Circulating Tumor DNA as a Biomarker for Precision Medicine in Prostate Cancer: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Inclusion and Exclusion Criteria
2.3. Data Extraction
2.4. Risk-of-Bias Assessment
3. Results and Discussion
3.1. Eligible Studies
3.2. Risk-of-Bias
3.3. Study and Patient Characteristics
3.4. Methods for Specimen Detected
3.5. Frequent Genes and Their Somatic/Germline and Genomic Alteration Aspect Type
3.6. Association of ctDNA Detection with Outcome Survival
3.7. Discussion
3.8. Practical Recommendations
- Sample type and handling: Plasma should be used rather than serum to minimize dilution by leukocyte cfDNA and to improve detection sensitivity. Pre-analytical variables (time to centrifugation, storage temperature, and number of freeze–thaw cycles) must be standardized to avoid degradation.
- DNA extraction: Bead-based kits generally yield higher recovery rates than column-based methods, particularly when ctDNA concentration is low. We recommend reporting recovery efficiency in each study to facilitate comparability.
- Sequencing approaches: Next-generation sequencing (NGS) with unique molecular identifiers (UMIs) and error-suppression algorithms should be preferred for detecting low-frequency variants (<1% VAF). Targeted panels focusing on recurrent alterations (TP53, BRCA2, AR, and PTEN) are currently the most practical for clinical monitoring.
- Interpretation of results: Variants associated with clonal hematopoiesis (e.g., ATM, CHEK2, and DNMT3A) should be carefully interpreted in parallel with matched white blood cell sequencing to reduce false positives.
- Clinical integration: At present, ctDNA analysis is most useful to (i) identify resistance mechanisms to androgen receptor signaling inhibitors (ARSI), (ii) guide the addition of PARP inhibitors in patients with Homologous Recombination Repair (HRR) alterations, and (iii) monitor emerging mutations during treatment.
- Reporting standards: Studies should systematically report the variant allele fraction (VAF), copy number thresholds, and assay sensitivity. Uniform reporting will accelerate meta-analyses and guideline development.
3.9. Limitations
4. Conclusions and Clinical Implications
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Medori, M.C.; Micheletti, C.; Gadler, M.; Benedetti, S.; Guerri, G.; Cristofoli, F.; Generali, D.; Donofrio, C.A.; Cominetti, M.; Fioravanti, A.; et al. Omics sciences and precision medicine in prostate cancer. Clin. Ter. 2023, 174 (Suppl. S2), 95–103. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Krilaviciute, A.; Becker, N.; Lakes, J.; Radtke, J.P.; Kuczyk, M.; Peters, I.; Harke, N.N.; Debus, J.; Koerber, S.A.; Herkommer, K.; et al. Digital rectal examination is not a useful screening test for prostate cancer. Eur. Urol. Oncol. 2023, 6, 566–573. [Google Scholar] [CrossRef]
- Han, C.; Zhu, L.; Liu, X.; Ma, S.; Liu, Y.; Wang, X. Differential diagnosis of uncommon prostate diseases: Combining mpMRI and clinical information. Insights Imaging 2021, 12, 79. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zhao, T.; Dong, B.; Chen, W.; Yang, G.; Xie, J.; Guo, C.; Wang, R.; Wang, H.; Huang, L.; et al. Circulating tumor DNA and tissue complementarily detect genomic alterations in metastatic hormone-sensitive prostate cancer. iScience 2024, 27, 108931. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, F.; Zhang, J.; Wang, L. Clinical applications of liquid biopsy in prostate cancer: From screening to predictive biomarker. Cancers 2022, 14, 1728. [Google Scholar] [CrossRef] [PubMed]
- Foser, S.; Maiese, K.; Digumarthy, S.R.; Puig-Butille, J.A.; Rebhan, C. Looking to the future of early detection in cancer: Liquid biopsies, imaging, and artificial intelligence. Clin. Chem. 2024, 70, 27–32. [Google Scholar] [CrossRef]
- Gezer, U.; Bronkhorst, A.J.; Holdenrieder, S. The utility of repetitive cell-free DNA in cancer liquid biopsies. Diagnostics 2022, 12, 1363. [Google Scholar] [CrossRef]
- Bin Riaz, I.; Wang, L.; Kohli, M. Liquid biopsy approach in the management of prostate cancer. Transl. Res. 2018, 201, 60–70. [Google Scholar] [CrossRef]
- Fonseca, N.M.; Maurice-Dror, C.; Herberts, C.; Tu, W.; Fan, W.; Murtha, A.J.; Kollmannsberger, C.; Kwan, E.M.; Parekh, K.; Schönlau, E.; et al. Prediction of plasma ctDNA fraction and prognostic implications of liquid biopsy in advanced prostate cancer. Nat. Commun. 2024, 15, 1828. [Google Scholar] [CrossRef]
- Dong, B.; Fan, L.; Yang, B.; Chen, W.; Li, Y.; Wu, K.; Zhang, F.; Dong, H.; Cheng, H.; Pan, J.; et al. Use of circulating tumor DNA for the clinical management of metastatic castration-resistant prostate cancer: A multicenter, real-world study. J. Natl. Compr. Cancer Netw. 2021, 19, 905–914. [Google Scholar] [CrossRef]
- Beltran, H.; Romanel, A.; Conteduca, V.; Casiraghi, N.; Sigouros, M.; Franceschini, G.M.; Orlando, F.; Fedrizzi, T.; Ku, S.-Y.; Dann, E.; et al. Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J. Clin. Investig. 2020, 130, 1653–1668. [Google Scholar] [CrossRef]
- Romanel, A.; Tandefelt, D.G.; Conteduca, V.; Jayaram, A.; Casiraghi, N.; Wetterskog, D.; Salvi, S.; Amadori, D.; Zafeiriou, Z.; Rescigno, P.; et al. Plasma AR and abiraterone-resistant prostate cancer. Sci. Transl. Med. 2015, 7, 312re10. [Google Scholar] [CrossRef] [PubMed]
- Annala, M.; Vandekerkhove, G.; Khalaf, D.; Taavitsainen, S.; Beja, K.; Warner, E.W.; Sunderland, K.; Kollmannsberger, C.; Eigl, B.J.; Finch, D.; et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018, 8, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Kwan, E.M.; Wyatt, A.W.; Chi, K.N. Towards clinical implementation of circulating tumor DNA in metastatic prostate cancer: Opportunities for integration and pitfalls to interpretation. Front. Oncol. 2022, 12, 1054497. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lopez-Valcarcel, M.; Lopez-Campos, F.; Zafra, J.; Cienfuegos, I.; Ferri, M.; Barrado, M.; Hernando, S.; Counago, F. Liquid biopsy to personalize treatment for metastatic prostate cancer. Am. J. Transl. Res. 2024, 16, 1531–1549. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, A.; Annala, M.; Beja, K.; Parimi, S.; Vandekerkhove, G.; Warner, E.; Zulfiqar, M.; Finch, D.; Oja, C.; Vergidis, J.; et al. Genomic alterations in circulating tumor DNA (ctDNA) are associated with clinical outcomes in treatment-naive metastatic castration-resistant prostate cancer (mCRPC) patients commencing androgen receptor (AR)-targeted therapy. Ann. Oncol. 2016, 27 (Suppl. S6), vi17. [Google Scholar] [CrossRef]
- Shaya, J.; Nonato, T.; Cabal, A.; Randall, J.M.; Millard, F.; Stewart, T.; McKay, R.R. Analysis of the prognostic significance of circulating tumor DNA in metastatic castrate-resistant prostate cancer. Clin. Genitourin. Cancer 2021, 19, 564.e1–564.e10. [Google Scholar] [CrossRef]
- Reimers, M.A.; Yip, S.M.; Zhang, L.; Cieslik, M.; Dhawan, M.; Montgomery, B.; Wyatt, A.W.; Chi, K.N.; Small, E.J.; Chinnaiyan, A.M.; et al. Clinical outcomes in cyclin-dependent kinase 12 mutant advanced prostate cancer. Eur. Urol. 2020, 77, 333–341. [Google Scholar] [CrossRef]
- Hemenway, G.; Tierno, M.B.; Nejati, R.; Sosa, R.; Zibelman, M. Clinical utility of liquid biopsy to identify genomic heterogeneity and secondary cancer diagnoses: A case report. Case Rep. Oncol. 2022, 15, 78–85. [Google Scholar] [CrossRef]
- Torquato, S.; Pallavajjala, A.; Goldstein, A.; Toro, P.V.; Silberstein, J.L.; Lee, J.; Nakazawa, M.; Waters, I.; Chu, D.; Shinn, D.; et al. Genetic alterations detected in cell-free DNA are associated with enzalutamide and abiraterone resistance in castration-resistant prostate cancer. JCO Precis. Oncol. 2019, 3, PO.18.00227. [Google Scholar] [CrossRef]
- Barata, P.; Agarwal, N.; Nussenzveig, R.; Gerendash, B.; Jaeger, E.; Hatton, W.; Ledet, E.; Lewis, B.; Layton, J.; Babiker, H.; et al. Clinical activity of pembrolizumab in metastatic prostate cancer with microsatellite instability high (MSI-H) detected by circulating tumor DNA. J. Immunother. Cancer 2020, 8, e001065. [Google Scholar] [CrossRef]
- Dang, H.X.; Chauhan, P.S.; Ellis, H.; Feng, W.; Harris, P.K.; Smith, G.; Qiao, M.; Dienstbach, K.; Beck, R.; Atkocius, A.; et al. Cell-free DNA alterations in the AR enhancer and locus predict resistance to AR-directed therapy in patients with metastatic prostate cancer. JCO Precis. Oncol. 2020, 4, 680–713. [Google Scholar] [CrossRef] [PubMed]
- Ravindranathan, D.; Russler, G.A.; Yantorni, L.; Drusbosky, L.M.; Bilen, M.A. Detection of microsatellite instability via circulating tumor DNA and response to immunotherapy in metastatic castration-resistant prostate cancer: A case series. Case Rep. Oncol. 2021, 14, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, B.A.; Pamarthy, S.; Shah, A.N.; Sagar, V.; Unno, K.; Han, H.; Yang, X.J.; Costa, R.B.; Nagy, R.J.; Lanman, R.B.; et al. Anaplastic lymphoma kinase mutation (ALK F1174C) in small cell carcinoma of the prostate and molecular response to alectinib. Clin. Cancer Res. 2018, 24, 2732–2739. [Google Scholar] [CrossRef]
- Ledet, E.M.; Lilly, M.B.; Sonpavde, G.; Lin, E.; Nussenzveig, R.H.; Barata, P.C.; Yandell, M.; Nagy, R.J.; Kiedrowski, L.; Agarwal, N.; et al. Comprehensive analysis of AR alterations in circulating tumor DNA from patients with advanced prostate cancer. Oncologist 2020, 25, 327–333. [Google Scholar] [CrossRef]
- Dincman, T.A.; Karam, J.A.Q.; Giordano, A.; Li, H.; Drusbosky, L.M.; Gourdin, T.S.; Howe, P.H.; Lilly, M.B. Genomic amplifications identified by circulating tumor DNA analysis guide prognosis in metastatic castration-resistant prostate cancer. Front. Oncol. 2024, 13, 1202277. [Google Scholar] [CrossRef] [PubMed]
- Moses, M.; Koksal, U.; Ledet, E.; Manogue, C.; Cotogno, P.; Lewis, B.; Layton, J.; Sartor, A.O.; Barata, P. Evaluation of the genomic alterations in the androgen receptor gene during treatment with high-dose testosterone for metastatic castrate-resistant prostate cancer. Oncotarget 2020, 11, 15–21. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wyatt, A.W.; Annala, M.; Aggarwal, R.; Beja, K.; Feng, F.; Youngren, J.; Foye, A.; Lloyd, P.; Nykter, M.; Beer, T.M.; et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J. Natl. Cancer Inst. 2017, 109, djx118. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Jia, G.; Chao, F.; Xie, F.; Zhang, Y.; Hou, C.; Huang, Y.; Tang, H.; Yu, J.; Zhang, J.; et al. Urine- and blood-based molecular profiling of human prostate cancer. Front. Oncol. 2022, 12, 759791. [Google Scholar] [CrossRef]
- Yu, J.; Cho, E.; Choi, J.; Lim, J.E.; Lee, J.; Kang, M.; Sung, H.H.; Jeong, B.C.; Seo, S.I.; Jeon, S.S.; et al. Genomic mutation profiling using liquid biopsy in Korean patients with prostate cancer: Circulating tumor DNA mutation predicts the development of castration resistance. Investig. Clin. Urol. 2021, 62, 224–232. [Google Scholar] [CrossRef]
- Fan, L.; Fei, X.; Zhu, Y.; Pan, J.; Sha, J.; Chi, C.; Gong, Y.; Du, X.; Zhou, L.; Dong, B.; et al. Comparative analysis of genomic alterations across castration sensitive and castration resistant prostate cancer via circulating tumor DNA sequencing. J. Urol. 2021, 205, 461–469. [Google Scholar] [CrossRef]
- Bang, S.; Won, D.; Shin, S.; Cho, K.S.; Park, J.W.; Lee, J.; Choi, Y.D.; Kang, S.; Lee, S.-T.; Choi, J.R.; et al. Circulating tumor DNA analysis on metastatic prostate cancer with disease progression. Cancers 2023, 15, 3998. [Google Scholar] [CrossRef]
- Du, X.; Fei, X.; Wang, J.; Dong, Y.; Fan, L.; Yang, B.; Chen, W.; Gong, Y.; Xia, B.; Zhu, H.; et al. Early serial circulating tumor DNA sequencing predicts the efficacy of chemohormonal therapy in patients with metastatic hormone-sensitive prostate cancer. Transl. Oncol. 2023, 34, 101701. [Google Scholar] [CrossRef]
- Fei, X.; Du, X.; Gong, Y.; Liu, J.; Fan, L.; Wang, J.; Wang, Y.; Zhu, Y.; Pan, J.; Dong, B.; et al. Early plasma circulating tumor DNA as a potential biomarker of disease recurrence in non-metastatic prostate cancer. Cancer Res. Treat. 2023, 55, 969–977. [Google Scholar] [CrossRef]
- Pan, J.; Zhao, J.; Ni, X.; Gan, H.; Wei, Y.; Wu, J.; Zhang, T.; Wang, Q.; Freedland, S.J.; Wang, B.; et al. The prevalence and prognosis of next-generation therapeutic targets in metastatic castration-resistant prostate cancer. Mol. Oncol. 2022, 16, 4011–4022. [Google Scholar] [CrossRef]
- Yuan, F.; Liu, N.; Yang, M.-Z.; Zhang, X.-T.; Luo, H.; Zhou, H. Circulating tumor DNA genomic profiling reveals the complicated olaparib-resistance mechanism in prostate cancer salvage therapy: A case report. World J. Clin. Cases 2022, 10, 3461–3472. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; He, L.; Huang, Q.; Zheng, S.; Zhang, Z.; Li, H.; Liu, S. Response to olaparib in metastatic castration-resistant prostate cancer with germline BRCA2 mutation: A case report. BMC Med. Genet. 2018, 19, 185. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Xu, Q.; Guo, H.; Yang, G.; Zhang, J.; Wang, H.; Xu, T.; Guo, C.; Yuan, J.; He, Y.; et al. Concordance and clinical significance of genomic alterations in progressive tumor tissue and matched circulating tumor DNA in aggressive-variant prostate cancer. Cancer Res. Commun. 2023, 3, 2221–2232. [Google Scholar] [CrossRef] [PubMed]
- De Laere, B.; Oeyen, S.; Mayrhofer, M.; Whitington, T.; van Dam, P.-J.; Van Oyen, P.; Ghysel, C.; Ampe, J.; Ost, P.; Demey, W.; et al. TP53 outperforms other androgen receptor biomarkers to predict abiraterone or enzalutamide outcome in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2019, 25, 1766–1773. [Google Scholar] [CrossRef]
- Conteduca, V.; Wetterskog, D.; Sharabiani, M.T.A.; Grande, E.; Fernandez-Pérez, M.P.; Jayaram, A.; Salvi, S.; Castellano, D.; Romanel, A.; Lolli, C.; et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: A multi-institution correlative biomarker study. Ann. Oncol. 2017, 28, 1508–1516. [Google Scholar] [CrossRef]
- Loehr, A.; Hussain, A.; Patnaik, A.; Bryce, A.H.; Castellano, D.; Font, A.; Shapiro, J.; Zhang, J.; Sautois, B.; Vogelzang, N.J.; et al. Emergence of BRCA reversion mutations in patients with metastatic castration-resistant prostate cancer after treatment with rucaparib. Eur. Urol. 2023, 83, 200–209. [Google Scholar] [CrossRef]
- Chi, K.N.; Barnicle, A.; Sibilla, C.; Lai, Z.; Corcoran, C.; Barrett, J.C.; Adelman, C.A.; Qiu, P.; Easter, A.; Dearden, S.; et al. Detection of BRCA1, BRCA2, and ATM alterations in matched tumor tissue and circulating tumor DNA in patients with prostate cancer screened in PROfound. Clin. Cancer Res. 2023, 29, 81–91. [Google Scholar] [CrossRef]
- Kohli, M.; Tan, W.; Zheng, T.; Wang, A.; Montesinos, C.; Wong, C.; Du, P.; Jia, S.; Yadav, S.; Horvath, L.G.; et al. Clinical and genomic insights into circulating tumor DNA-based alterations across the spectrum of metastatic hormone-sensitive and castrate-resistant prostate cancer. EBioMedicine 2020, 54, 102728. [Google Scholar] [CrossRef]
- Jayaram, A.; Wingate, A.; Wetterskog, D.; Wheeler, G.; Sternberg, C.; Jones, R.; Berruti, A.; Lefresne, F.; Lahaye, M.; Thomas, S.; et al. Plasma tumor gene conversions after one cycle abiraterone acetate for metastatic castration-resistant prostate cancer: A biomarker analysis of a multicenter international trial. Ann. Oncol. 2021, 32, 726–735. [Google Scholar] [CrossRef]
- Oya, M.; Armstrong, A.J.; Thiery-Vuillemin, A.; Shore, N.; Procopio, G.; Arslan, C.; Mehra, N.; Parnis, F.; Brown, E.; Schlurmann, F.C.; et al. Biomarker analysis and updated results from the phase III PROpel trial of abiraterone and olaparib vs abiraterone and placebo as first-line therapy for patients with metastatic castration-resistant prostate cancer. Ann. Oncol. 2022, 33 (Suppl. S9), S1495. [Google Scholar] [CrossRef]
- Clarke, N.W.; Armstrong, A.J.; Thiery-Vuillemin, A.; Oya, M.; Shore, N.D.; Procopio, G.; Guedes, J.D.C.; Arslan, C.; Mehra, N.; Parnis, F.; et al. Final overall survival in PROpel: Abiraterone and olaparib versus abiraterone and placebo as first-line therapy for metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2023, 41 (Suppl. S6), LBA16. [Google Scholar] [CrossRef]
- Agarwal, N.; Lucas, J.; Aguilar-Bonavides, C.; Thomas, S.; Gormley, M.; Chowdhury, S.; Merseburger, A.S.; Bjartell, A.; Uemura, H.; Özgüroğlu, M.; et al. Genomic aberrations associated with overall survival in metastatic castration-sensitive prostate cancer treated with apalutamide or placebo plus androgen deprivation therapy in TITAN. J. Clin. Oncol. 2022, 40 (Suppl. S16), 5066. [Google Scholar] [CrossRef]
- Belic, J.; Graf, R.; Bauernhofer, T.; Cherkas, Y.; Ulz, P.; Waldispuehl-Geigl, J.; Perakis, S.; Gormley, M.; Patel, J.; Li, W.; et al. Genomic alterations in plasma DNA from patients with metastasized prostate cancer receiving abiraterone or enzalutamide. Int. J. Cancer 2018, 143, 1236–1248. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sautois, B.; Loehr, A.; Watkins, S.P.; Schroeder, H.; Abida, W. A case study of clinical response to rucaparib in a patient with metastatic castration-resistant prostate cancer and a RAD51B alteration. Curr. Oncol. 2022, 29, 4178–4184. [Google Scholar] [CrossRef]
- Dong, X.; Zheng, T.; Zhang, M.; Dai, C.; Wang, L.; Wang, L.; Zhang, R.; Long, Y.; Wen, D.; Xie, F.; et al. Circulating cell-free DNA-based detection of tumor suppressor gene copy number loss and its clinical implication in metastatic prostate cancer. Front. Oncol. 2021, 11, 720727. [Google Scholar] [CrossRef]
- Carr, T.H.; Adelman, C.; Barnicle, A.; Kozarewa, I.; Luke, S.; Lai, Z.; Hollis, S.; Dougherty, B.; Harrington, E.A.; Kang, J.; et al. Homologous recombination repair gene mutation characterization by liquid biopsy: A phase II trial of olaparib and abiraterone in metastatic castrate-resistant prostate cancer. Cancers 2021, 13, 5830. [Google Scholar] [CrossRef] [PubMed]
- Necchi, A.; Cucchiara, V.; Grivas, P.; Bratslavsky, G.; Jacob, J.; Spiess, P.E.; Sokol, E.S.; Killian, J.K.; Lin, D.; Ramkissoon, S.; et al. Contrasting genomic profiles from metastatic sites, primary tumors, and liquid biopsies of advanced prostate cancer. Cancer 2021, 127, 4557–4564. [Google Scholar] [CrossRef] [PubMed]
- Goodall, J.; Assaf, Z.J.; Shi, Z.; Seed, G.; Zhang, L.; Lauffer, B.; Yuan, W.; Wongchenko, M.; Oliveira, F.; Carreira, S.; et al. Circulating tumor DNA (ctDNA) dynamics associate with treatment response and radiological progression-free survival (rPFS): Analyses from a randomized phase II trial in metastatic castration-resistant prostate cancer (mCRPC). J. Clin. Oncol. 2020, 38 (Suppl. S15), 5508. [Google Scholar] [CrossRef]
- Knutson, T.P.; Luo, B.; Kobilka, A.; Lyman, J.; Guo, S.; Munro, S.A.; Li, Y.; Heer, R.; Gaughan, L.; Morris, M.J.; et al. AR alterations inform circulating tumor DNA detection in metastatic castration-resistant prostate cancer patients. Nat. Commun. 2024, 15, 10648. [Google Scholar] [CrossRef]
- Saad, F.; Armstrong, A.J.; Thiery-Vuillemin, A.; Oya, M.; Shore, N.; Procopio, G.; Guedes, J.D.; Arslan, C.; Mehra, N.; Parnis, F.; et al. MP11-16 Prostate-specific antigen analyses in PROpel: Abiraterone and olaparib versus abiraterone and placebo as first-line therapy for metastatic castration-resistant prostate cancer (abstract MP11-16). J. Urol. 2023, 209 (Suppl. S4), e131. [Google Scholar] [CrossRef]
- Kristiansen, A.; Sautois, B.; Crippa, A.; Mortezavi, A.; Discacciati, A.; Karlsson, C.; Ullén, A.; Jänes, E.; Enblad, G.; Oldenburg, J.; et al. Efficacy of carboplatin in patients with metastatic castration-resistant prostate cancer: Results from the biomarker-driven, randomised, outcome-adaptive ProBio trial (abstract 1619P). Ann. Oncol. 2024, 35 (Suppl. S2), S978. [Google Scholar] [CrossRef]
- De Bono, J.S.; Morris, M.J.; Sartor, O.; Wei, X.X.; Fizazi, K.; Herrmann, K.; Piulats, J.M.; Mahammedi, H.; Logothetis, C.; George, D.J.; et al. Baseline ctDNA analyses and associations with outcomes in taxane-naive patients with mCRPC treated with 177Lu-PSMA-617 versus change of ARPI in PSMAfore (abstract 5008). J. Clin. Oncol. 2024, 42 (Suppl. S16), 5008. [Google Scholar] [CrossRef]
- Lin, H.-M.; Mak, B.; Yeung, N.; Huynh, K.; Meikle, T.G.; Mellett, N.A.; Kwan, E.M.; Fettke, H.; Tran, B.; Davis, I.D.; et al. Overcoming enzalutamide resistance in metastatic prostate cancer by targeting sphingosine kinase. EBioMedicine 2021, 72, 103625. [Google Scholar] [CrossRef]
- Fettke, H.; Kwan, E.M.; Bukczynska, P.; Steen, J.A.; Docanto, M.; Ng, N.; Parente, P.; Mant, A.; Foroughi, S.; Pezaro, C.; et al. Independent prognostic impact of plasma NCOA2 alterations in metastatic castration-resistant prostate cancer. Prostate 2021, 81, 992–1001. [Google Scholar] [CrossRef]
- Annala, M.; Taavitsainen, S.; Khalaf, D.J.; Vandekerkhove, G.; Beja, K.; Sipola, J.; Warner, E.W.; Herberts, C.; Wong, A.; Fu, S.; et al. Evolution of Castration-Resistant Prostate Cancer in ctDNA during Sequential Androgen Receptor Pathway Inhibition. Clin. Cancer Res. 2021, 27, 4610–4623. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, N.; Sirico, M.; Generali, D.; Zanconati, F.; Scaggiante, B. Circulating cell-free nucleic acids as prognostic and therapy predictive tools for metastatic castrate-resistant prostate cancer. World J. Clin. Oncol. 2020, 11, 450–463. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, J.-S.; Kim, M.; Seong, M.-W.; Kim, H.-S.; Lee, Y.K.; Kang, H.J. Plasma vs. serum in circulating tumor DNA measurement: Characterization by DNA fragment sizing and digital droplet polymerase chain reaction. Clin. Chem. Lab. Med. 2020, 58, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.A.; Yeung, S.-W.; Lui, W.-B.; Rainer, T.H.; Lo, Y.D. Effects of preanalytical factors on the molecular size of cell-free DNA in blood. Clin. Chem. 2005, 51, 781–784. [Google Scholar] [CrossRef] [PubMed]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): A statement paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic–implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Pittella-Silva, F.; Chin, Y.M.; Chan, H.T.; Nagayama, S.; Miyauchi, E.; Low, S.-K.; Nakamura, Y. Plasma or serum: Which is preferable for mutation detection in liquid biopsy? Clin. Chem. 2020, 66, 946–957. [Google Scholar] [CrossRef]
- Park, J.J.; Chu, A.; Li, J.; Ali, A.; McKay, R.R.; Hwang, C.; Labriola, M.K.; Jang, A.; Kilari, D.; Mo, G.; et al. Repeat next-generation sequencing testing on progression in men with metastatic prostate cancer can identify new actionable alterations. JCO Precis. Oncol. 2024, 8, e2300567. [Google Scholar] [CrossRef]
- Tukachinsky, H.; Madison, R.W.; Chung, J.H.; Gjoerup, O.V.; Severson, E.A.; Dennis, L.; Fendler, B.J.; Morley, S.; Zhong, L.; Graf, R.P.; et al. Genomic analysis of circulating tumor DNA in 3334 patients with advanced prostate cancer identifies targetable BRCA alterations and AR resistance mechanisms. Clin. Cancer Res. 2021, 27, 3094–3105. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.K.; Mohamad, N.-V.; Giaze, T.R.; Chin, K.-Y.; Mohamed, N.; Ima-Nirwana, S. Prostate cancer and bone metastases: The underlying mechanisms. Int. J. Mol. Sci. 2019, 20, 2587. [Google Scholar] [CrossRef]
- Gandaglia, G.; Abdollah, F.; Schiffmann, J.; Trudeau, V.; Shariat, S.F.; Kim, S.P.; Perrotte, P.; Montorsi, F.; Briganti, A.; Trinh, Q.; et al. Distribution of metastatic sites in patients with prostate cancer: A population-based analysis. Prostate 2014, 74, 210–216. [Google Scholar] [CrossRef]
- Schweizer, M.T.; Gulati, R.; Bs, M.B.; Konnick, E.Q.; Cheng, H.H.; Klemfuss, N.; De Sarkar, N.; Yu, E.Y.; Montgomery, R.B.; Nelson, P.S.; et al. Clinical determinants for successful circulating tumor DNA analysis in prostate cancer. Prostate 2019, 79, 701–708. [Google Scholar] [CrossRef]
- Herberts, C.; Wyatt, A.W. Technical and biological constraints on ctDNA-based genotyping. Trends Cancer 2021, 7, 995–1009. [Google Scholar] [CrossRef]
- Batool, S.M.; Hsia, T.; Beecroft, A.; Lewis, B.; Ekanayake, E.; Rosenfeld, Y.; Escobedo, A.K.; Gamblin, A.S.; Rawal, S.; Cote, R.J.; et al. Extrinsic and intrinsic preanalytical variables affecting liquid biopsy in cancer. Cell Rep. Med. 2023, 4, 101196. [Google Scholar] [CrossRef]
- Armstrong, A.J.; Taylor, A.; Haffner, M.C.; Abida, W.; Bryce, A.H.; Karsh, L.I.; Tagawa, S.T.; Twardowski, P.; Serritella, A.V.; Lang, J.M. Germline and somatic testing for homologous repair deficiency in patients with prostate cancer (part 1 of 2). Prostate Cancer Prostatic Dis. 2024, 28, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Huffman, B.; Parikh, A. The Emerging Role of Circulating Tumor DNA: Will Tissue Become Obsolete? 1 December 2021. Available online: https://dailynews.ascopubs.org/do/emerging-role-circulating-tumor-dna-tissue-become-obsolete (accessed on 1 May 2025).
- Alix-Panabières, C.; Pantel, K. Liquid biopsy: From discovery to clinical application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef]
- Bayle, A.; Peyraud, F.; Belcaid, L.; Brunet, M.; Aldea, M.; Clodion, R.; Dubos, P.; Vasseur, D.; Nicotra, C.; Geraud, A.; et al. Liquid versus tissue biopsy for detecting actionable alterations according to the ESMO Scale for Clinical Actionability of molecular Targets in patients with advanced cancer: A study from the French National Center for Precision Medicine (PRISM). Ann. Oncol. 2022, 33, 1328–1331. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, B.; Wu, A.; Wetterskog, D.; Attard, G. Blood-based liquid biopsies for prostate cancer: Clinical opportunities and challenges. Br. J. Cancer 2022, 127, 1394–1402. [Google Scholar] [CrossRef]
- Taavitsainen, S.; Annala, M.; Ledet, E.; Beja, K.; Miller, P.J.; Moses, M.; Nykter, M.; Chi, K.N.; Sartor, O.; Wyatt, A.W. Evaluation of commercial circulating tumor DNA test in metastatic prostate cancer. JCO Precis. Oncol. 2019, 3, PO. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.; de la Fuente, R.P.; Lichtman, S.; Gomez, J.; Doroshow, D.; Hirsch, F.; Veluswamy, R.; Marron, T.; Rohs, N.; Smith, C.; et al. Prognostic value of variant allele frequency in circulating tumor DNA for survival outcomes in metastatic non-small cell lung cancer. J. Thorac. Oncol. 2024, 19 (Suppl. S2), S502–S503. [Google Scholar] [CrossRef]
- Kato, T.; Shiota, M.; Nishimoto, K.; Matsubara, N.; Osawa, T.; Abe, T.; Yasumizu, Y.; Tanaka, N.; Yamamoto, Y.; Ishizuya, Y.; et al. Prognostic significance of circulating tumor DNA alterations in advanced renal cell carcinoma from SCRUM-Japan MONSTAR-SCREEN: A nationwide genomic profiling project. Br. J. Cancer 2025, 133, 111–120. [Google Scholar] [CrossRef]
- Podany, E.L.; Foffano, L.; Gerratana, L.; Medford, A.J.; Clifton, K.; Tapiavala, S.; Velimirovic, M.; Lipsyc-Sharf, M.; Reduzzi, C.; Bubie, A.; et al. Racial differences in ctDNA profiles, targeted therapy use, and outcomes in metastatic breast cancer. JAMA Netw. Open 2025, 8, e2461899. [Google Scholar] [CrossRef]
- Lee, S.-B.; Kim, H.-G.; Hwang, S.-H.; Kim, K.-J.; Lee, J.H.; Seo, J.; Kang, M.; Jung, E.H.; Suh, K.J.; Kim, S.H.; et al. Longitudinal comparative analysis of circulating tumor DNA and matched tumor tissue DNA in patients with metastatic colorectal cancer receiving palliative first-line systemic anti-cancer therapy. Cancer Res. Treat. 2024, 56, 1171–1182. [Google Scholar] [CrossRef]
- Mandel, P.; Hoeh, B.; Humke, C.; Doering, C.; Wenzel, M.; Garcia, C.C.; Fuhr, N.; Koll, F.; Fassl, A.; Tilki, D.; et al. Feasibility of next-generation sequencing of liquid biopsy (circulating tumor DNA) samples and tumor tissue from patients with metastatic prostate cancer in a real-world clinical setting in Germany. Eur. Urol. Focus 2024, 10, 339–345. [Google Scholar] [CrossRef]
- Shiota, M.; Matsubara, N.; Kato, T.; Eto, M.; Osawa, T.; Abe, T.; Shinohara, N.; Nishimoto, K.; Yasumizu, Y.; Tanaka, N.; et al. Genomic profiling and clinical utility of circulating tumor DNA in metastatic prostate cancer: SCRUM-Japan MONSTAR SCREEN project. BJC Rep. 2024, 2, 28. [Google Scholar] [CrossRef]
- Zhang, J.; Dong, A.; Li, S.; Ren, X.; Zhang, X. Consistency of genotyping data from simultaneously collected plasma circulating tumor DNA and tumor-DNA in lung cancer patients. J. Thorac. Dis. 2020, 12, 7290–7297. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, N.; de Bono, J.; Olmos, D.; Procopio, G.; Kawakami, S.; Ürün, Y.; van Alphen, R.; Flechon, A.; Carducci, M.A.; Choi, Y.D.; et al. Olaparib Efficacy in Patients with Metastatic Castration-resistant Prostate Cancer and BRCA1, BRCA2, or ATM Alterations Identified by Testing Circulating Tumor DNA. Clin. Cancer Res. 2023, 29, 92–99. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sumiyoshi, T.; Chi, K.N.; Wyatt, A.W. Clinical implications of genomic alterations in metastatic prostate cancer. Prostate Cancer Prostatic Dis. 2021, 24, 310–322. [Google Scholar] [CrossRef]
- Lozano, R.; Castro, E.; Aragón, I.M.; Cendón, Y.; Cattrini, C.; López-Casas, P.P.; Olmos, D. Genetic aberrations in DNA repair pathways: A cornerstone of precision oncology in prostate cancer. Br. J. Cancer 2021, 124, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Seed, G.; Bertan, C.; Rescigno, P.; Dolling, D.; Figueiredo, I.; Miranda, S.; Nava Rodrigues, D.; Gurel, B.; Clarke, M.; et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. J. Clin. Investig. 2020, 130, 1743–1751. [Google Scholar] [CrossRef]
- Kloten, V.; Rüchel, N.; Brüchle, N.O.; Gasthaus, J.; Freudenmacher, N.; Steib, F.; Mijnes, J.; Eschenbruch, J.; Binnebösel, M.; Knüchel, R.; et al. Liquid biopsy in colon cancer: Comparison of different circulating DNA extraction systems following absolute quantification of KRAS mutations using Intplex allele-specific PCR. Oncotarget 2017, 8, 86253–86263. [Google Scholar] [CrossRef]
- Pérez-Barrios, C.; Nieto-Alcolado, I.; Torrente, M.; Jiménez-Sánchez, C.; Calvo, V.; Gutierrez-Sanz, L.; Palka, M.; Donoso-Navarro, E.; Provencio, M.; Romero, A. Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: Impact on biomarker testing. Transl. Lung Cancer Res. 2016, 5, 665–672. [Google Scholar] [CrossRef]
- Sorber, L.; Zwaenepoel, K.; Deschoolmeester, V.; Roeyen, G.; Lardon, F.; Rolfo, C.; Pauwels, P. A comparison of cell-free DNA isolation kits: Isolation and quantification of cell-free DNA in plasma. J. Mol. Diagn. 2017, 19, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, R.J.; Lee, J.H.; Kefford, R.F.; Rizos, H. Evaluation of commercial kits for purification of circulating free DNA. Cancer Genet. 2018, 228–229, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. Comparison of methods for the isolation of cell-free DNA from cell culture supernatant. Tumour Biol. 2020, 42, 1010428320916314. [Google Scholar] [CrossRef]
- Lee, J.-S.; Cho, E.H.; Kim, B.; Hong, J.; Kim, Y.-G.; Kim, Y.; Jang, J.-H.; Lee, S.-T.; Kong, S.-Y.; Lee, W.; et al. Clinical practice guideline for blood-based circulating tumor DNA assays. Ann. Lab. Med. 2024, 44, 195–209. [Google Scholar] [CrossRef]
- Dickinson, K.; Sharma, A.; Agnihotram, R.-K.V.; Altuntur, S.; Park, M.; Meterissian, S.; Burnier, J.V. Circulating tumor DNA and survival in metastatic breast cancer: A systematic review and meta-analysis. JAMA Netw. Open 2024, 7, e2431722. [Google Scholar] [CrossRef]
- Williams, P.M.; Forbes, T.; Lund, S.P.; Cole, K.D.; He, H.-J.; Karlovich, C.; Paweletz, C.P.; Stetson, D.; Yee, L.M.; Connors, D.E.; et al. Validation of ctDNA quality control materials through a precompetitive collaboration of the Foundation for the National Institutes of Health. JCO Precis. Oncol. 2021, 5, PO.20.00528. [Google Scholar] [CrossRef]
- Panet, F.; Papakonstantinou, A.; Borrell, M.; Vivancos, J.; Vivancos, A.; Oliveira, M. Use of ctDNA in early breast cancer: Analytical validity and clinical potential. npj Breast Cancer 2024, 10, 50. [Google Scholar] [CrossRef]
- Deveson, I.W.; Gong, B.; Lai, K.; LoCoco, J.S.; Richmond, T.A.; Schageman, J.; Zhang, Z.; Novoradovskaya, N.; Willey, J.C.; Jones, W.; et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nat. Biotechnol. 2021, 39, 1115–1128. [Google Scholar] [CrossRef]
- Mizuno, K.; Sumiyoshi, T.; Okegawa, T.; Terada, N.; Ishitoya, S.; Miyazaki, Y.; Kojima, T.; Katayama, H.; Fujimoto, N.; Hatakeyama, S.; et al. Clinical impact of detecting low-frequency variants in cell-free DNA on treatment of castration-resistant prostate cancer. Clin. Cancer Res. 2021, 27, 6164–6173. [Google Scholar] [CrossRef]
- Hamid, A.A.; Gray, K.P.; Shaw, G.; MacConaill, L.E.; Evan, C.; Bernard, B.; Loda, M.; Corcoran, N.M.; Van Allen, E.M.; Choudhury, A.D.; et al. Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. Eur. Urol. 2019, 76, 89–97. [Google Scholar] [CrossRef]
- Chen, J.; Shi, M.; Choi, S.Y.C.; Wang, Y.; Lin, D.; Zeng, H.; Wang, Y. Genomic alterations in neuroendocrine prostate cancer: A systematic review and meta-analysis. BJUI Compass 2023, 4, 256–265. [Google Scholar] [CrossRef]
- Aparicio, A.M.; Shen, L.; Tapia, E.L.N.; Lu, J.-F.; Chen, H.-C.; Zhang, J.; Wu, G.; Wang, X.; Troncoso, P.; Corn, P.; et al. Combined tumor suppressor defects characterize clinically defined aggressive variant prostate cancers. Clin. Cancer Res. 2016, 22, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Korn, J.M.; Gao, X.; Rakiec, D.P.; Ruddy, D.A.; Doshi, S.; Yuan, J.; Kovats, S.G.; Kim, S.; Cooke, V.G.; et al. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov. 2013, 3, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Abida, W.; Armenia, J.; Gopalan, A.; Brennan, R.; Walsh, M.; Barron, D.; Danila, D.; Rathkopf, D.; Morris, M.; Slovin, S.; et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis. Oncol. 2017, 2017, PO.17.00029. [Google Scholar] [CrossRef] [PubMed]
- Armenia, J.; Wankowicz, S.A.M.; Liu, D.; Gao, J.; Kundra, R.; Reznik, E.; Chatila, W.K.; Chakravarty, D.; Han, G.C.; Coleman, I.; et al. The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 2018, 50, 645–651. [Google Scholar] [CrossRef]

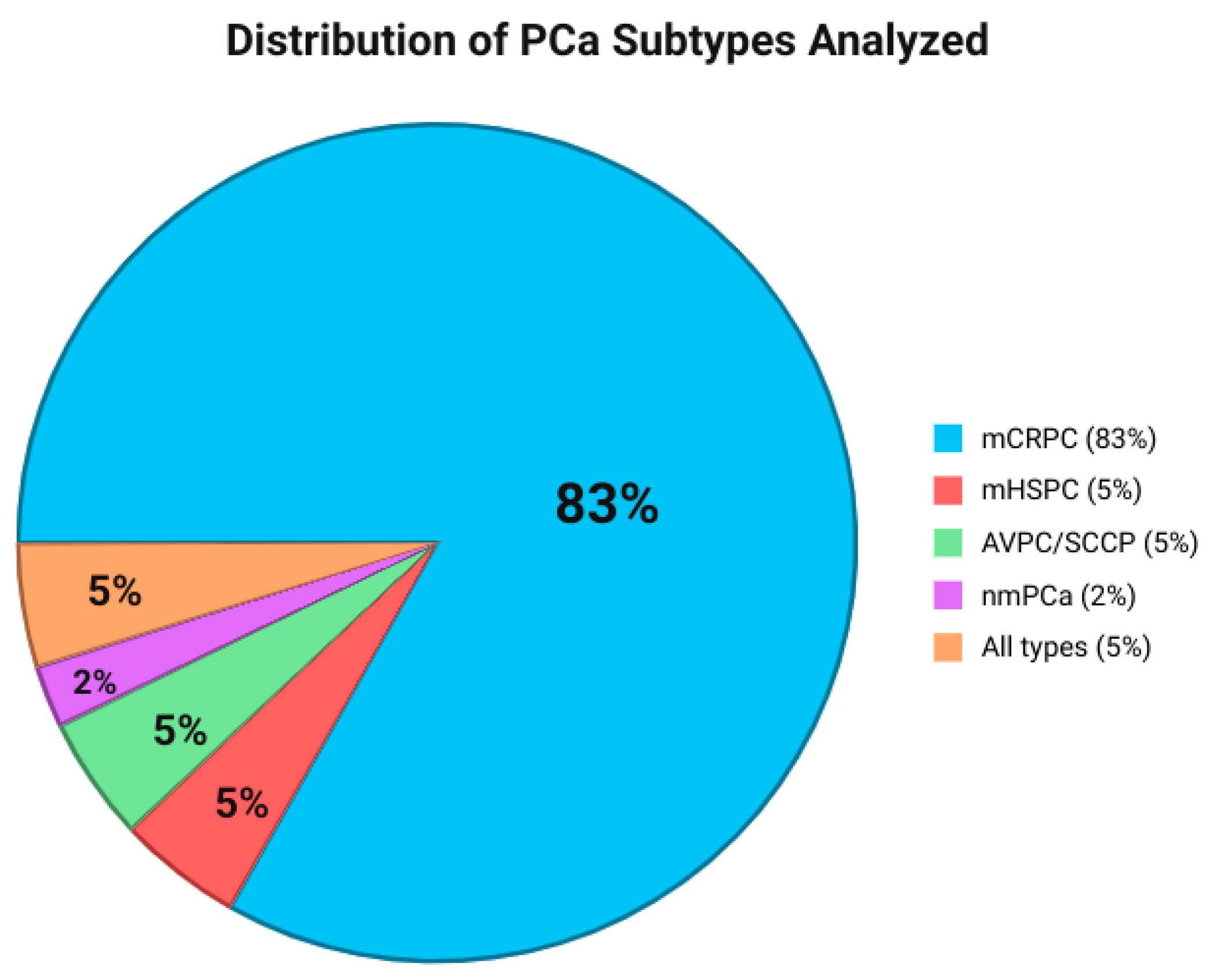
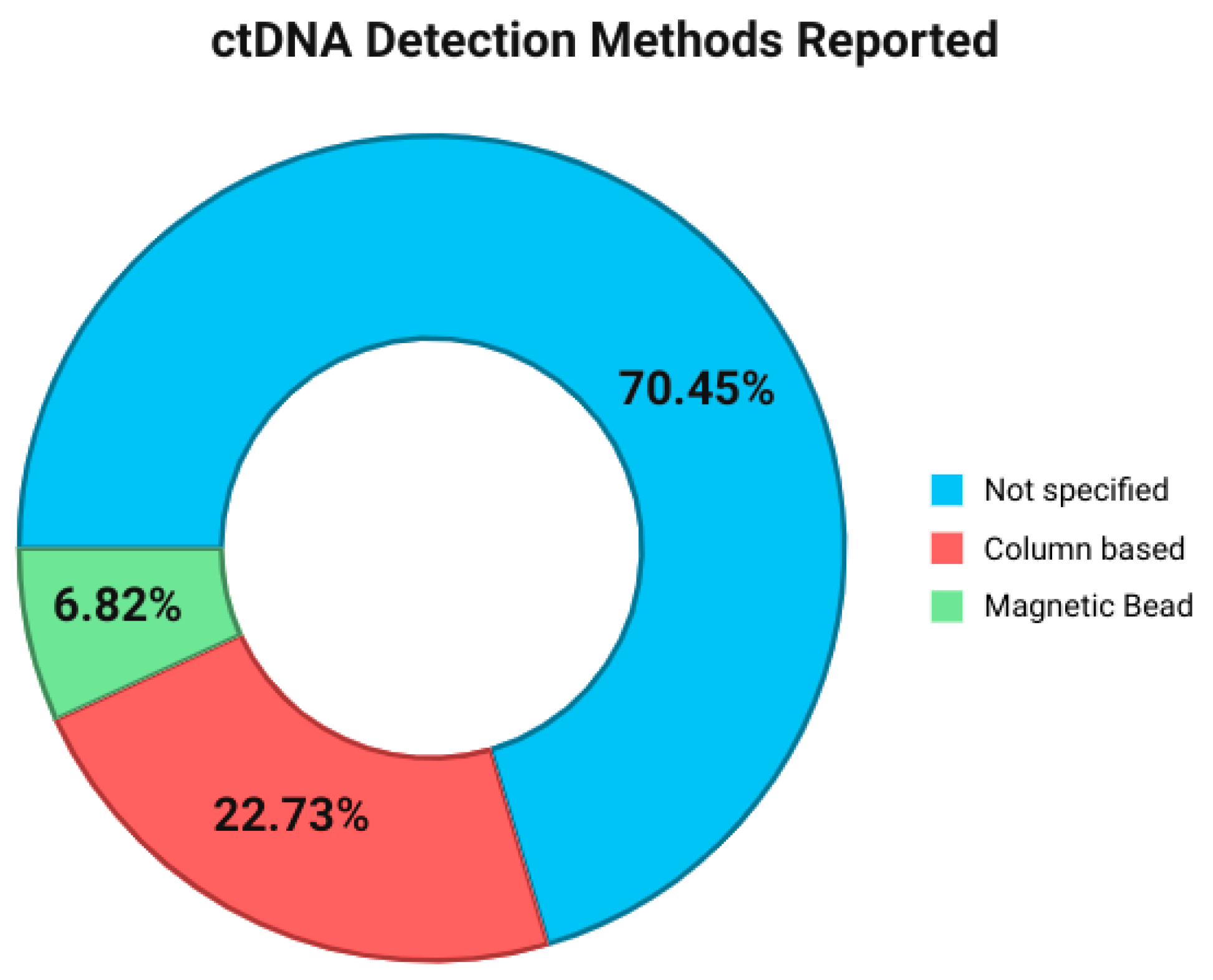
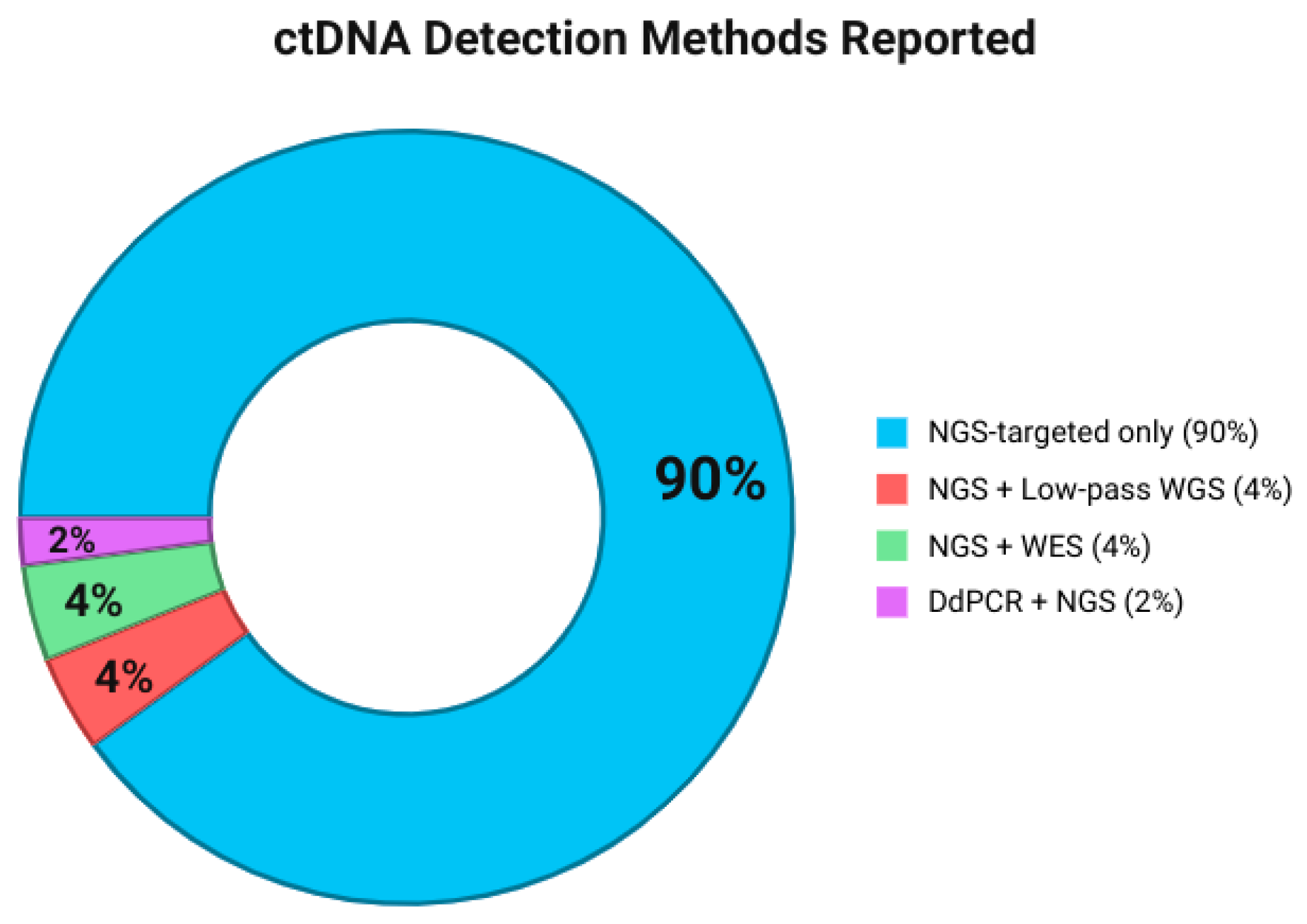
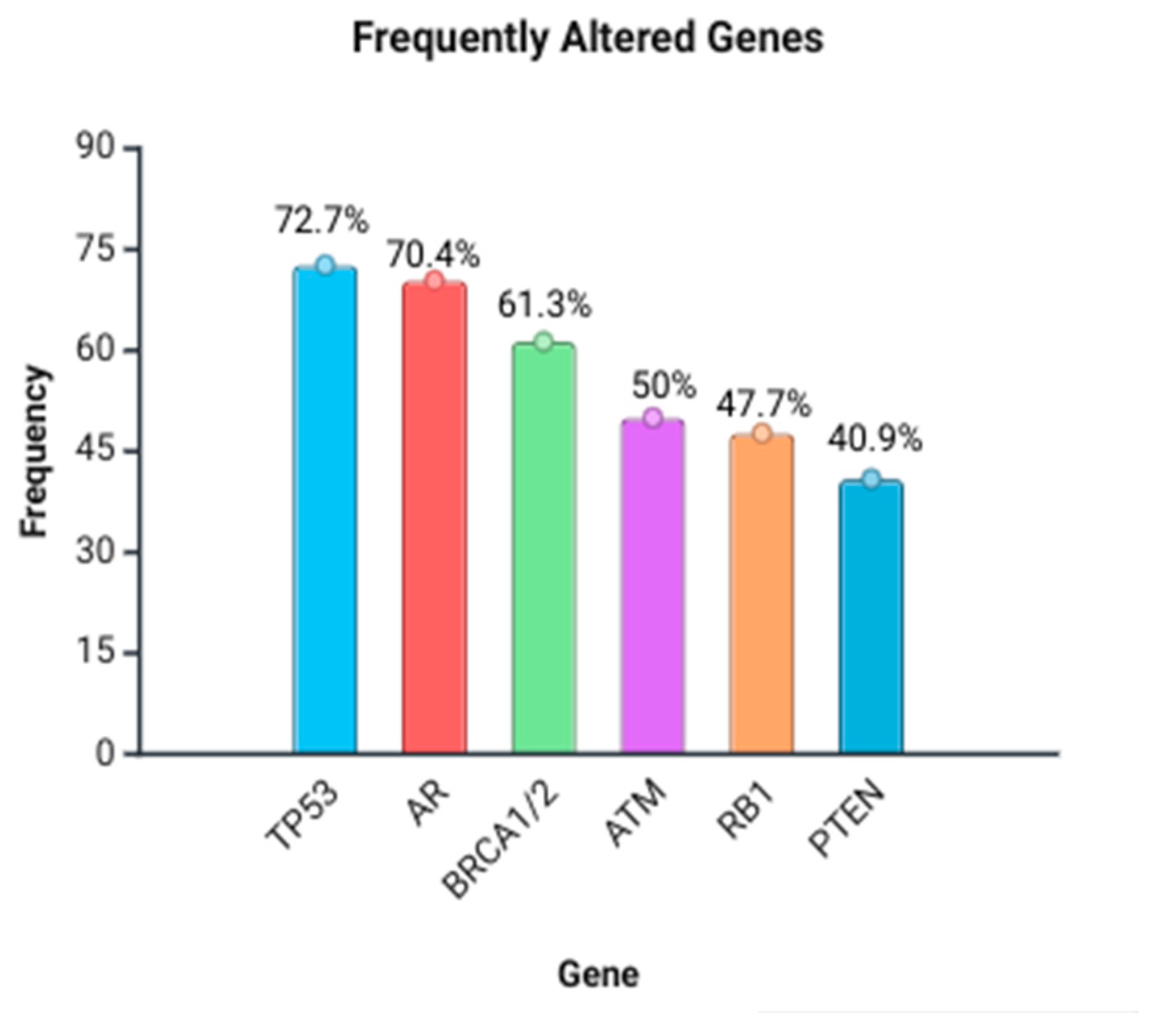

| Gene | Frequency of Reports (n Studies) | Common Alteration Types Reported |
|---|---|---|
| TP53 | 32/44 | Missense, deletions, loss of function, and copy number loss |
| AR | 31/44 | Amplifications (CNG), ligand-binding domain mutations (L702H, T878A, H875Y, W742C/L, and F877L), and rearrangements |
| BRCA1/2 | 27/44 | Germline and somatic truncating, frameshift, deletions, reversion mutations, pathogenic variants, and nonsense |
| ATM | 22/44 | Germline and somatic missense, truncations, and deletions |
| PTEN | 18/44 | Copy number loss, deletions, and inactivation |
| RB1 | 21/44 | Deletions, mutations, and rearrangements |
| CDK12 | 9/44 | Mutations and biallelic loss |
| PIK3CA | 13/44 | Missense mutations and amplifications |
| MSI-H/MMR genes (MSH2, MSH6, MLH1, and PMS2) | 6/44 | Frameshift, loss, and microsatellite instability |
| SPOP | 3/44 | Point mutations |
| MYC | 6/44 | Amplifications and mutations |
| APC | 7/44 | Mutations |
| Others (e.g., PALB2, CHEK2, FANCA, NCOR2, FOXA1, BRAF, EGFR, MET, FGFR1/2/3/4, ERBB2, IDH1, HOXB13, etc.) | ≤3/44 each | Various rare mutations or copy number changes |
| Gene/Pathway | Alteration Type | Clinical Association (OS/PFS) | References |
|---|---|---|---|
| TP53 | Mutations, deletions, and copy number loss; specific variants (e.g., c.665_672*11del) | Worse OS and/or PFS; some studies report platinum sensitivity despite poor prognosis | [18] (trend), [14,21,23,37,39,40,44,45,48,51,55,57] |
| PTEN | Loss/deletion and inactivation; frameshift (e.g., p.Y46Qfs*5) | Shorter OS/poor prognosis; aggressive phenotype | [39,45,48,51,60] |
| RB1 | Loss/deletion and mutations | Shorter OS; adverse prognosis; lineage plasticity features | [39,44,45,51,55] |
| AR (amplifications/CNV, SNVs, and GSRs) | CN gain; LBD SNVs (L702H, T878A, H875Y, W742C/L, and F877L); structural rearrangements | Shorter OS and/or rPFS in multiple studies; early progression on ARSIs in some cohorts | [14,18,37,40,41,44,55,59] |
| PIK3CA/PI3K pathway | Mutations and CN gain/amplification | Worse OS/PFS; aggressive disease biology | [21,45,48] |
| HRR genes (BRCA1/2, ATM, CDK12, CHEK2, PALB2, etc.) | Deleterious/truncating/germline and somatic | Worse OS/PFS on ARSI; prognostic effect heterogeneous across genes | [14,36,44] (worse PFS on Abiraterone HRRmt); (therapy benefit details placed in Table S8 Supplementary Material) |
| NCOA2 | Copy number gain; missense | Significantly shorter OS and PFS | [60] |
| MYC | Copy number gain/amplification | Poorer outcomes in some datasets; neutral in others | [55] (poorer); [44] (no clear link); [51] (N/A survival stated) |
| MYCN | Copy number gain | Associated with adverse outcomes/AVPC features | [55] |
| TMPRSS2-ERG | Fusion | No explicit OS/PFS link | [23] |
| CHD1 | Loss/deletion | Worse metastasis-free survival noted contextually; OS/PFS not clearly quantified | [37] |
| Gene/Pathway | Alteration Type | Therapy Association | References |
|---|---|---|---|
| AR | CN gain/amplification; LBD SNVs (L702H, T878A, F877L, H875Y, and W742C/L); GSRs; enhancer amplification | Resistance to ARSIs (Enzalutamide/Abiraterone); shorter response duration; primary resistance with AR-GSRs; enhancer/gene body amp linked to poor ARSI outcomes | [14,18,21,23,40,41,59] |
| TP53 + RB1 (±PTEN) co-alteration | Co-loss/combined alterations | Lineage plasticity/neuroendocrine-like features; ARSI resistance; highly aggressive biology | [21,39,51,55] |
| PTEN | Deletion/loss | Poor response to AR-targeted therapy; aggressive course | [51,58,60] (poor ARPI rPFS) |
| HRR genes (BRCA1/2, ATM, etc.) | Pathogenic/truncating (germline and somatic) | PARP inhibitor benefit; greatest with BRCA1/2; mixed/limited benefit with non-BRCA HRR | [14,46,47,52] (poor ARSI outcomes) |
| PALB2 | Pathogenic + reversion mutations | Initial PARPi sensitivity; reversion mutations → acquired PARPi resistance | [37] |
| PMS2/MMR | Pathogenic mutation/MSI-H | May benefit from checkpoint inhibitors (e.g., pembrolizumab) | [18,22] |
| CDK12 | Mutations/biallelic loss | High TMB → checkpoint inhibitor sensitivity; limited PARPi benefit | [34,52] |
| PI3K pathway (PIK3CA and PTEN loss context) | Mutations/CN gain | AR-targeted therapy resistance; rationale for PI3K/AKT/mTOR combinations | [14,21,48] |
| Therapy-modality signal (Lu-PSMA) | AR, TP53, and PTEN alterations | Poorer rPFS on ARPIs but better rPFS with 177Lu-PSMA-617 vs. changing ARPI | [58] |
| Platinum sensitivity signal | TP53 alterations | Poor OS overall but better response to platinum chemo in AVPC context | [39] |
| NCOA2 | CN gain/missense | Poor ARPI outcomes (no PSA Responses when gain present) | [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chanhih, N.; Laraqui, A.; Hassine, S.; Ameur, A.; Hamedoun, L.; El Annaz, H.; Abi, R.; Tagajdid, M.R.; Amine, I.L.; Ennibi, K.; et al. Circulating Tumor DNA as a Biomarker for Precision Medicine in Prostate Cancer: A Systematic Review. Int. J. Mol. Sci. 2025, 26, 11049. https://doi.org/10.3390/ijms262211049
Chanhih N, Laraqui A, Hassine S, Ameur A, Hamedoun L, El Annaz H, Abi R, Tagajdid MR, Amine IL, Ennibi K, et al. Circulating Tumor DNA as a Biomarker for Precision Medicine in Prostate Cancer: A Systematic Review. International Journal of Molecular Sciences. 2025; 26(22):11049. https://doi.org/10.3390/ijms262211049
Chicago/Turabian StyleChanhih, Nouhaila, Abdelilah Laraqui, Salma Hassine, Ahmed Ameur, Larbi Hamedoun, Hicham El Annaz, Rachid Abi, Mohamed Rida Tagajdid, Idriss Lahlou Amine, Khalid Ennibi, and et al. 2025. "Circulating Tumor DNA as a Biomarker for Precision Medicine in Prostate Cancer: A Systematic Review" International Journal of Molecular Sciences 26, no. 22: 11049. https://doi.org/10.3390/ijms262211049
APA StyleChanhih, N., Laraqui, A., Hassine, S., Ameur, A., Hamedoun, L., El Annaz, H., Abi, R., Tagajdid, M. R., Amine, I. L., Ennibi, K., Benjouad, A., & Belayachi, L. (2025). Circulating Tumor DNA as a Biomarker for Precision Medicine in Prostate Cancer: A Systematic Review. International Journal of Molecular Sciences, 26(22), 11049. https://doi.org/10.3390/ijms262211049







