Genome-Wide Identification and Functional Characterization of CesA10 and CesA11 Genes Involved in Cellulose Biosynthesis in Sugarcane
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification of CesAs Genes in S. officinarum
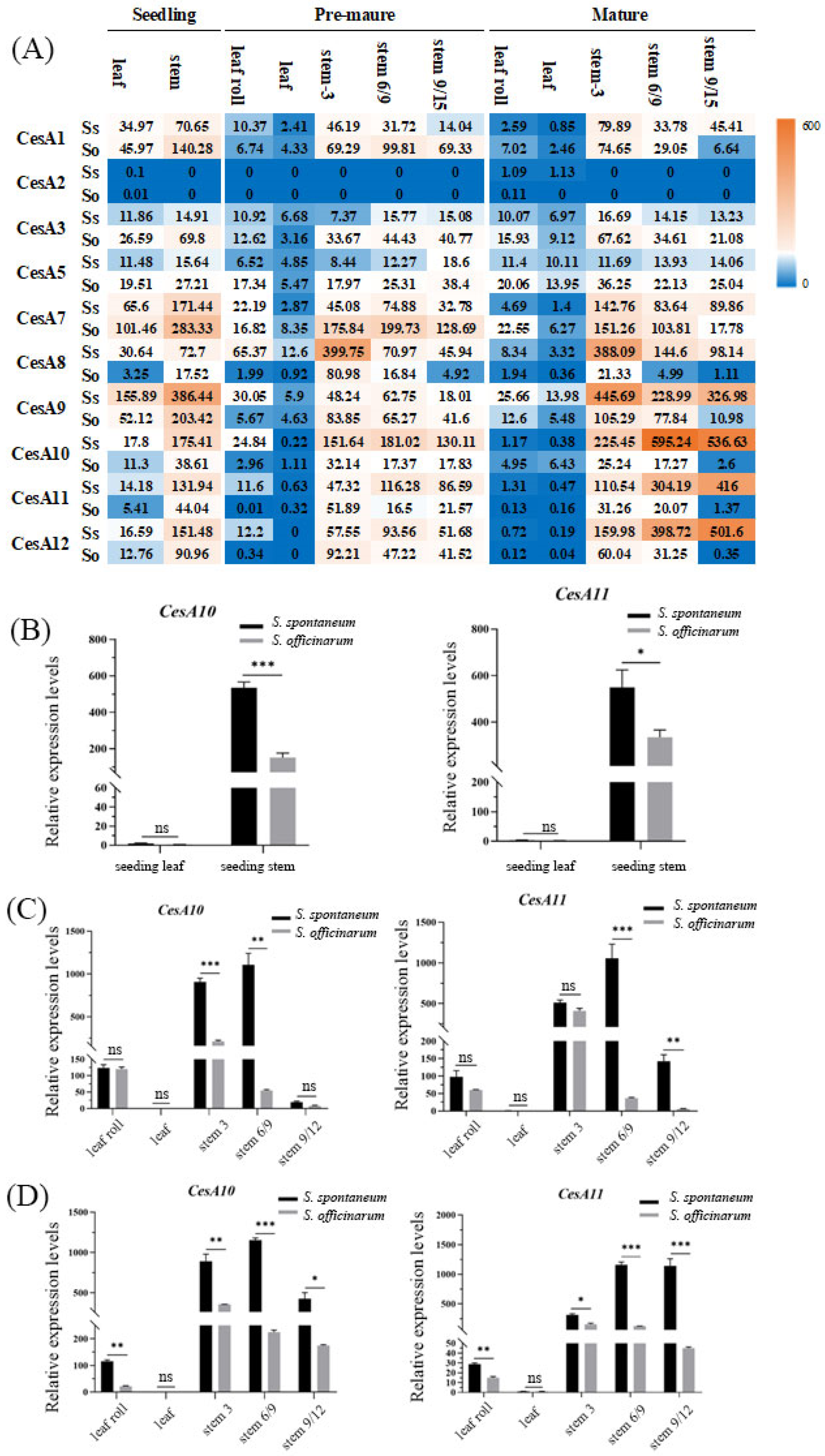
2.2. The Expression Patterns of CesA Genes at Different Developmental Stages in S. spontaneum and S. officinarum
2.3. The Subcellular Localization of SoCesA10 and SoCesA11
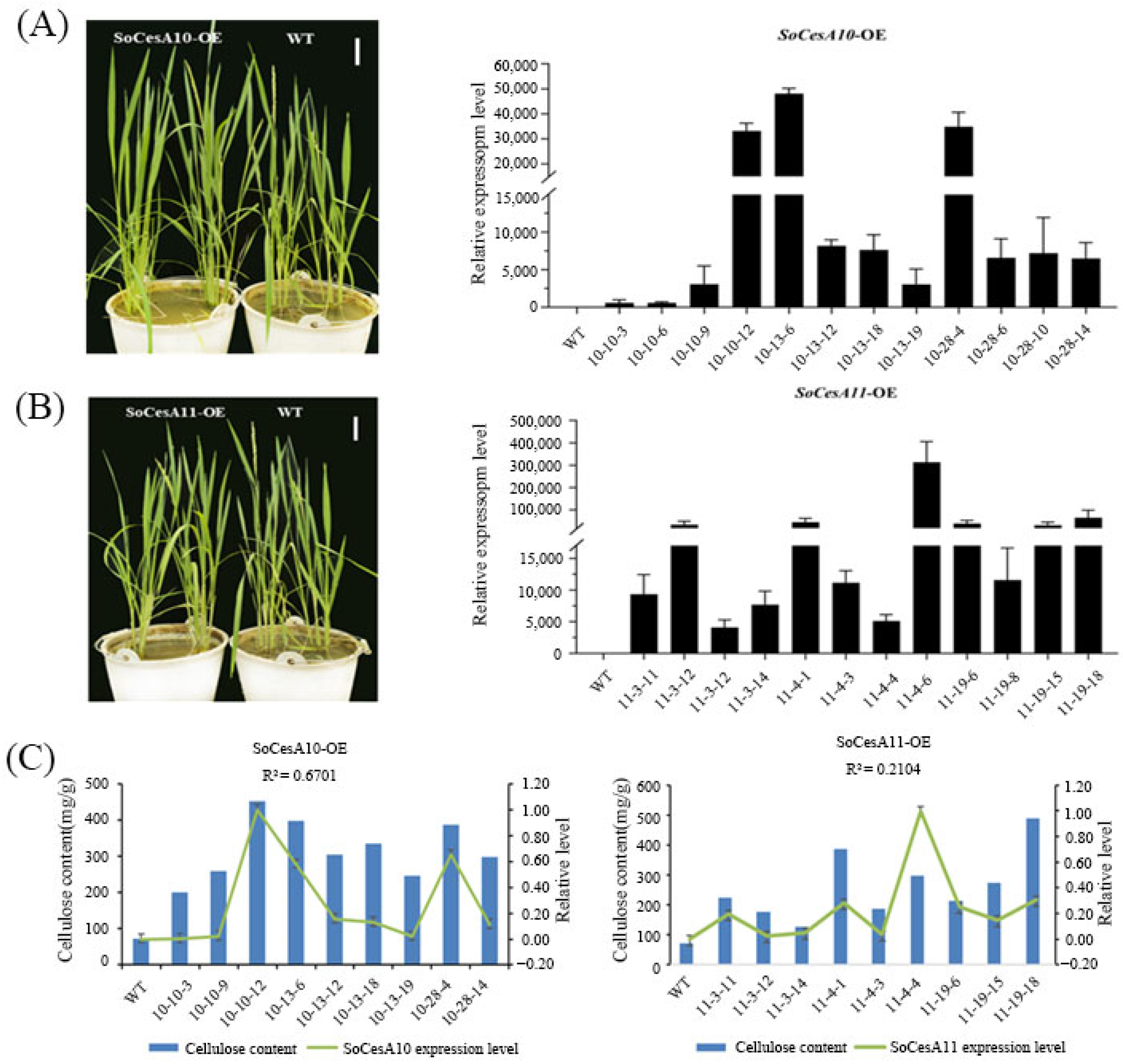
2.4. The Correlation Between the Expression Level of SoCesA and Cellulose Content
2.5. Genetic Transformation of SoCesA10 and SoCesA11 Genes in Rice
3. Discussion
3.1. Comparative Genomics Reveals Organ-Specific Expression and Interspecies Differential Expression of CesA Genes in Sugarcane
3.2. The Regulatory Role of the CesA Gene in Plant Cellulose Synthesis and Growth
4. Materials and Methods
4.1. Plant Materials, RNA Extraction, and Sequencing
4.2. Identification and Physicochemical Analysis of Cellulose Synthase Gene Family
4.3. Gene Structure, Protein Conserved Motifs, and Phylogenetic Analysis
4.4. Measurement of Cellulose Content in S. spontaneum and S. officinarum
4.5. Subcellular Localization and Genetic Transformation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thirugnanasambandam, P.P.; Hoang, N.V.; Furtado, A.; Botha, F.C.; Henry, R.J. Association of variation in the sugarcane transcriptome with sugar content. BMC Genom. 2017, 18, 909. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Y.; Feng, X.; Hua, X.; Dou, M.; Yao, W.; Zhang, M.; Zhang, J. Expression Profiling and MicroRNA Regulatory Networks of Homeobox Family Genes in Sugarcane Saccharum spontaneum L. Int. J. Mol. Sci. 2022, 23, 8724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Tang, H.; Zhang, Q.; Hua, X.; Ma, X.; Zhu, F.; Jones, T.; Zhu, X.; Bowers, J.; et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet 2018, 50, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Qi, B.; Hu, J.; Zhu, L.; Duan, Y.; Zhang, W.; Gou, L. Response of maize stalk to plant density on cellulose accumulation by modulating enzymes activities. Field Crops Res. 2023, 304, 109152. [Google Scholar] [CrossRef]
- Liu, F.; Li, Z.; Cai, W.; Wu, W. Effects of planting densities and planting depths on yield and lodging resistance in sugarcane. Chin. J. Trop. Crops 2023, 45, 330–339. [Google Scholar]
- Wei, J.; Shen, H.; Luo, J.; Wu, D.; Zhu, G. Empirical Analysis on Influencing Factors of Sugarcane Growing Income in Guangxi. Sugar Crops China 2021, 43, 79–86. [Google Scholar]
- Li, K.; Yang, X.; Liu, X.-G.; Hu, X.-J.; Wu, Y.-J.; Wang, Q.; Ma, F.-Q.; Li, S.-Q.; Wang, H.-W.; Liu, Z.-F.; et al. QTL analysis of the developmental changes in cell wall components and forage digestibility in maize (Zea mays L.). J. Integr. Agric. 2022, 21, 3501–3513. [Google Scholar] [CrossRef]
- Xu, J.-J.; Zhou, J.; Cai, Z.; Sun, J.-L.; Li, Y.-Z.; Fan, X.-W. ZmPGIP1 regulates stem strength by enhancing lignin and cellulose biosynthesis in Arabidopsis thaliana. Biotechnol. Biotechnol. Equip. 2024, 38, 2356867. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Z.; Sakamoto, S.; Mitsuda, N.; Ren, A.; Persson, S.; Zhang, D. ETHYLENE RESPONSE FACTOR 34 promotes secondary cell wall thickening and strength of rice peduncles. Plant Physiol. 2022, 190, 1806–1820. [Google Scholar] [CrossRef]
- Zhong, R.; Cui, D.; Ye, Z.H. Secondary cell wall biosynthesis. New Phytol. 2019, 221, 1703–1723. [Google Scholar] [CrossRef]
- McFarlane, H.E.; Doring, A.; Persson, S. The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 2014, 65, 69–94. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, H.; Li, X.; Ge, Y.; Lu, X.; Li, H. Characterization of ZmCesAs for Secondary Cell Wall Biosynthesis in Maize. J. Plant Biol. 2024, 67, 161–174. [Google Scholar] [CrossRef]
- Polko, J.K.; Kieber, J.J. The Regulation of Cellulose Biosynthesis in Plants. Plant Cell 2019, 31, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Sheng, J.; Zhu, F.; Zhao, L.; Hu, X.; Zheng, X.; Zhou, F.; Hu, Z.; Diao, Y.; Jin, S. Differential expression patterns reveal the roles of cellulose synthase genes (CesAs) in primary and secondary cell wall biosynthesis in Miscanthus × giganteus. Ind. Crops Prod. 2020, 145, 112129. [Google Scholar] [CrossRef]
- Casu, R.E.; Rae, A.L.; Nielsen, J.M.; Perroux, J.M.; Bonnett, G.D.; Manners, J.M. Tissue-specific transcriptome analysis within the maturing sugarcane stalk reveals spatial regulation in the expression of cellulose synthase and sucrose transporter gene families. Plant Mol. Biol. 2015, 89, 607–628. [Google Scholar] [CrossRef]
- Somerville, C. Cellulose Synthesis in Higher Plants. Annu. Rev. Cell Dev. Biol. 2006, 22, 53–78. [Google Scholar] [CrossRef]
- Guo, B.; Huang, X.; Qi, J.; Sun, H.; Lv, C.; Wang, F.; Zhu, J.; Xu, R. Brittle culm 3, encoding a cellulose synthase subunit 5, is required for cell wall biosynthesis in barley (Hordeum vulgare L.). Front. Plant Sci. 2022, 13, 989406. [Google Scholar] [CrossRef]
- Taylor, N.G.; Howells, R.M.; Huttly, A.K.; Vickers, K.; Turner, S.R. Interactions among three distinct CesA proteinsessential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 2003, 100, 1450–1455. [Google Scholar] [CrossRef]
- Wang, L.; Guo, K.; Li, Y.; Tu, Y.; Hu, H.; Wang, B.; Cui, X.; Peng, L. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 2010, 10, 282–306. [Google Scholar] [CrossRef]
- Kumar, M.; Wightman, R.; Atanassov, I.; Gupta, A.; Hurst, C.; Hemsley, P.; Turner, S. S-Acylation of the cellulose synthase complex is essential for its plasma membrane localization. Science 2016, 353, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Su, J.; Wang, H.; Zhang, Z.; Zhang, X.; Van de Peer, Y.; Chen, F.; Fang, W.; Guan, Z.; Zhang, F.; et al. Analyses of a chromosome-scale genome assembly reveal the origin and evolution of cultivated chrysanthemum. Nat. Commun. 2023, 14, 2021. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Fan, J.; Fang, A.; Li, Y.; Tariqjaveed, M.; Li, D.; Hu, D.; Wang, W.-M. Ustilaginoidea virens: Insights into an Emerging Rice Pathogen. Annu. Rev. Phytopathol. 2020, 58, 363–385. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.A.; Shirley, N.J.; King, B.J.; Harvey, A.J.; Fincher, G.B. The CesA Gene Family of Barley. Quantitative Analysis of Transcripts Reveals Two Groups of Co-Expressed Genes. Plant Physiol. 2004, 134, 224–236. [Google Scholar] [CrossRef]
- Yin, Y.; Huang, J.; Xu, Y. The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 2009, 9, 99. [Google Scholar] [CrossRef]
- Neta, H.; Doron, H.; Tim, H.; Kanwarpal, S.D.; Beatriz, X.-C.; Deborah, P.D. A Comparative Analysis of the Plant Cellulose Synthase (CesA) Gene Family. Plant Physiol. 2000, 123, 1313–1323. [Google Scholar] [CrossRef]
- Niu, J.; Miao, X.; Wang, D.; Huang, W.; Yang, L.; Li, Y. Analysis of sugar accumulation characteristics and metabolism-relateed enzymes activities in the high and low sugar sugarcane at elongation stage. Jiangsu J. Agr. Sci. 2019, 35, 537–543. [Google Scholar]
- Hosaka, G.K.; Correr, F.H.; da Silva, C.C.; Sforça, D.A.; Barreto, F.Z.; Balsalobre, T.W.A.; Pasha, A.; de Souza, A.P.; Provart, N.J.; Carneiro, M.S.; et al. Temporal Gene Expression in Apical Culms Shows Early Changes in Cell Wall Biosynthesis Genes in Sugarcane. Front. Plant Sci. 2021, 12, 736797. [Google Scholar] [CrossRef]
- Zhang, B.; Zhou, Y. Plant Cell Wall Formation and Regulation. Sci. Sin. Vitae 2015, 45, 544–556. [Google Scholar] [CrossRef]
- Mengistie, E.; McDonald, A.G. Effect of cell wall compositions on lodging resistance of cereal crops: Review. J. Agric. Sci. 2024, 161, 794–807. [Google Scholar] [CrossRef]
- Park, S.; Ding, S.-Y. Five amino acid mismatches in the zinc-finger domains of Cellulose Synthase 5 and Cellulose Synthase 6 cooperatively modulate their functional properties by controlling homodimerization in Arabidopsis. Plant Mol. Biol. 2024, 114, 76. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, C.; Huang, R.; Zhang, K.; Wang, Q.; Fu, C.; Liu, W.; Sun, C.; Wang, P.; Wang, F.; et al. Rice Brittle Culm19 Encoding Cellulose Synthase Subunit CESA4 Causes Dominant Brittle Phenotype But has No Distinct Influence on Growth and Grain Yield. Rice 2021, 14, 95. [Google Scholar] [CrossRef]
- Zhang, L.; Thapa Magar, M.S.; Wang, Y.; Cheng, Y. Tip growth defective1 interacts with cellulose synthase A3 to regulate cellulose biosynthesis in Arabidopsis. Plant Mol. Biol. 2022, 110, 1–12. [Google Scholar] [CrossRef]
- Yan, C.; Yan, S.; Zeng, X.; Zhang, Z.; Gu, M. Fine Mapping and Isolation of Bc7(t), Allelic to OsCesA4. J. Genet. Genom. 2007, 34, 1019–1027. [Google Scholar] [CrossRef]
- Zhang, B.; Deng, L.; Qian, Q.; Xiong, G.; Zeng, D.; Li, R.; Guo, L.; Li, J.; Zhou, Y. A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice. Plant Mol. Biol. 2009, 71, 509–524. [Google Scholar] [CrossRef]
- Chen, F.; Wang, D.; Qin, Y.; Fang, J.; Yuan, S.; Peng, L.; Zhao, J.; Li, X. A Missense Mutation in the Zinc Finger Domain of OsCESA7 Deleteriously Affects Cellulose Biosynthesis and Plant Growth in Rice. PLoS ONE 2016, 11, e0153993. [Google Scholar]
- Wang, D.; Yuan, S.; Yin, L.; Zhao, J.; Guo, B.; Lan, J.; Li, X. A missense mutation in the transmembrane domain of CESA9 affects cell wall biosynthesis and plant growth in rice. Plant Sci. 2012, 196, 117–124. [Google Scholar] [CrossRef]
- Niu, Z.; Bai, Q.; Lv, J.; Tian, W.; Mao, K.; Wei, Q.; Zheng, Y.; Yang, H.; Gao, C.; Wan, D. The fasciclin-like arabinogalactan protein FLA11 of Ostrya rehderiana impacts wood formation and salt stress in Populus. Environ. Exp. Bot. 2024, 219, 105651. [Google Scholar] [CrossRef]
- Shen, Y.; Ma, F.; Wang, M.; Li, X.; Zhang, M.; Jiangfeng, H. Accurate Evaluation and Mechanism Analysis of Mechanical Strength of Sugarcane Stalk. Chin. J. Trop. Crops 2022, 43, 207–215. [Google Scholar]
- Lima, D.U.; Santos, H.P.; Tiné, M.A.; Molle, F.R.D.; Buckeridge, M.S. Patterns of expression of cell wall related genes in sugarcane. Genet. Mol. Biol. 2001, 24, 191–198. [Google Scholar] [CrossRef][Green Version]
- Kuang, B.; Zhao, J.; Li, S.; Wei, N.; Feng, M.; Yang, X. Bioinformatics Analysis and Function Prediction of CesA7 Gene in Sugarcane. Chin. J. Trop. Crops 2023, 40, 1337–1347. [Google Scholar][Green Version]
- Zhang, J.; Ming, R.; Wang, Z.; Wang, Y.; Yuan, Y.; Zhong, W.; Hua, X.; Li, Z. Genome-Wide Identification and Expression Profile Analysis of WRKY Family Genes in the Autopolyploid Saccharum spontaneum. Plant Cell Physiol. 2020, 61, 616–630. [Google Scholar][Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Battistuzzi, F.U. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | Number of Alleles | Number of Amino Acids | Theoretical pI | Molecular Weight | Aliphatic Index | Grand Average of Hydropathicity | Instability Index | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| SoCesA1 | Soffic.09G0009150 | 5 | 1076 | 6.6 | 121,187.11 | 84.89 | −0.195 | 39.89 | Plasma membrane |
| SoCesA2 | Soffic.10G0017310 | 6 | 937 | 6.7 | 102,705.03 | 87.39 | −0.126 | 52.04 | Plasma membrane |
| SoCesA3 | Soffic.03G0004580 | 9 | 1094 | 6.12 | 123,061.17 | 88.92 | −0.154 | 36.93 | Plasma membrane |
| SoCesA5 | Soffic.01G0017790 | 14 | 1317 | 8.47 | 147,389.74 | 83.35 | −0.261 | 41.1 | Plasma membrane |
| SoCesA7 | Soffic.02G0008760 | 8 | 1087 | 6.52 | 122,887.03 | 81.15 | −0.245 | 39.78 | Plasma membrane |
| SoCesA8 | Soffic.02G0007110 | 3 | 1095 | 7.42 | 122,601.02 | 82.55 | −0.186 | 44.61 | Plasma membrane |
| SoCesA9 | Soffic.02G0032990 | 8 | 1081 | 8.04 | 120,986.5 | 81.34 | −0.231 | 37.39 | Plasma membrane |
| SoCesA10 | Soffic.01G0033860 | 3 | 1374 | 8.69 | 154,548.22 | 76.8 | −0.316 | 39.45 | Plasma membrane |
| SoCesA11 | Soffic.02G0033140 | 6 | 981 | 6.22 | 110,255.67 | 83.9 | −0.115 | 39.84 | Plasma membrane |
| SoCesA12 | Soffic.02G0014650 | 6 | 1051 | 6.36 | 118,141.03 | 84.67 | −0.158 | 36.17 | Plasma membrane |
| Primer Name | Left Primer | Right Primer |
|---|---|---|
| GAPDH | CACGGCCACTGGAAGCA | TCCTCAGGGTTCCTGATGCC |
| 25S rRNA | GCAGCCAAGCGTTCATAGC | CCTATTGGTGGGTGAACAATCC |
| SoCesA10 | GTCATCAGCTGCGGATACGA | GTGCAGTACACCGACTTCCA |
| SoCesA11 | CATGGCCTGGGAACAATCCT | TGTCAGAACAGCGGACACTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Qi, N.; Han, Y.; Cai, L.; Wang, X.; Shang, H.; Zhang, Q.; Zhang, J. Genome-Wide Identification and Functional Characterization of CesA10 and CesA11 Genes Involved in Cellulose Biosynthesis in Sugarcane. Int. J. Mol. Sci. 2025, 26, 11046. https://doi.org/10.3390/ijms262211046
Xu Y, Qi N, Han Y, Cai L, Wang X, Shang H, Zhang Q, Zhang J. Genome-Wide Identification and Functional Characterization of CesA10 and CesA11 Genes Involved in Cellulose Biosynthesis in Sugarcane. International Journal of Molecular Sciences. 2025; 26(22):11046. https://doi.org/10.3390/ijms262211046
Chicago/Turabian StyleXu, Yi, Nameng Qi, Yi Han, Liying Cai, Xue Wang, Heyang Shang, Qing Zhang, and Jisen Zhang. 2025. "Genome-Wide Identification and Functional Characterization of CesA10 and CesA11 Genes Involved in Cellulose Biosynthesis in Sugarcane" International Journal of Molecular Sciences 26, no. 22: 11046. https://doi.org/10.3390/ijms262211046
APA StyleXu, Y., Qi, N., Han, Y., Cai, L., Wang, X., Shang, H., Zhang, Q., & Zhang, J. (2025). Genome-Wide Identification and Functional Characterization of CesA10 and CesA11 Genes Involved in Cellulose Biosynthesis in Sugarcane. International Journal of Molecular Sciences, 26(22), 11046. https://doi.org/10.3390/ijms262211046






