Epigenetic and Post-Translational Regulation of Schlafen Family Expression and Their Differential Methods of Regulating Proteins
Abstract
1. Introduction
1.1. Origin and Derivation
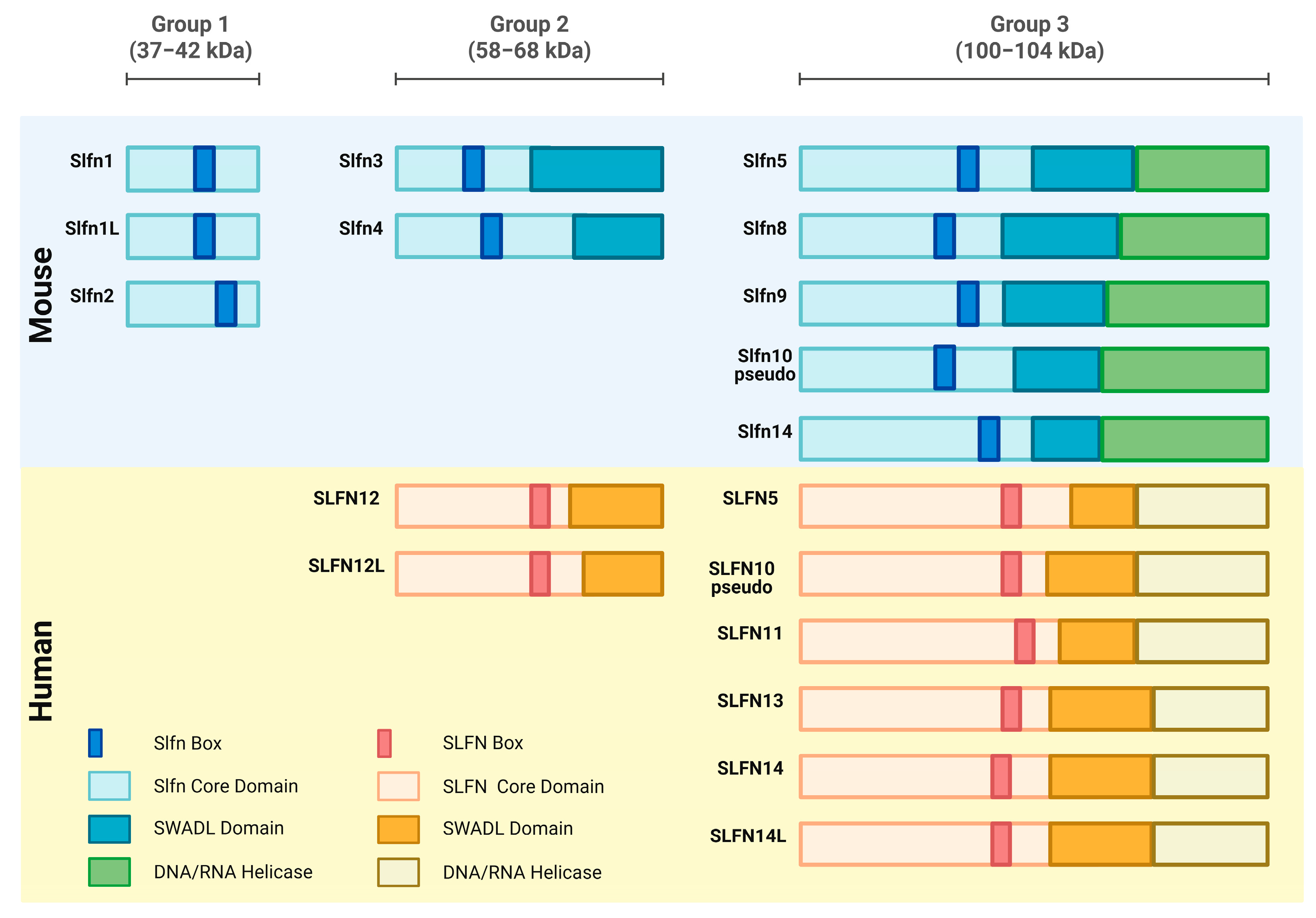
1.2. Schlafen Gene Characteristics and Subcellular Localization
2. Schlafen Function in Mice and Humans
2.1. Role in Immune Cell Differentiation
2.2. Role in Malignant Cell Differentiation
2.3. Role in Viral Replication
3. Emerging Importance of Epigenetic Regulation in Schlafen-Mediated Immunity and Cancer
4. Role of Regulatory Influence on the Expression of Schlafen Family Members
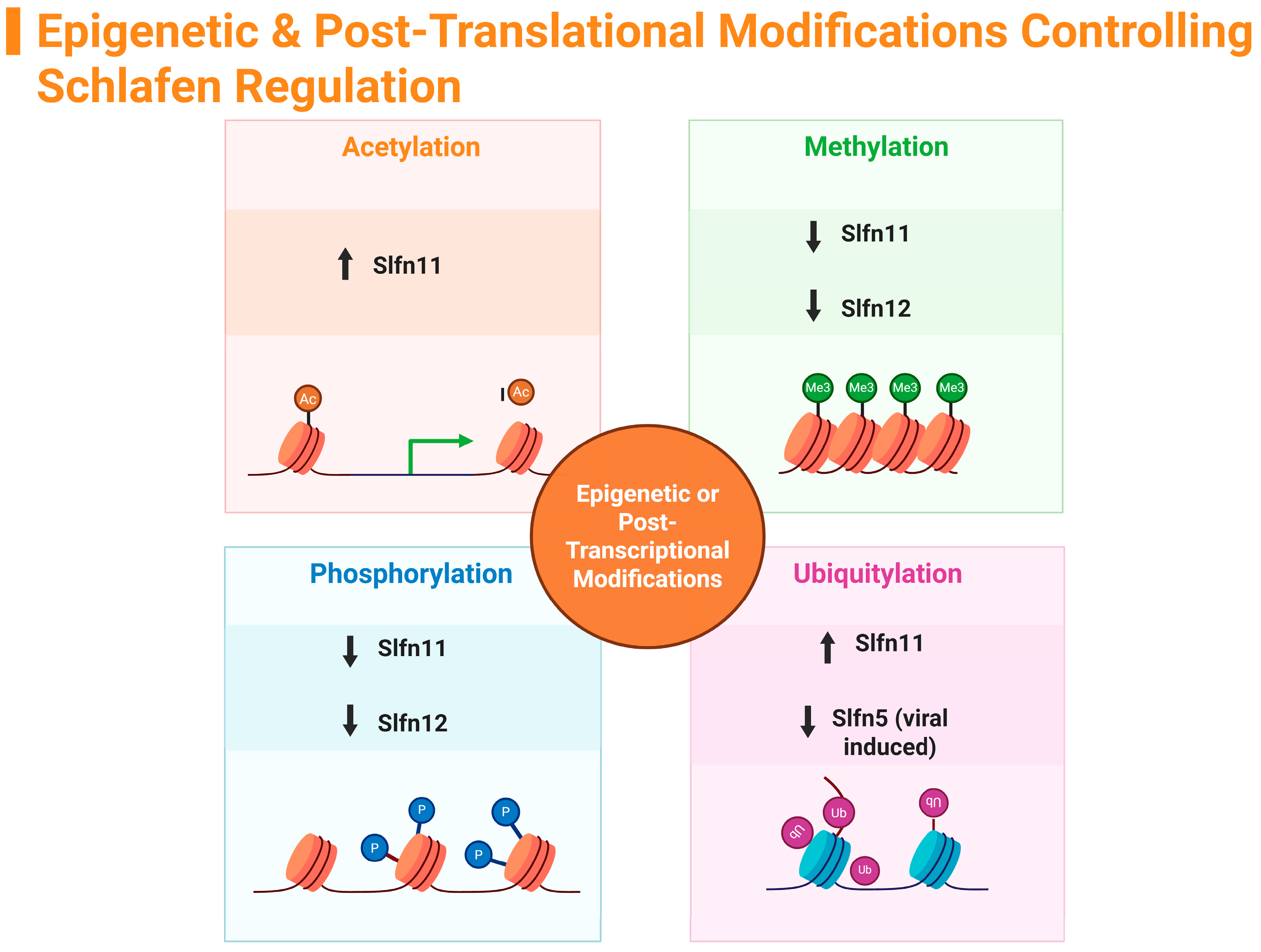
5. Regulation of Schlafen Family Members by Epigenetic Modifications
5.1. Brief Overview of Epigenetic Modifications
5.2. Acetylation
5.3. Methylation
5.4. Phosphorylation—Epigenetic and Post-Translation Modifications
5.5. Ubiquitylation
| Type of Modification | Which SLFN is Modified | Location of Modification | SLFN RNA Expression | References |
|---|---|---|---|---|
| Acetylation | ||||
| Histone Acetylation (↓ acetylation) | SLFN11 | H3K18Ac (CD47-null cells). | ↓ Downregulated | [40] |
| Histone Deacetylation (epigenetic inactivation) | SLFN11 | H3K9Ac (K562 cells) | ↓ Downregulated | [21] |
| Histone Acetylation (↑ via HDAC inhibitor FK228) | SLFN11 | H3K9Ac and H3K27Ac (SCLC small cell lung cancer cell lines: H82, H69, DMS273, and H526) | ↑ Upregulated (increased acetylation, reduced DNA methylation) | [22] |
| Histone Acetylation (↑ via HDAC inhibitors FK228 and Panobinostat) | SLFN11 | H3K9Ac (B-cell-derived lymphoma cell lines: FL18 and FL318) | ↑ Upregulated | [41] |
| Methylation | ||||
| DNA Methylation (hypermethylation) | SLFN11 | SLFN11 CpG promoter island hypermethylation (NCI-60 human ovarian and non-small cell lung cancer (NSCLC) line | ↓ Downregulated | [43] |
| DNA Methylation (reduced via HDAC inhibitors FK228, SAHA) | SLFN11 | H3K9Ac and H3K27Ac (SCLC small cell lung cancer cell lines: H82, H69, DMS273, and H526) | ↑ Upregulated after demethylation | [22] |
| DNA Methylation (hypermethylation) | SLFN11 | SLFN11 promoter CpG island methylation (gastric cancer cell lines: SNU16, MGC803, and NUGC3) | ↓ Downregulated | [19] |
| DNA Methylation (hypermethylation) | SLFN11 | SLFN11 promoter CpG island methylation (colorectal cancer cell lines: RKO, DLD1, SW620, LOVO—complete; Ls180—partial; DKO—unmethylated) | ↓ Downregulated | [18] |
| DNA Methylation (reversible via 5-Aza treatment) | SLFN11 | SLFN11 promoter CpG methylation (bladder urothelial carcinoma cells) | ↓ Downregulated; ↑ Upregulated after demethylation | [44] |
| DNA Methylation (hypermethylation) | SLFN11 | Promoter region in ovarian cancers (HGSC, CCC) | ↓ Downregulated | [20] |
| DNA Methylation (exercise-induced demethylation) | SLFN12 | SLFN12 promoter CpG methylation (leukocytes) | ↓ Downregulated; ↑ Upregulated after exercise | [45] |
| DNA Methylation (hypermethylation) | SLFN12 | SLFN12 promoter CpG methylation (CD4+ and CD8+ T cells in naïve multiple sclerosis) | ↓ Downregulated | [24,46] |
| DNA Methylation (hypomethylation) | SLFN12 | SLFN12 promoter CpG methylation (hashimoto’s thyroiditis whole blood) | ↑ Upregulated | [25] |
| Phosphorylation (Epigenetic and Post-Translation Modification) | ||||
| Phosphorylation | SLFN11 | S214, S219, T230, S753 | ↓ Downregulated; Inhibits RNase and DNA binding | [48,49] |
| Dephosphorylation (via protein phosphatase 2A) | SLFN11 | S180, S219, T230, S750, S753 | ↑ Upregulated; SLFN11, increases drug sensitivity | [51] |
| Phosphorylation/Dephosphorylation (via PDE3A interaction) | SLFN12 | S368, S573 (RNA-binding domain) | ↓ Downregulated; Inhibits RNase; ↑ Dephosphorylation activates RNase function | [28,30,31,52] |
| Drug-induced Dephosphorylation (BAY 2666605) | SLFN12 | PDE3A–SLFN12 complex (heterotetrameric interface) | ↑ Upregulated; induces cancer-selective cytotoxicity | [27,32] |
| Ubiquitylation (Post-Translation Modification) | ||||
| Ubiquitylation | SLFN11 | Global ubiquitylation (SLFN11-KO cells) | ↑ Upregulated; increases drug sensitivity | [53] |
| Ubiquitylation | SLFN5 | K residues (HFFs, HeLa, HEK293T lines) | ↓ Downregulated; viral countermeasure | [54] |
6. Conclusions
Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Slfn | Murine Schlafen |
| SLFN | Human Schlafen |
| TNBC | Triple negative breast cancer |
| IFN-α2 | Interferon-α2 |
| SCLC | Small-cell lung cancer |
| MS | Multiple sclerosis |
References
- Schwarz, D.A.; Katayama, C.D.; Hedrick, S.M. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity 1998, 9, 657–668. [Google Scholar] [CrossRef]
- Mavrommatis, E.; Fish, E.N.; Platanias, L.C. The Schlafen Family of Proteins and Their Regulation by Interferons. J. Interf. Cytokine Res. 2013, 33, 206–210. [Google Scholar] [CrossRef]
- Chen, J.; Kuhn, L.A. Deciphering the three-domain architecture in schlafens and the structures and roles of human schlafen12 and serpinB12 in transcriptional regulation. J. Mol. Graph. Model. 2019, 90, 59–76. [Google Scholar] [CrossRef]
- Seong, R.-K.; Seo, S.-W.; Kim, J.-A.; Fletcher, S.J.; Morgan, N.V.; Kumar, M.; Choi, Y.-K.; Shin, O.S. Schlafen 14 (SLFN14) is a novel antiviral factor involved in the control of viral replication. Immunobiology 2017, 222, 979–988. [Google Scholar] [CrossRef] [PubMed]
- de La Casa-Espero’n, E. From mammals to viruses: The Schlafen genes in developmental, proliferative and immune processes. Biomol. Concepts 2011, 2, 159–169. [Google Scholar] [CrossRef]
- Abdel-Mohsen, M.; Raposo, R.A.S.; Deng, X.; Li, M.; Liegler, T.; Sinclair, E.; Salama, M.S.; A Ghanem, H.E.-D.; Hoh, R.; Wong, J.K.; et al. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology 2013, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.T.; Weitzman, M.D. Schlafens Can Put Viruses to Sleep. Viruses 2022, 14, 442. [Google Scholar] [CrossRef]
- Geserick, P.; Kaiser, F.; Klemm, U.; Kaufmann, S.H.E.; Zerrahn, J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int. Immunol. 2004, 16, 1535–1548. [Google Scholar] [CrossRef]
- Neumann, B.; Zhao, L.; Murphy, K.; Gonda, T.J. Subcellular localization of the Schlafen protein family. Biochem. Biophys. Res. Commun. 2008, 370, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Al-Marsoummi, S.; Vomhof-DeKrey, E.E.; Basson, M.D. Schlafens: Emerging Proteins in Cancer Cell Biology. Cells 2021, 10, 2238. [Google Scholar] [CrossRef]
- Mavrommatis, E.; Arslan, A.D.; Sassano, A.; Hua, Y.; Kroczynska, B.; Platanias, L.C. Expression and regulatory effects of Murine Schlafen (Slfn) genes in malignant melanoma and renal cell carcinoma. J. Biol. Chem. 2013, 288, 33006–33015. [Google Scholar] [CrossRef]
- Lund, S.; Christensen, K.V.; Hedtjärn, M.; Mortensen, A.L.; Hagberg, H.; Falsig, J.; Hasseldam, H.; Schrattenholz, A.; Pörzgen, P.; Leist, M. The dynamics of the LPS triggered inflammatory response of murine microglia under different culture and in vivo conditions. J. Neuroimmunol. 2006, 180, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Bustos, O.; Naik, S.; Ayers, G.; Casola, C.; Perez-Lamigueiro, M.A.; Chippindale, P.T.; Pritham, E.J.; de la Casa-Esperón, E. Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene 2009, 447, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Warsi, S.; Dahl, M.; Smith, E.M.K.; Rydstrom, A.; Mansell, E.; Sigurdsson, V.; Sjoberg, J.; Soneji, S.; Rorby, E.; Siva, K.; et al. Schlafen2 is a regulator of quiescence in adult murine hematopoietic stem cells. Haematologica 2022, 107, 2884–2896. [Google Scholar] [CrossRef]
- Gubser, C.; Goodbody, R.; Ecker, A.; Brady, G.; O’Neill, L.A.; Jacobs, N.; Smith, G.L. Camelpox virus encodes a schlafen-like protein that affects orthopoxvirus virulence. J. Gen. Virol. 2007, 88, 1667–1676. [Google Scholar] [CrossRef]
- Valdez, F.; Salvador, J.; Palermo, P.M.; Mohl, J.E.; Hanley, K.A.; Watts, D.; Llano, M. Schlafen 11 Restricts Flavivirus Replication. J. Virol. 2019, 93, e00104-19. [Google Scholar] [CrossRef]
- Al-Marsoummi, S.; Vomhof-DeKrey, E.; Basson, M.D. Schlafen12 Reduces the Aggressiveness of Triple Negative Breast Cancer through Post-Transcriptional Regulation of ZEB1 That Drives Stem Cell Differentiation. Cell. Physiol. Biochem. 2019, 53, 999–1014. [Google Scholar] [CrossRef]
- He, T.; Zhang, M.; Zheng, R.; Zheng, S.; Linghu, E.; Herman, J.G.; Guo, M. Methylation of SLFN11 is a marker of poor prognosis and cisplatin resistance in colorectal cancer. Epigenomics 2017, 9, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, L.; Wu, L.; Zhang, L.; Nie, G.; Guo, M. Methylation of SLFN11 promotes gastric cancer growth and increases gastric cancer cell resistance to cisplatin. J. Cancer 2019, 10, 6124–6134. [Google Scholar] [CrossRef]
- Akashi, H.; Yachida, N.; Ueda, H.; Yamaguchi, M.; Yamawaki, K.; Tamura, R.; Suda, K.; Ishiguro, T.; Adachi, S.; Nagase, Y.; et al. SLFN11 is a BRCA independent biomarker for the response to platinum-based chemotherapy in high-grade serous ovarian cancer and clear cell ovarian carcinoma. Mol. Cancer Ther. 2024, 23, 106–116. [Google Scholar] [CrossRef]
- Tang, S.-W.; Thomas, A.; Murai, J.; Trepel, J.B.; Bates, S.E.; Rajapakse, V.N.; Pommier, Y. Overcoming resistance to DNA-targeted agents by epigenetic activation of Schlafen 11 (SLFN11) expression with class I histone deacetylase inhibitors. Clin. Cancer Res. 2018, 24, 1944–1953. [Google Scholar] [CrossRef]
- Yin, Y.-P.; Ma, L.-Y.; Cao, G.-Z.; Hua, J.-H.; Lv, X.-T.; Lin, W.-C. FK228 potentiates topotecan activity against small cell lung cancer cells via induction of SLFN11. Acta Pharmacol. Sin. 2021, 43, 2119–2127. [Google Scholar] [CrossRef]
- Brown, S.R.; Vomhof-DeKrey, E.E.; Al-Marsoummi, S.; Brown, N.D.; Hermanson, K.; Basson, M.D. Schlafen Family Intra-Regulation by IFN-α2 in Triple-Negative Breast Cancer. Cancers 2023, 15, 5658. [Google Scholar] [CrossRef] [PubMed]
- Rhead, B.; Brorson, I.S.; Berge, T.; Adams, C.; Quach, H.; Moen, S.M.; Berg-Hansen, P.; Celius, E.G.; Sangurdekar, D.P.; Bronson, P.G.; et al. Increased DNA methylation of SLFN12 in CD4+ and CD8+ T cells from multiple sclerosis patients. PLoS ONE 2018, 13, e0206511. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, J.; Chen, Y.; Ren, B.; Wan, S.; Chen, Y.; He, Y.; Wei, Q.; Gao, H.; Liu, L.; et al. Genome-wide DNA methylation pattern in whole blood of patients with Hashimoto thyroiditis. Front. Endocrinol. 2023, 14, 1259903. [Google Scholar] [CrossRef] [PubMed]
- Vomhof-DeKrey, E.E.; Umthun, J.; Basson, M.D. Loss of Schlafen3 influences the expression levels of Schlafen family members in ileum, thymus, and spleen tissue. PeerJ 2020, 8, e8461. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, N.; Huang, Y.; Wang, Y.; Sun, Y.; Wu, Q.; Li, D.; Gao, S.; Wang, H.-W.; Huang, N.; et al. Structure of PDE3A–SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells. Nat. Commun. 2021, 12, 6204. [Google Scholar] [CrossRef] [PubMed]
- Garvie, C.W.; Wu, X.; Papanastasiou, M.; Lee, S.; Fuller, J.; Schnitzler, G.R.; Horner, S.W.; Baker, A.; Zhang, T.; Mullahoo, J.P.; et al. Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase. Nat. Commun. 2021, 12, 4375. [Google Scholar] [CrossRef]
- de Waal, L.; Lewis, T.A.; Rees, M.G.; Tsherniak, A.; Wu, X.; Choi, P.S.; Gechijian, L.; Hartigan, C.; Faloon, P.W.; Hickey, M.J.; et al. Identification of cancer-cytotoxic modulators of PDE3A by predictive chemogenomics. Nat. Chem. Biol. 2015, 12, 102–108. [Google Scholar] [CrossRef]
- Yan, B.; Ding, Z.; Zhang, W.; Cai, G.; Han, H.; Ma, Y.; Cao, Y.; Wang, J.; Chen, S.; Ai, Y. Multiple PDE3A modulators act as molecular glues promoting PDE3A-SLFN12 interaction and induce SLFN12 dephosphorylation and cell death. Cell Chem. Biol. 2022, 29, 958–969.e5. [Google Scholar] [CrossRef]
- Greulich, H. A complex puzzle: Regulation of SLFN12 RNase activity by phosphorylation. Cell Chem. Biol. 2022, 29, 925–927. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, K.P.; McKean, M.; Goldoni, S.; Genvresse, I.; Garrido, M.F.; Li, R.; Wilkinson, G.; Kneip, C.; Yap, T.A. First-in-Human Dose-Escalation Study of the First-in-Class PDE3A–SLFN12 Complex Inducer BAY 2666605 in Patients with Advanced Solid Tumors Coexpressing SLFN12 and PDE3A. Clin. Cancer Res. 2024, 30, 5568–5576. [Google Scholar] [CrossRef]
- Vomhof-DeKrey, E.E.; Lee, J.; Lansing, J.; Brown, C.; Darland, D.; Basson, M.D. Schlafen 3 knockout mice display gender-specific differences in weight gain, food efficiency, and expression of markers of intestinal epithelial differentiation, metabolism, and immune cell function. PLoS ONE 2019, 14, e0219267. [Google Scholar] [CrossRef]
- Brown, S.R.; Vomhof-DeKrey, E.E.; Al-Marsoummi, S.; Beyer, T.; Lauckner, B.; Samson, M.; Sattar, S.; Brown, N.D.; Basson, M.D. SLFN12 Expression Significantly Effects the Response to Chemotherapy Drugs in Triple-Negative Breast Cancer. Cancers 2024, 16, 3848. [Google Scholar] [CrossRef]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic Modifications: Basic Mechanisms and Role in Cardiovascular Disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef]
- Zhou, K.; Li, Y.; Wang, W.; Chen, Y.; Qian, B.; Liang, Y.; Li, H.; Xu, R.; Zhuang, L. SLFN11: A pan-cancer biomarker for DNA-targeted drugs sensitivity and therapeutic strategy guidance. Front. Oncol. 2025, 15, 1582738. [Google Scholar] [CrossRef] [PubMed]
- Ramazi, S.; Zahiri, J. Post-translational modifications in proteins: Resources, tools and prediction methods. Database 2021, 2021, baab012. [Google Scholar] [CrossRef] [PubMed]
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone Acetyltransferases. Annu. Rev. Biochem. 2001, 70, 81–120. [Google Scholar] [CrossRef]
- Herceg, Z.; Murr, R. Mechanisms of histone modifications. In Handbook of Epigenetics; Academic Press: Cambridge, MA, USA, 2011; pp. 25–45. [Google Scholar] [CrossRef]
- Kaur, S.; Schwartz, A.L.; Jordan, D.G.; Soto-Pantoja, D.R.; Kuo, B.; Elkahloun, A.G.; Griner, L.M.; Thomas, C.J.; Ferrer, M.; Thomas, A.; et al. Identification of Schlafen-11 as a Target of CD47 Signaling That Regulates Sensitivity to Ionizing Radiation and Topoisomerase Inhibitors. Front. Oncol. 2019, 9, 994. [Google Scholar] [CrossRef]
- Moribe, F.; Nishikori, M.; Takashima, T.; Taniyama, D.; Onishi, N.; Arima, H.; Sasanuma, H.; Akagawa, R.; Elloumi, F.; Takeda, S.; et al. Epigenetic suppression of SLFN11 in germinal center B-cells during B-cell development. PLoS ONE 2021, 16, e0237554. [Google Scholar] [CrossRef]
- Miller, J.L.; Grant, P.A. The Role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell. Biochem. 2013, 61, 289–317. [Google Scholar] [CrossRef]
- Nogales, V.; Reinhold, W.C.; Varma, S.; Martinez-Cardus, A.; Moutinho, C.; Moran, S.; Heyn, H.; Sebio, A.; Barnadas, A.; Pommier, Y.; et al. Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget 2016, 7, 3084–3097. [Google Scholar] [CrossRef]
- Xu, Z.; Ye, B.; Li, M.; Fan, Z.; Sun, J.; Huang, Z.; Ma, X.; Shi, P. The expression of SLFN11 is related to the sensitivity of bladder cancer cells to DNA damage agents. Gene Rep. 2024, 34, 101881. [Google Scholar] [CrossRef]
- Noronha, N.Y.; Rodrigues, G.d.S.; Noma, I.H.Y.; Brandao, C.F.C.; Rodrigues, K.P.; Bruno, A.C.; Sae-Lee, C.; Watanabe, L.M.; Pinhel, M.A.d.S.; Schineider, I.M.; et al. 14-weeks combined exercise epigenetically modulated 118 genes of menopausal women with prediabetes. Front. Endocrinol. 2022, 13, 895489. [Google Scholar] [CrossRef] [PubMed]
- Perez, R.E.; Eckerdt, F.; Platanias, L.C. Schlafens: Emerging Therapeutic Targets. Cancers 2024, 16, 1805. [Google Scholar] [CrossRef]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics 2012, 7, 1098–1108. [Google Scholar] [CrossRef]
- Malone, D.; Lardelli, R.M.; Li, M.; David, M. Dephosphorylation activates the interferon-stimulated Schlafen family member 11 in the DNA damage response. J. Biol. Chem. 2019, 294, 14674–14685. [Google Scholar] [CrossRef]
- Hou, P.; Hao, W.; Qin, B.; Li, M.; Zhao, R.; Cui, S. Structural and biochemical characterization of Schlafen11 N-terminal domain. Nucleic Acids Res. 2023, 51, 7053–7070. [Google Scholar] [CrossRef]
- Metzner, F.J.; Wenzl, S.J.; Kugler, M.; Krebs, S.; Hopfner, K.-P.; Lammens, K. Mechanistic understanding of human SLFN11. Nat. Commun. 2022, 13, 5464. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, K.; Maekawa, M.; Iimori, Y.; Ogawa, A.; Urano, T.; Kono, N.; Takeda, H.; Higashiyama, S.; Arita, M.; Murai, J. The crucial role of single-stranded DNA binding in enhancing sensitivity to DNA-damaging agents for Schlafen 11 and Schlafen 13. iScience 2023, 26, 108529. [Google Scholar] [CrossRef]
- Jo, U.; Pommier, Y. Structural, molecular, and functional insights into Schlafen proteins. Exp. Mol. Med. 2022, 54, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Murai, Y.; Jo, U.; Murai, J.; Jenkins, L.M.; Huang, S.-Y.N.; Chakka, S.; Chen, L.; Cheng, K.; Fukuda, S.; Takebe, N.; et al. SLFN11 Inactivation Induces Proteotoxic Stress and Sensitizes Cancer Cells to Ubiquitin Activating Enzyme Inhibitor TAK-243. Cancer Res. 2021, 81, 3067–3078. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.T.; Dybas, J.M.; Kulej, K.; Reyes, E.D.; Price, A.M.; Akhtar, L.N.; Orr, A.; Garcia, B.A.; Boutell, C.; Weitzman, M.D. Comparative proteomics identifies Schlafen 5 (SLFN5) as a herpes simplex virus restriction factor that suppresses viral transcription. Nat. Microbiol. 2021, 6, 234–245. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajpathy, O.; Vomhof-DeKrey, E.E. Epigenetic and Post-Translational Regulation of Schlafen Family Expression and Their Differential Methods of Regulating Proteins. Int. J. Mol. Sci. 2025, 26, 11058. https://doi.org/10.3390/ijms262211058
Rajpathy O, Vomhof-DeKrey EE. Epigenetic and Post-Translational Regulation of Schlafen Family Expression and Their Differential Methods of Regulating Proteins. International Journal of Molecular Sciences. 2025; 26(22):11058. https://doi.org/10.3390/ijms262211058
Chicago/Turabian StyleRajpathy, Odele, and Emilie E. Vomhof-DeKrey. 2025. "Epigenetic and Post-Translational Regulation of Schlafen Family Expression and Their Differential Methods of Regulating Proteins" International Journal of Molecular Sciences 26, no. 22: 11058. https://doi.org/10.3390/ijms262211058
APA StyleRajpathy, O., & Vomhof-DeKrey, E. E. (2025). Epigenetic and Post-Translational Regulation of Schlafen Family Expression and Their Differential Methods of Regulating Proteins. International Journal of Molecular Sciences, 26(22), 11058. https://doi.org/10.3390/ijms262211058






