Hope, T.A. 671P LuMIERE: A phase I/II study evaluating safety, dosimetry, and preliminary activity of [177Lu]Lu-FAP-2286 in patients with advanced solid tumors. Ann. Oncol. 2024, 35, S526. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 November 2025).
- Picard Toolkit; Broad Institute: Cambridge, MA, USA, 2019.
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef]
- Wolock, S.L.; Lopez, R.; Klein, A.M. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 2019, 8, 281–291.e9. [Google Scholar] [CrossRef]
- McGinnis, C.S.; Murrow, L.M.; Gartner, Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019, 8, 329–337.e4. [Google Scholar] [CrossRef]
- iCMS Single-Sample Classifier. GitHub Repository. 2024. Available online: https://github.com/CRCrepository/iCMS.SSC (accessed on 8 November 2025).
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Peter, W.; Eide, J.B.; Ragnhild, A. Lothe, Anita Sveen CMScaller: An R Package for Consensus Molecular Subtyping of Colorectal Cancer Pre-Clinical Models; GitHub: San Francisco, CA, USA, 2017. [Google Scholar]
- Eide, P.W.; Bruun, J.; Lothe, R.A.; Sveen, A. CMScaller: An R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci. Rep. 2017, 7, 16618. [Google Scholar] [CrossRef]
- Borelli, B.; Fontana, E.; Giordano, M.; Antoniotti, C.; Lonardi, S.; Bergamo, F.; Pietrantonio, F.; Morano, F.; Tamburini, E.; Boccaccino, A.; et al. Prognostic and predictive impact of consensus molecular subtypes and CRCAssigner classifications in metastatic colorectal cancer: A translational analysis of the TRIBE2 study. ESMO Open 2021, 6, 100073. [Google Scholar] [CrossRef] [PubMed]
- Baena-Del Valle, J.A.; Zheng, Q.; Hicks, J.L.; Fedor, H.; Trock, B.J.; Morrissey, C.; Corey, E.; Cornish, T.C.; Sfanos, K.S.; De Marzo, A.M. Rapid Loss of RNA Detection by In Situ Hybridization in Stored Tissue Blocks and Preservation by Cold Storage of Unstained Slides. Am. J. Clin. Pathol. 2017, 148, 398–415. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Liu, H.; Ge, Y.; Wang, X. Integrated single-cell and bulk RNA sequencing analysis identifies a cancer associated fibroblast-related signature for predicting prognosis and therapeutic responses in colorectal cancer. Cancer Cell Int. 2021, 21, 552. [Google Scholar] [CrossRef] [PubMed]
- Chandekar, K.R.; Prashanth, A.; Vinjamuri, S.; Kumar, R. FAPI PET/CT Imaging—An Updated Review. Diagnostics 2023, 13, 2018. [Google Scholar] [CrossRef]
- Farin, H.F.; Mosa, M.H.; Ndreshkjana, B.; Grebbin, B.M.; Ritter, B.; Menche, C.; Kennel, K.B.; Ziegler, P.K.; Szabo, L.; Bollrath, J.; et al. Colorectal Cancer Organoid-Stroma Biobank Allows Subtype-Specific Assessment of Individualized Therapy Responses. Cancer Discov. 2023, 13, 2192–2211. [Google Scholar] [CrossRef]
- Ooft, S.N.; Weeber, F.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van Werkhoven, E.; Schipper, L.; Hoes, L.; Vis, D.J.; van de Haar, J.; et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019, 11, eaay2574. [Google Scholar] [CrossRef]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernandez-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef]
- Bartolomucci, A.; Nobrega, M.; Ferrier, T.; Dickinson, K.; Kaorey, N.; Nadeau, A.; Castillo, A.; Burnier, J.V. Circulating tumor DNA to monitor treatment response in solid tumors and advance precision oncology. NPJ Precis. Oncol. 2025, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Gianni, C.; Palleschi, M.; Merloni, F.; Di Menna, G.; Sirico, M.; Sarti, S.; Virga, A.; Ulivi, P.; Cecconetto, L.; Mariotti, M.; et al. Cell-Free DNA Fragmentomics: A Promising Biomarker for Diagnosis, Prognosis and Prediction of Response in Breast Cancer. Int. J. Mol. Sci. 2022, 23, 14197. [Google Scholar] [CrossRef]
- Dong, W.; Hu, W.; Lu, Y.; Zheng, Q. Cell-free DNA fragmentomics: A universal framework for early cancer detection and monitoring. Am. J. Clin. Exp. Immunol. 2025, 14, 237–240. [Google Scholar] [CrossRef]
- Conway, A.M.; Pearce, S.P.; Clipson, A.; Hill, S.M.; Chemi, F.; Slane-Tan, D.; Ferdous, S.; Hossain, A.; Kamieniecka, K.; White, D.J.; et al. A cfDNA methylation-based tissue-of-origin classifier for cancers of unknown primary. Nat. Commun. 2024, 15, 3292. [Google Scholar] [CrossRef] [PubMed]
- Moon, G.Y.; Dalkiran, B.; Park, H.S.; Shin, D.; Son, C.; Choi, J.H.; Bang, S.; Lee, H.; Doh, I.; Kim, D.H.; et al. Dual Biomarker Strategies for Liquid Biopsy: Integrating Circulating Tumor Cells and Circulating Tumor DNA for Enhanced Tumor Monitoring. Biosensors 2025, 15, 74. [Google Scholar] [CrossRef] [PubMed]
| Clinical Scenario | Line/Setting | Suggested Regimen | Key Evidence (Trial; Statistics) | Limitations/Notes |
|---|---|---|---|---|
| MSI-H/dMMR metastatic CRC (mCRC) | First-line | PD-1 monotherapy (pembrolizumab) | KEYNOTE-177: PFS 16.5 vs. 8.2 mo (HR 0.60, 95% CI 0.45–0.79); OS 77.5 vs. 36.7 mo (HR 0.73, 95% CI 0.53–0.99); 5-yr OS 54.8% vs. 44.2%; grade ≥ 3 AEs 22% vs. 67% | OS may be diluted by a 62% crossover (effective crossover); the evidence is mainly based on PD-1 monotherapy |
| BRAF V600E-mutated mCRC (often pMMR/MSS) | First-line | Encorafenib + Cetuximab + mFOLFOX6 | BREAKWATER: PFS 12.8 vs. 7.1 mo (HR 0.53, 95% CI 0.41–0.68; p < 0.001); interim OS 30.3 vs. 15.1 mo (HR 0.49, 95% CI 0.38–0.63; p < 0.001) | The indication is limited to BRAF V600E; with a 46.1% incidence of serious adverse events, safety management is required. For MSI-H, first-line therapy should still prioritize PD-1. |
| RAS-WT, left-sided mCRC (MSS/pMMR) | First-line | Anti-EGFR + doublet chemo (e.g., panitumumab + mFOLFOX6) | PARADIGM: Left-sided OS 37.9 vs. 34.3 mo (HR 0.82; p = 0.03); ORR 80.2% vs. 68.6%; PFS 13.1 vs. 11.9 mo (HR 1.00). Meta analysis study: Left sided OS HR 0.80 (95% CI 0.71–0.90), PFS NS (HR 0.93) | Left-sided tumors showed concentrated benefit; no OS gain on the right, with PFS often favoring bevacizumab. EGFR-related toxicities (skin rash, hypomagnesemia) require management. |
| pMMR/MSS—immune checkpoint inhibitor (ICI) combination trial | Post-standard (pretreated) | (mainly negative results) | LEAP-017: lenvatinib + pembrolizumab vs. SOC, OS 9.8 vs. 9.3 mo (HR 0.83) IMblaze370: atezolizumab + cobimetinib vs. regorafenib, OS 8.87 vs. 8.51 mo (HR 1.00, p = 0.99) | Unselected pMMR/MSS: no OS benefit in phase III trials; trial enrollment advised considering biomarkers and organ context |
| Ligand/Program | Modality and Key Features | Development Status (as of 2025) | Key Data/Notes |
|---|---|---|---|
| FAP-2286 | Peptide binder; comparatively longer tumor residence and internalization | Phase I/II ongoing (LuMIERE) | First-in-human studies reported acceptable tolerability and favorable dosimetry; early signals of activity. Ongoing trial evaluates safety, dosimetry, and preliminary efficacy. |
| OncoFAP-23 | Multivalent small molecule designed to improve tumor uptake and retention | Preclinical (clinical entry in preparation) | 2024 preclinical work showed higher tumor retention with reduced normal-organ uptake and improved in vivo antitumor effects—addresses short-residence limitation of first-generation tracers. |
| FAPI tetramers/multimeric optimization series | Multimeric/high-avidity designs to enhance affinity and residence time | Preclinical/translational | Structure-activity optimization reports describe improved kinetics (improved residence and target binding) relative to early monomers; clinical translation pending. |
| FAPI-46 family | Early-generation small molecules | Case series/small exploratory studies | Compassionate-use and small cohorts suggest feasibility and manageable safety; in some tumors, rapid washout limits delivered dose—motivates next-gen ligands. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, S. From CMS to iCMS/IMF: Developing Roadmap to Precision Therapy in Colorectal Cancer. Int. J. Mol. Sci. 2025, 26, 11086. https://doi.org/10.3390/ijms262211086
Jung S. From CMS to iCMS/IMF: Developing Roadmap to Precision Therapy in Colorectal Cancer. International Journal of Molecular Sciences. 2025; 26(22):11086. https://doi.org/10.3390/ijms262211086
Chicago/Turabian StyleJung, Sungwon. 2025. "From CMS to iCMS/IMF: Developing Roadmap to Precision Therapy in Colorectal Cancer" International Journal of Molecular Sciences 26, no. 22: 11086. https://doi.org/10.3390/ijms262211086
APA StyleJung, S. (2025). From CMS to iCMS/IMF: Developing Roadmap to Precision Therapy in Colorectal Cancer. International Journal of Molecular Sciences, 26(22), 11086. https://doi.org/10.3390/ijms262211086





