HSPA5, a Host Cellular Heat-Shock Protein Required for Influenza a Virus Replication
Abstract
1. Introduction
2. Results
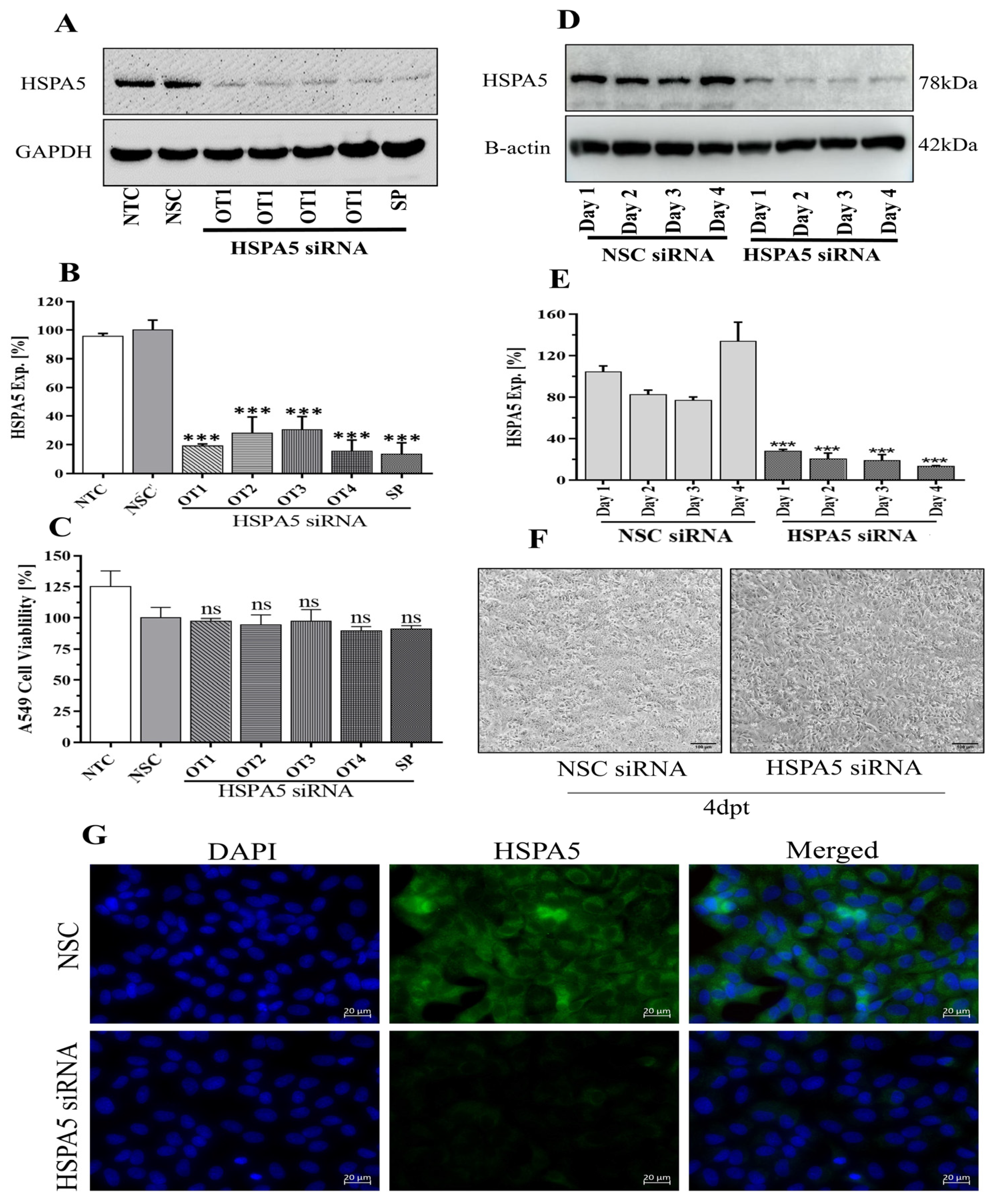
2.1. Optimization of HSPA5 Knockdown by siRNA Treatment
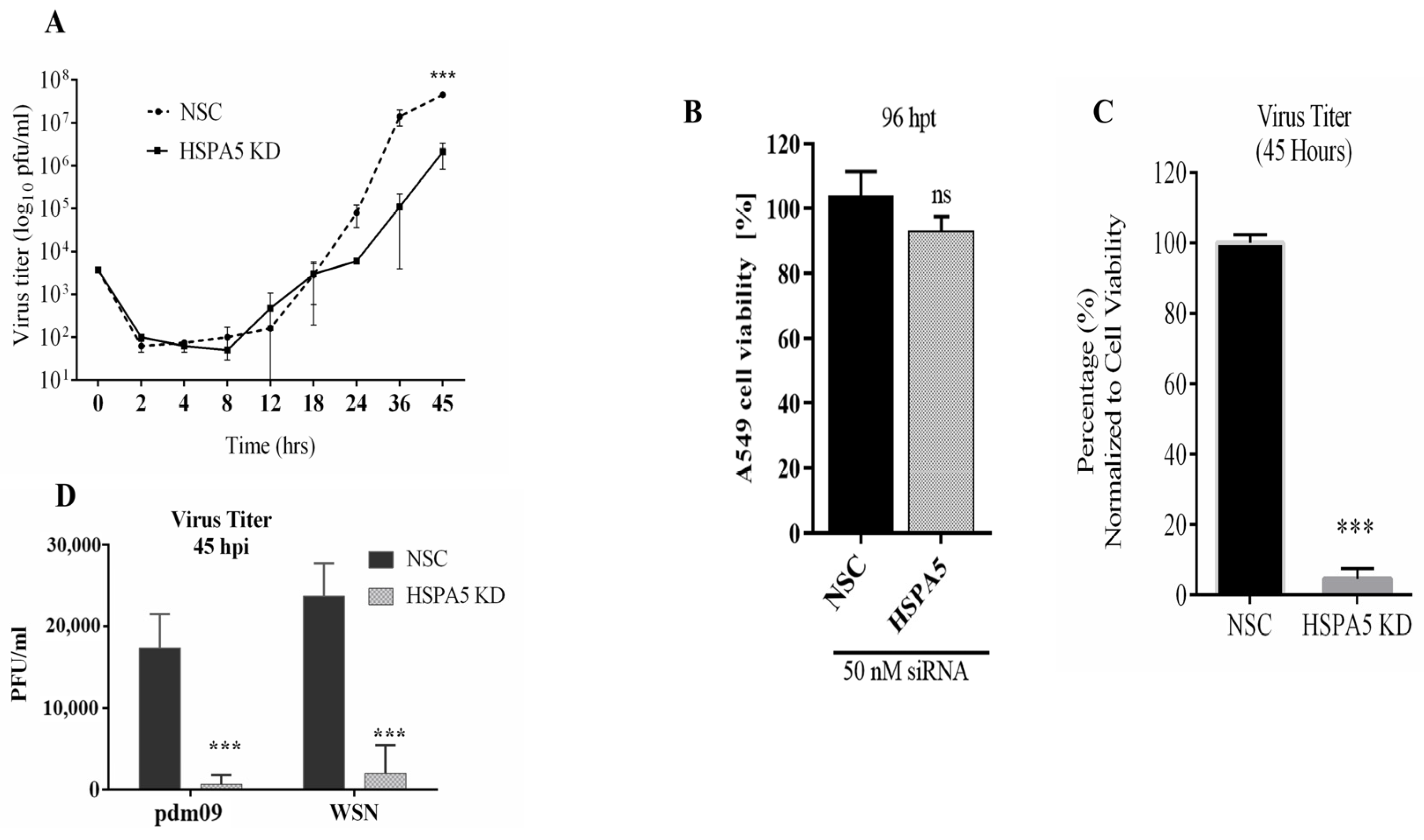
2.2. Impact of HSPA5 KD on IAV Replication
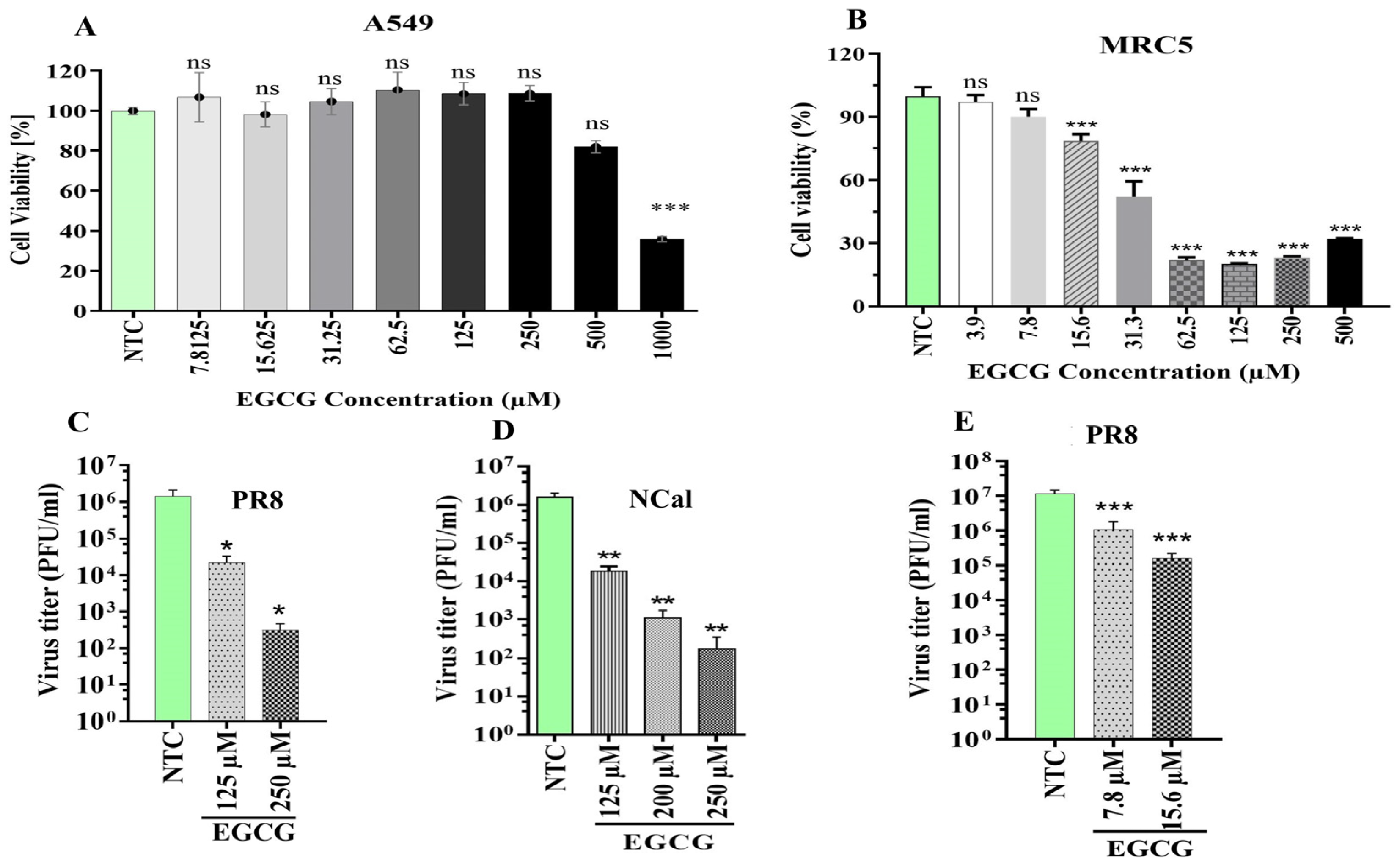
2.3. Inhibition of HSPA5 by Epigallocatechin-3-Gallate (EGCG) Suppresses IAV Replication
2.4. Effect of HSPA5 Silencing on IAV Viral Protein Synthesis and vRNA Transcription
2.5. Impact of HSPA5 Knockdown on Proteome Regulation During IAV Infection
3. Discussion
3.1. HSPA5 KD Alters IAV-Mediated Host Proteomic Responses
3.2. HSPA5 KD Causes Differential Regulation of Cellular Functions and Signaling Pathways During IAV Infection
3.3. HSPA5 Protein Is Required for Maturation of IAV During Replication
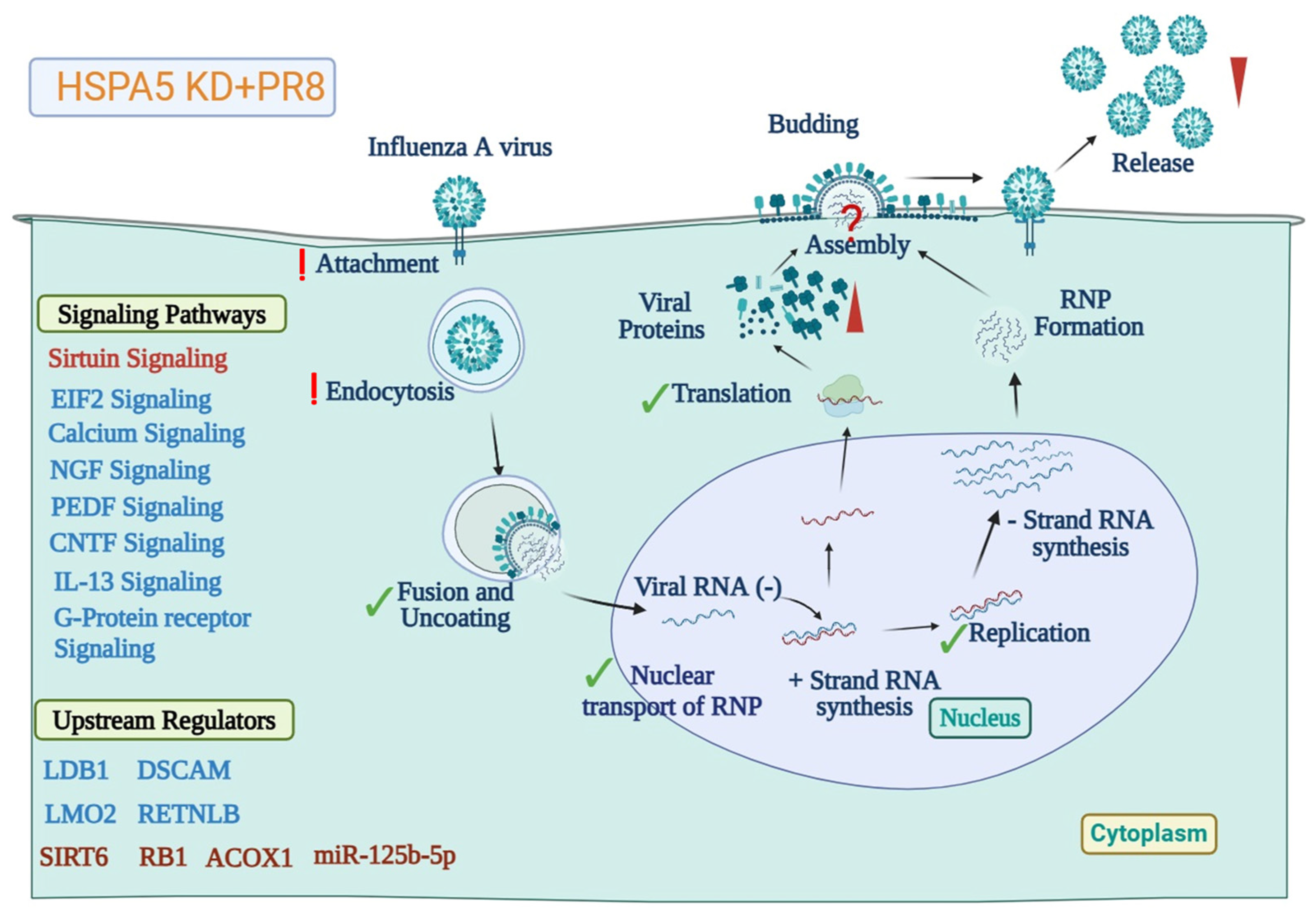
4. Materials and Methods
4.1. Cells and Viruses
4.2. Infection and Plaque Assay
4.3. Cell Viability
4.4. siRNA Transfection
4.5. Protein Extraction and Quantification
4.6. SomaScan Analyses
4.7. Immunoblotting
4.8. RNA Extraction and Real-Time PCR
4.9. Impact of HSPA5 Inhibitors on IAV Replication
4.10. Photomicrography
4.11. Immunofluorescent Microscopy
4.12. Statistical and Bioinformatics Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glezen, W.P. Clinical practice. Prevention and treatment of seasonal influenza. N. Engl. J. Med. 2008, 359, 2579–2585. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Morens, D.M. Influenza: The once and future pandemic. Public Health Rep. 2010, 125 (Suppl. S3), 16–26. [Google Scholar] [CrossRef]
- Johnson, N.P.; Mueller, J. Updating the accounts: Global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med. 2002, 76, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.L.; Palese, P. Orthomyxoviridae: The viruses and their replication. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1691–1740. [Google Scholar]
- Tong, S.; Li, Y.; Rivailler, P.; Conrardy, C.; Castillo, D.A.A.; Chen, L.-M.; Recuenco, S.; Ellison, J.A.; Davis, C.T.; York, I.A.; et al. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA 2012, 109, 4269–4274. [Google Scholar] [CrossRef]
- Sorrell, E.M.; Ramirez-Nieto, G.C.; Gomez-Osorio, I.G.; Perez, D.R. Genesis of pandemic influenza. Cytogenet. Genome Res. 2007, 117, 394–402. [Google Scholar] [CrossRef]
- Rashid, M.U.; Gao, A.; Coombs, K.M. Influenza A Virus Uses PSMA2 for Downregulation of the NRF2-Mediated Oxidative Stress Response. J. Virol. 2022, 96, e0199021. [Google Scholar] [CrossRef]
- Hendershot, L.M.; Valentine, V.A.; Lee, A.S.; Morris, S.W.; Shapiro, D.N. Localization of the gene encoding human BiP/GRP78, the endoplasmic reticulum cognate of the HSP70 family, to chromosome 9q34. Genomics 1994, 20, 281–284. [Google Scholar] [CrossRef]
- Brocchieri, L.; Conway de Macario, E.; Macario, A.J. hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 2008, 8, 19. [Google Scholar] [CrossRef]
- Cunnea, P.M.; Miranda-Vizuete, A.; Bertoli, G.; Simmen, T.; Damdimopoulos, A.E.; Hermann, S.; Leinonen, S.; Huikko, M.P.; Gustafsson, J.; Sitia, R.; et al. ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J. Biol. Chem. 2003, 278, 1059–1066. [Google Scholar] [CrossRef]
- Yang, J.; Nune, M.; Zong, Y.; Zhou, L.; Liu, Q. Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP. Structure 2015, 23, 2191–2203. [Google Scholar] [CrossRef] [PubMed]
- Simons, J.F.; Ferro-Novick, S.; Rose, M.D.; Helenius, A. BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J. Cell Biol. 1995, 130, 41–49. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Law, D.T.; Williams, D.B. Binding protein BiP is required for translocation of secretory proteins into the endoplasmic reticulum in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1991, 88, 1565–1569. [Google Scholar] [CrossRef]
- Vogel, J.P.; Misra, L.M.; Rose, M.D. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J. Cell Biol. 1990, 110, 1885–1895. [Google Scholar] [CrossRef]
- Chapman, R.; Sidrauski, C.; Walter, P. Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu. Rev. Cell Dev. Biol. 1998, 14, 459–485. [Google Scholar] [CrossRef]
- Korennykh, A.; Walter, P. Structural basis of the unfolded protein response. Annu. Rev. Cell Dev. Biol. 2012, 28, 251–277. [Google Scholar] [CrossRef]
- Okamura, K.; Kimata, Y.; Higashio, H.; Tsuru, A.; Kohno, K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 2000, 279, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.M.; Park, S.; Kim, S.J.; Kim, H.; Lee, B.; Kim, J.; Park, J.; Kim, S.T.; Yang, H.-K.; Kim, W.H.; et al. KIAA1324 Suppresses Gastric Cancer Progression by Inhibiting the Oncoprotein GRP78. Cancer Res. 2015, 75, 3087–3097. [Google Scholar] [CrossRef] [PubMed]
- Khongwichit, S.; Sornjai, W.; Jitobaom, K.; Greenwood, M.; Greenwood, M.P.; Hitakarun, A.; Wikan, N.; Murphy, D.; Smith, D.R. A functional interaction between GRP78 and Zika virus E protein. Sci. Rep. 2021, 11, 393. [Google Scholar] [CrossRef] [PubMed]
- Turpin, J.; Frumence, E.; Harrabi, W.; Haddad, J.G.; El Kalamouni, C.; Desprès, P.; Krejbich-Trotot, P.; Viranaïcken, W. Zika virus subversion of chaperone GRP78/BiP expression in A549 cells during UPR activation. Biochimie 2020, 175, 99–105. [Google Scholar] [CrossRef]
- Nain, M.; Mukherjee, S.; Karmakar, S.P.; Paton, A.W.; Paton, J.C.; Abdin, M.Z.; Basu, A.; Kalia, M.; Vrati, S. GRP78 Is an Important Host Factor for Japanese Encephalitis Virus Entry and Replication in Mammalian Cells. J. Virol. 2017, 91, e02274-16. [Google Scholar] [CrossRef]
- Jindadamrongwech, S.; Thepparit, C.; Smith, D.R. Identification of GRP 78 (BiP) as a liver cell expressed receptor element for dengue virus serotype 2. Arch. Virol. 2004, 149, 915–927. [Google Scholar] [CrossRef]
- Triantafilou, K.; Fradelizi, D.; Wilson, K.; Triantafilou, M. GRP78, a coreceptor for coxsackievirus A9, interacts with major histocompatibility complex class I molecules which mediate virus internalization. J. Virol. 2002, 76, 633–643. [Google Scholar] [CrossRef]
- Elfiky, A.A. SARS-CoV-2 Spike-Heat Shock Protein A5 (GRP78) Recognition may be Related to the Immersed Human Coronaviruses. Front. Pharmacol. 2020, 11, 577467. [Google Scholar] [CrossRef]
- Carlos, A.J.; Ha, D.P.; Yeh, D.-W.; Van Krieken, R.; Tseng, C.-C.; Zhang, P.; Gill, P.; Machida, K.; Lee, A.S. The chaperone GRP78 is a host auxiliary factor for SARS-CoV-2 and GRP78 depleting antibody blocks viral entry and infection. J. Biol. Chem. 2021, 296, 100759. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.; Park, R.; Park, Y.-I.; Cha, Y.-E.; Yamamoto, A.; Lee, J.I.; Park, J. EGCG, a green tea polyphenol, inhibits human coronavirus replication in vitro. Biochem. Biophys. Res. Commun. 2021, 547, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Ning, M.; Chen, B.; Li, X.; Sun, H.; Pu, J.; Liu, J.; Wang, N.; Huang, Y. Comparative IP-MS Reveals HSPA5 and HSPA8 Interacting with Hemagglutinin Protein to Promote the Replication of Influenza A Virus. Pathogens 2025, 14, 535. [Google Scholar] [CrossRef]
- Liu, J.; Chen, K.; Wu, W.; Pang, Z.; Zhu, D.; Yan, X.; Wang, B.; Qiu, J.; Fang, Z. GRP78 exerts antiviral function against influenza A virus infection by activating the IFN/JAK-STAT signaling. Virology 2024, 600, 110249. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Wang, R.; Zou, W.; Sun, X.; Liu, X.; Zhao, L.; Wang, S.; Jin, M. The Influenza Virus H5N1 Infection Can Induce ROS Production for Viral Replication and Host Cell Death in A549 Cells Modulated by Human Cu/Zn Superoxide Dismutase (SOD1) Overexpression. Viruses 2016, 8, 13. [Google Scholar] [CrossRef]
- Zhong, M.Z.; Xu, M.N.; Zheng, S.Q.; Cheng, S.Q.; Zeng, K.; Huang, X.W. Manipulating host secreted protein gene expression: An indirect approach by HPV11/16 E6/E7 to suppress PBMC cytokine secretion. Virol. J. 2024, 21, 172. [Google Scholar] [CrossRef]
- Sekaran, K.; Varghese, R.P.; Krishnan, S.; Zayed, H.; El Allali, A.; Doss, G.P.C. Dissecting Crucial Gene Markers Involved in HPV-Associated Oropharyngeal Squamous Cell Carcinoma from RNA-Sequencing Data through Explainable Artificial Intelligence. Front. Biosci. (Landmark Ed.) 2024, 29, 220. [Google Scholar] [CrossRef]
- Peng, P.; Li, N.; Zhang, N.; Fu, X.; Peng, S.; Zhao, Y.; Ai, B. Identifying Luteolin as a Potential Drug for Treating Lung Adenocarcinoma with COVID-19 Affection based on Integration Analysis of Pharmacology and Transcriptome. Curr. Med. Chem. 2024, 31, 5432–5447. [Google Scholar] [CrossRef]
- Chan, K.K.; Hon, T.; Au, K.; Choi, H.; Wong, D.K.; Chan, A.C.; Yuen, M.; Lai, C.; Lo, R.C. Stanniocalcin 1 is a serum biomarker and potential therapeutic target for HBV-associated liver fibrosis. J. Pathol. 2022, 257, 227–238. [Google Scholar] [CrossRef]
- Schmitz, T.; Freuer, D.; Goßlau, Y.; Warm, T.D.; Hyhlik-Dürr, A.; Linseisen, J.; Meisinger, C.; Kirchberger, I. Can inflammatory plasma proteins predict Long COVID or Fatigue severity after SARS-CoV-2 infection? Virus Res. 2024, 344, 199363. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, A.; Salentin, R.; Meyer, R.G.; Bussfeld, D.; Pauligk, C.; Fesq, H.; Hofmann, P.; Nain, M.; Gemsa, D.; Sprenger, H. Defense against influenza A virus infection: Essential role of the chemokine system. Immunobiology 2001, 204, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Li, N.; Yang, Z.; Li, H.; Zheng, H.; Yang, J.; Chen, Y.; Zhao, X.; Mei, J.; Shi, H.; et al. Role of CXCL5 in Regulating Chemotaxis of Innate and Adaptive Leukocytes in Infected Lungs Upon Pulmonary Influenza Infection. Front. Immunol. 2021, 12, 785457. [Google Scholar] [CrossRef]
- Betakova, T.; Kostrabova, A.; Lachova, V.; Turianova, L. Cytokines Induced During Influenza Virus Infection. Curr. Pharm. Des. 2017, 23, 2616–2622. [Google Scholar] [CrossRef]
- Hinshaw, V.S.; Olsen, C.W.; Dybdahl-Sissoko, N.; Evans, D. Apoptosis: A mechanism of cell killing by influenza A and B viruses. J. Virol. 1994, 68, 3667–3673. [Google Scholar] [CrossRef] [PubMed]
- Mori, I.; Komatsu, T.; Takeuchi, K.; Nakakuki, K.; Sudo, M.; Kimura, Y. In vivo induction of apoptosis by influenza virus. J. Gen. Virol. 1995, 76 Pt 11, 2869–2873. [Google Scholar] [CrossRef]
- Atkin-Smith, G.K.; Duan, M.; Chen, W.; Poon, I.K.H. The induction and consequences of Influenza A virus-induced cell death. Cell Death Dis. 2018, 9, 1002. [Google Scholar] [CrossRef]
- To, K.-F.; Chan, P.K.; Chan, K.-F.; Lee, W.-K.; Lam, W.-Y.; Wong, K.-F.; Tang, N.L.; Tsang, D.N.; Sung, R.Y.; Buckley, T.A.; et al. Pathology of fatal human infection associated with avian influenza A H5N1 virus. J. Med. Virol. 2001, 63, 242–246. [Google Scholar] [CrossRef]
- Guarner, J.; Falcón-Escobedo, R. Comparison of the pathology caused by H1N1, H5N1, and H3N2 influenza viruses. Arch. Med. Res. 2009, 40, 655–661. [Google Scholar] [CrossRef]
- Cheng, J.; Tao, J.; Li, B.; Shi, Y.; Liu, H. The tyrosine 73 and serine 83 dephosphorylation of H1N1 swine influenza virus NS1 protein attenuates virus replication and induces high levels of beta interferon. Virol. J. 2019, 16, 152. [Google Scholar] [CrossRef]
- Cui, L.; Zheng, W.; Li, M.; Bai, X.; Yang, W.; Li, J.; Fan, W.; Gao, G.F.; Sun, L.; Liu, W. Phosphorylation Status of Tyrosine 78 Residue Regulates the Nuclear Export and Ubiquitination of Influenza A Virus Nucleoprotein. Front. Microbiol. 2019, 10, 1816. [Google Scholar] [CrossRef]
- Mecate-Zambrano, A.; Sukumar, S.; Seebohm, G.; Ciminski, K.; Schreiber, A.; Anhlan, D.; Greune, L.; Wixler, L.; Grothe, S.; Stein, N.C.; et al. Discrete spatio-temporal regulation of tyrosine phosphorylation directs influenza A virus M1 protein towards its function in virion assembly. PLoS Pathog. 2020, 16, e1008775. [Google Scholar] [CrossRef]
- Hsiang, T.Y.; Zhou, L.; Krug, R.M. Roles of the phosphorylation of specific serines and threonines in the NS1 protein of human influenza A viruses. J. Virol. 2012, 86, 10370–10376. [Google Scholar] [CrossRef] [PubMed]
- Orr-Burks, N.; Murray, J.; Todd, K.V.; Bakre, A.; Tripp, R.A. G-Protein-Coupled Receptor and Ion Channel Genes Used by Influenza Virus for Replication. J. Virol. 2021, 95. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, M.; Cheng, A.; Yang, Q.; Wu, Y.; Jia, R.; Liu, M.; Zhu, D.; Chen, S.; Zhang, S.; et al. The role of host eIF2α in viral infection. Virol. J. 2020, 17, 112. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Zhang, F.; Liu, X.; Qin, S.; Yang, X.; Kong, D.; Pan, X.; You, H.; Zheng, K.; Tang, R. Calcium signaling in hepatitis B virus infection and its potential as a therapeutic target. Cell Commun. Signal. 2021, 19, 82. [Google Scholar] [CrossRef]
- Fujioka, Y.; Tsuda, M.; Nanbo, A.; Hattori, T.; Sasaki, J.; Sasaki, T.; Miyazaki, T.; Ohba, Y. A Ca2+-dependent signalling circuit regulates influenza A virus internalization and infection. Nat. Commun. 2013, 4, 2763. [Google Scholar] [CrossRef]
- Cheshenko, N.; Del Rosario, B.; Woda, C.; Marcellino, D.; Satlin, L.M.; Herold, B.C. Herpes simplex virus triggers activation of calcium-signaling pathways. J. Cell Biol. 2003, 163, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Sun, Y.; Yang, Z.; Ding, C. Calcium Ions Signaling: Targets for Attack and Utilization by Viruses. Front. Microbiol. 2022, 13, 889374. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, J.R.; Viejo-Borbolla, A.; Martinez-Martín, N.; Blanco, S.; Wandosell, F.; Alcamí, A. Secreted herpes simplex virus-2 glycoprotein G modifies NGF-TrkA signaling to attract free nerve endings to the site of infection. PLoS Pathog. 2015, 11, e1004571. [Google Scholar] [CrossRef] [PubMed]
- Othumpangat, S.; Gibson, L.F.; Samsell, L.; Piedimonte, G. NGF is an essential survival factor for bronchial epithelial cells during respiratory syncytial virus infection. PLoS ONE 2009, 4, e6444. [Google Scholar] [CrossRef]
- Barman, T.K.; Huber, V.C.; Bonin, J.L.; Califano, D.; Salmon, S.L.; McKenzie, A.N.J.; Metzger, D.W. Viral PB1-F2 and host IFN-γ guide ILC2 and T cell activity during influenza virus infection. Proc. Natl. Acad. Sci. USA 2022, 119, e2118535119. [Google Scholar] [CrossRef]
- Rynda-Apple, A.; Harmsen, A.; Erickson, A.S.; Larson, K.; Morton, R.V.; Richert, L.E.; Harmsen, A.G. Regulation of IFN-γ by IL-13 dictates susceptibility to secondary postinfluenza MRSA pneumonia. Eur. J. Immunol. 2014, 44, 3263–3272. [Google Scholar] [CrossRef]
- Simon, P.F.; McCorrister, S.; Hu, P.; Chong, P.; Silaghi, A.; Westmacott, G.; Coombs, K.M.; Kobasa, D. Highly Pathogenic H5N1 and Novel H7N9 Influenza A Viruses Induce More Profound Proteomic Host Responses than Seasonal and Pandemic H1N1 Strains. J. Proteome Res. 2015, 14, 4511–4523. [Google Scholar] [CrossRef]
- Ninomiya, M.; Kondo, Y.; Kimura, O.; Funayama, R.; Nagashima, T.; Kogure, T.; Morosawa, T.; Tanaka, Y.; Nakayama, K.; Shimosegawa, T. The expression of miR-125b-5p is increased in the serum of patients with chronic hepatitis B infection and inhibits the detection of hepatitis B virus surface antigen. J. Viral Hepat. 2016, 23, 330–339. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Zhou, Q.; Pan, J.; Xu, J. Potential Predictive Value of miR-125b-5p, miR-155-5p and Their Target Genes in the Course of COVID-19. Infect. Drug Resist. 2022, 15, 4079–4091. [Google Scholar] [CrossRef]
- Huang, C.W.; Tsai, K.N.; Chen, Y.S.; Chang, R.Y. Differential miRNA Expression Profiling Reveals Correlation of miR125b-5p with Persistent Infection of Japanese Encephalitis Virus. Int. J. Mol. Sci. 2021, 22, 4218. [Google Scholar] [CrossRef]
- Shwetha, S.; Sharma, G.; Raheja, H.; Goel, A.; Aggarwal, R.; Das, S. Interaction of miR-125b-5p with Human antigen R mRNA: Mechanism of controlling HCV replication. Virus Res. 2018, 258, 1–8. [Google Scholar] [CrossRef]
- You, L.; Chen, J.; Liu, W.; Xiang, Q.; Luo, Z.; Wang, W.; Xu, W.; Wu, K.; Zhang, Q.; Liu, Y.; et al. Enterovirus 71 induces neural cell apoptosis and autophagy through promoting ACOX1 downregulation and ROS generation. Virulence 2020, 11, 537–553. [Google Scholar] [CrossRef]
- Hesbacher, S.; Pfitzer, L.; Wiedorfer, K.; Angermeyer, S.; Borst, A.; Haferkamp, S.; Scholz, C.-J.; Wobser, M.; Schrama, D.; Houben, R. RB1 is the crucial target of the Merkel cell polyomavirus Large T antigen in Merkel cell carcinoma cells. Oncotarget 2016, 7, 32956–32968. [Google Scholar] [CrossRef]
- Pascual-Pasto, G.; Bazan-Peregrino, M.; Olaciregui, N.G.; Restrepo-Perdomo, C.A.; Mato-Berciano, A.; Ottaviani, D.; Weber, K.; Correa, G.; Paco, S.; Vila-Ubach, M.; et al. Therapeutic targeting of the RB1 pathway in retinoblastoma with the oncolytic adenovirus VCN-01. Sci. Transl. Med. 2019, 11, eaat9321. [Google Scholar] [CrossRef]
- Li, P.; Jin, Y.; Qi, F.; Wu, F.; Luo, S.; Cheng, Y.; Montgomery, R.R.; Qian, F. SIRT6 Acts as a Negative Regulator in Dengue Virus-Induced Inflammatory Response by Targeting the DNA Binding Domain of NF-κB p65. Front. Cell Infect. Microbiol. 2018, 8, 113. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Cheng, S.-T.; Ren, J.-H.; Ren, F.; Yu, H.-B.; Wang, Q.; Huang, A.-L.; Chen, J. SIRT6 Inhibitor, OSS_128167 Restricts Hepatitis B Virus Transcription and Replication Through Targeting Transcription Factor Peroxisome Proliferator-Activated Receptors α. Front. Pharmacol. 2019, 10, 1270. [Google Scholar] [CrossRef]
- Zhou, C.; Liu, Y.; Wei, Q.; Chen, Y.; Yang, S.; Cheng, A.; Zhang, G. HSPA5 Promotes Attachment and Internalization of Porcine Epidemic Diarrhea Virus through Interaction with the Spike Protein and the Endo-/Lysosomal Pathway. J. Virol. 2023, 97, e0054923. [Google Scholar] [CrossRef]
- Han, B.; Lv, Y.; Moser, D.; Zhou, X.; Woehrle, T.; Han, L.; Osterman, A.; Rudelius, M.; Choukér, A.; Lei, P. ACE2-independent SARS-CoV-2 virus entry through cell surface GRP78 on monocytes—Evidence from a translational clinical and experimental approach. EBioMedicine 2023, 98, 104869. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, E.P.; Eraso, P.; Mazón, M.J.; Santos, V.; Moreno-Bueno, G.; Cano, A.; Portillo, F. LOXL2 drives epithelial-mesenchymal transition via activation of IRE1-XBP1 signalling pathway. Sci. Rep. 2017, 7, 44988. [Google Scholar] [CrossRef]
- Dana, R.C.; Welch, W.J.; Deftos, L.J. Heat shock proteins bind calcitonin. Endocrinology 1990, 126, 672–674. [Google Scholar] [CrossRef] [PubMed]
- Evensen, N.A.; Kuscu, C.; Nguyen, H.-L.; Zarrabi, K.; Dufour, A.; Kadam, P.; Hu, Y.-J.; Pulkoski-Gross, A.; Bahou, W.F.; Zucker, S.; et al. Unraveling the role of KIAA1199, a novel endoplasmic reticulum protein, in cancer cell migration. J. Natl. Cancer Inst. 2013, 105, 1402–1416. [Google Scholar] [CrossRef]
- Oka, O.B.; Pringle, M.A.; Schopp, I.M.; Braakman, I.; Bulleid, N.J. ERdj5 is the ER reductase that catalyzes the removal of non-native disulfides and correct folding of the LDL receptor. Mol. Cell 2013, 50, 793–804. [Google Scholar] [CrossRef]
- Ng, D.T.; Watowich, S.S.; Lamb, R.A. Analysis in vivo of GRP78-BiP/substrate interactions and their role in induction of the GRP78-BiP gene. Mol. Biol. Cell 1992, 3, 143–155. [Google Scholar] [CrossRef]
- Oikawa, D.; Kimata, Y.; Kohno, K.; Iwawaki, T. Activation of mammalian IRE1alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp. Cell Res. 2009, 315, 2496–2504. [Google Scholar] [CrossRef]
- Macario, A.J.; Conway de Macario, E. Molecular chaperones: Multiple functions, pathologies, and potential applications. Front. Biosci. 2007, 12, 2588–2600. [Google Scholar] [CrossRef]
- Weisburg, J.H.; Weissman, D.B.; Sedaghat, T.; Babich, H. In vitro cytotoxicity of epigallocatechin gallate and tea extracts to cancerous and normal cells from the human oral cavity. Basic Clin. Pharmacol. Toxicol. 2004, 95, 191–200. [Google Scholar] [CrossRef]
- Isbrucker, R.A.; Bausch, J.; Edwards, J.A.; Wolz, E. Safety studies on epigallocatechin gallate (EGCG) preparations. Part 1: Genotoxicity. Food Chem. Toxicol. 2006, 44, 626–635. [Google Scholar] [CrossRef]
- Herold, S.; Becker, C.; Ridge, K.M.; Budinger, G.R. Influenza virus-induced lung injury: Pathogenesis and implications for treatment. Eur. Respir. J. 2015, 45, 1463–1478. [Google Scholar] [CrossRef] [PubMed]
- Coombs, K.M.; Berard, A.; Xu, W.; Krokhin, O.; Meng, X.; Cortens, J.P.; Kobasa, D.; Wilkins, J.; Brown, E.G. Quantitative proteomic analyses of influenza virus-infected cultured human lung cells. J. Virol. 2010, 84, 10888–10906. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.U.; Coombs, K.M. Serum-reduced media impacts on cell viability and protein expression in human lung epithelial cells. J. Cell Physiol. 2019, 234, 7718–7724. [Google Scholar] [CrossRef] [PubMed]
- Candia, J.; Cheung, F.; Kotliarov, Y.; Fantoni, G.; Sellers, B.; Griesman, T.; Huang, J.; Stuccio, S.; Zingone, A.; Ryan, B.M.; et al. Assessment of Variability in the SOMAscan Assay. Sci. Rep. 2017, 7, 14248. [Google Scholar] [CrossRef] [PubMed]
- Brody, E.N.; Gold, L.; Lawn, R.M.; Walker, J.J.; Zichi, D. High-content affinity-based proteomics: Unlocking protein biomarker discovery. Expert Rev. Mol. Diagn. 2010, 10, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.-U.; Zahedi-Amiri, A.; Glover, K.K.M.; Gao, A.; Nickol, M.N.; Kindrachuk, J.; Wilkins, J.A.; Coombs, K.M. Zika virus dysregulates human Sertoli cell proteins involved in spermatogenesis with little effect on tight junctions. PLoS Negl. Trop. Dis. 2020, 14, e0008335. [Google Scholar] [CrossRef] [PubMed]
- Rahim, N.; Selman, M.; Sauder, P.J.; Forbes, N.E.; Stecho, W.; Xu, W.; Lebar, M.; Brown, E.G.; Coombs, K.M. Generation and characterization of a new panel of broadly reactive anti-NS1 mAbs for detection of influenza A virus. J. Gen. Virol. 2013, 94 Pt 3, 593–605. [Google Scholar] [CrossRef]
- Rashid, M.U.; Coombs, K.M. Chloride Intracellular Channel Protein 1 (CLIC1) Is a Critical Host Cellular Factor for Influenza A Virus Replication. Viruses 2024, 16, 129. [Google Scholar] [CrossRef]
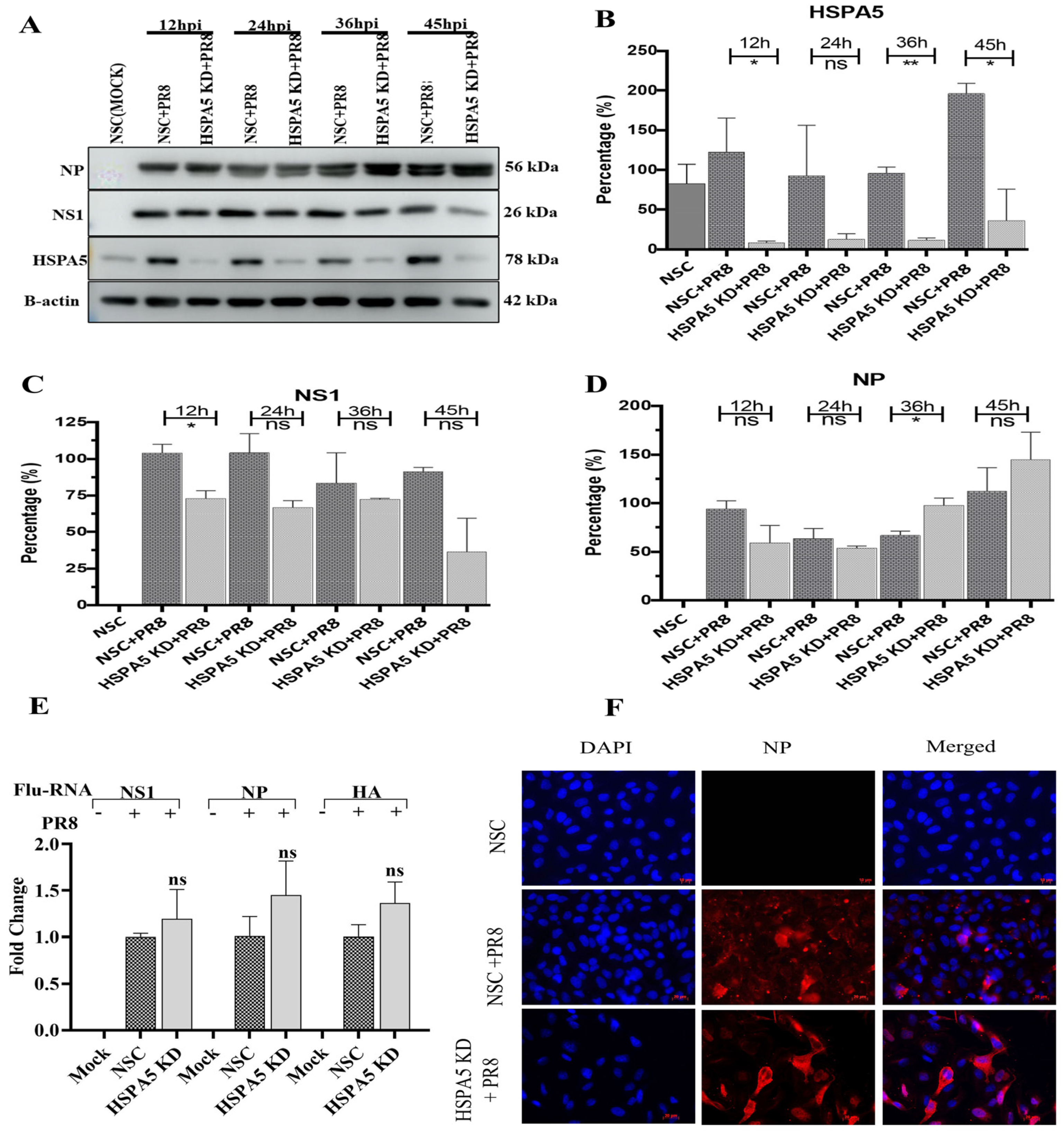
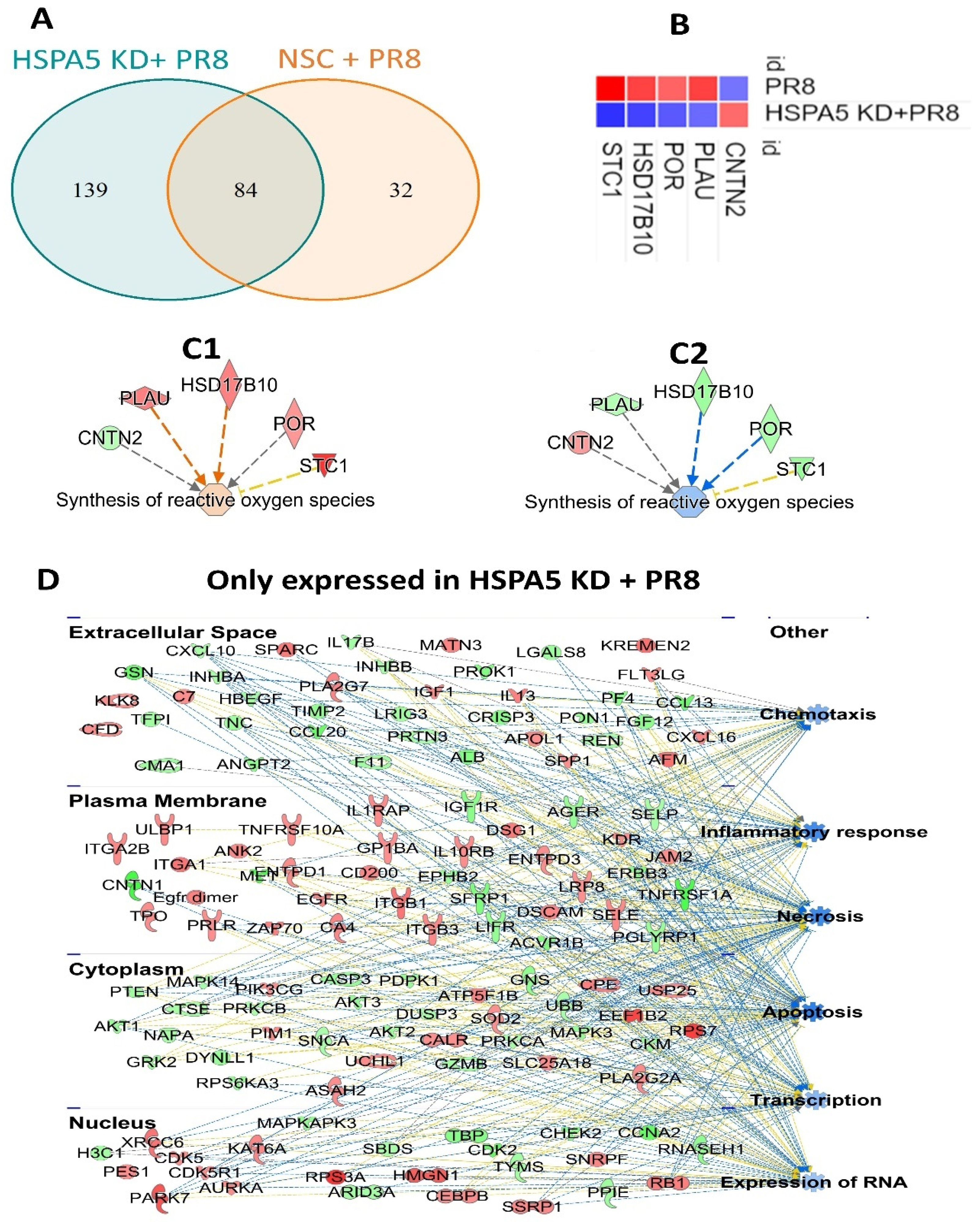
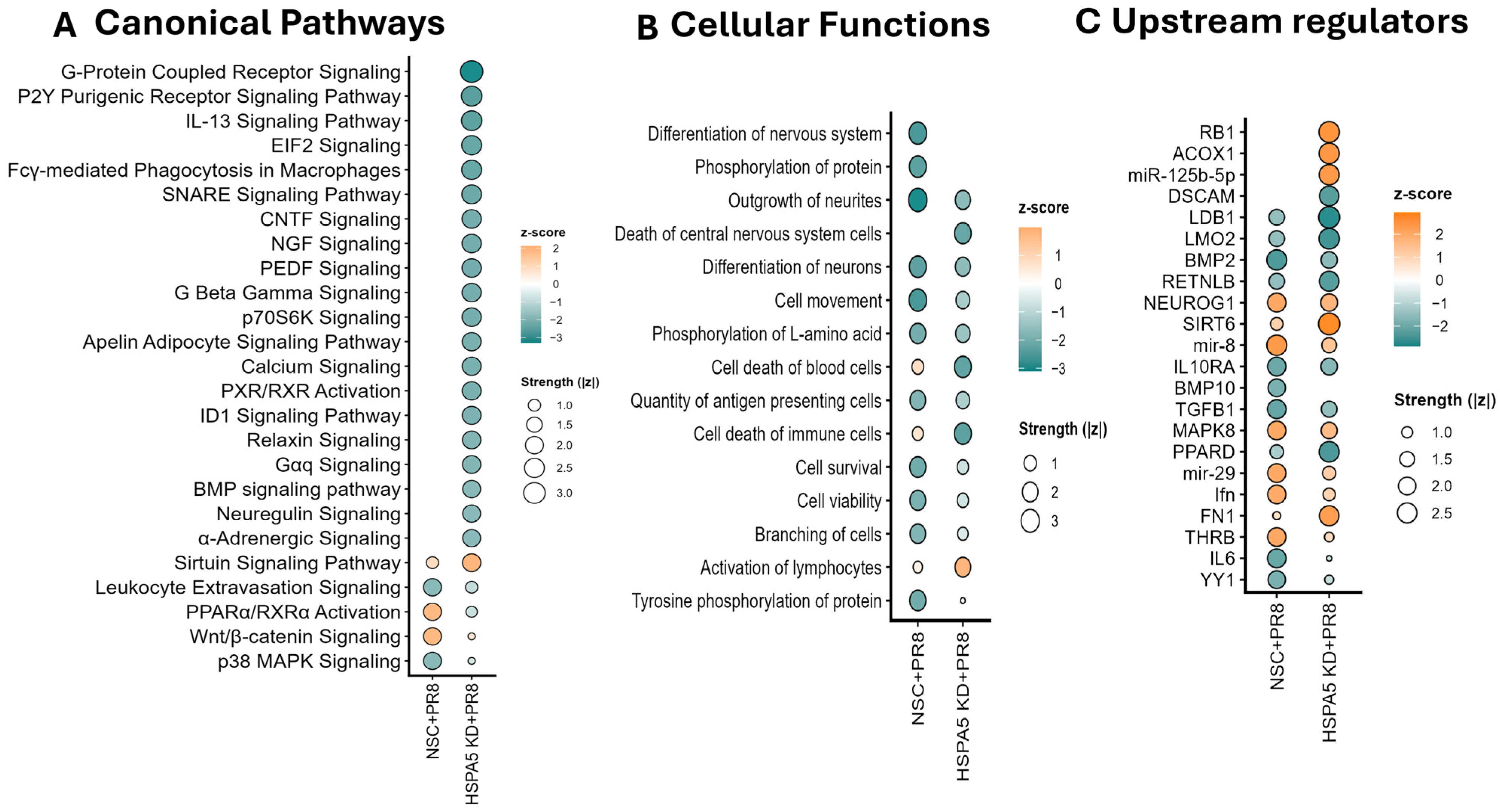
| Range of Fold Change | PR8 | Total Significant | HSPA5 KD + PR8 | Total Significant |
|---|---|---|---|---|
| (Protein No.) | (Protein No.) | (Protein No.) | (Protein No.) | |
| and F.C. > 1.00 | 76 | 218 | 277 | 503 |
| and F.C. < 1.00 | 142 | 226 | ||
| and F.C. > 1.10 | 68 | 197 | 260 | 478 |
| and F.C. < −1.10 | 129 | 218 | ||
| and F.C. > 1.20 | 43 | 144 | 170 | 347 |
| and F.C. < −1.20 | 101 | 177 | ||
| and F.C. > 1.30 | 31 | 116 | 78 | 223 |
| and F.C. < −1.30 | 85 | 145 | ||
| and F.C. > 1.50 | 20 | 68 | 23 | 112 |
| and F.C. < −1.50 | 48 | 89 | ||
| and F.C. > 1.60 | 17 | 57 | 20 | 92 |
| and F.C. < −1.60 | 40 | 72 | ||
| and F.C. > 2.00 | 11 | 33 | 12 | 40 |
| and F.C. < −2.00 | 22 | 28 | ||
| and F.C. > 2.50 | 8 | 22 | 6 | 19 |
| and F.C. < −2.50 | 14 | 13 |
| Type(s) | Symbols | Entrez Gene Name | NSC + PR8 (FC) | p-Value | HSPA5 KD + PR8 (FC) | p-Value |
|---|---|---|---|---|---|---|
| Cytokine | CXCL8 | C-X-C motif chemokine ligand 8 | 10.267 | 2.21 × 10−5 | 4.228 | 2.08 × 10−4 |
| CCL13 | C-C motif chemokine ligand 13 | −1.419 | 9.43 × 10−2 | −2.282 | 2.29 × 10−3 | |
| PF4 | platelet factor 4 | −1.261 | 1.02 × 10−1 | −1.821 | 2.70 × 10−3 | |
| IFNL1 | interferon lambda 1 | 2.868 | 3.46 × 10−2 | 2.007 | 3.08 × 10−3 | |
| CCL5 | C-C motif chemokine ligand 5 | 6.386 | 3.59 × 10−3 | 5.011 | 3.87 × 10−3 | |
| CCL20 | C-C motif chemokine ligand 20 | −1.275 | 2.61 × 10−1 | −2.346 | 1.22 × 10−2 | |
| Enzyme | PARK7 | Parkinsonism associated deglycase | 1.102 | 4.97 × 10−1 | 1.993 | 1.34 × 10−3 |
| TOP1 | DNA topoisomerase I | 1.5 | 1.82 × 10−3 | 3.771 | 1.39 × 10−3 | |
| HAT1 | histone acetyltransferase 1 | −2.37 | 2.10 × 10−4 | −2.612 | 1.67 × 10−3 | |
| CNTN1 | contactin 1 | −2.928 | 1.58 × 10−2 | −2.908 | 1.70 × 10−3 | |
| XRCC6 | X-ray repair cross complementing 6 | 1.248 | 1.29 × 10−1 | 1.636 | 3.55 × 10−3 | |
| EFEMP1 | EGF containing fibulin extracellular matrix protein 1 | −1.495 | 1.75 × 10−2 | −1.619 | 8.03 × 10−3 | |
| GNS | glucosamine (N-acetyl)-6-sulfatase | −1.223 | 7.69 × 10−2 | −1.723 | 1.99 × 10−2 | |
| ENTPD5 | ectonucleoside triphosphate diphosphohydrolase 5 | −1.49 | 4.88 × 10−3 | −1.505 | 2.01 × 10−2 | |
| AKR1A1 | aldo-keto reductase family 1 member A1 | −1.717 | 3.82 × 10−2 | −1.619 | 2.03 × 10−2 | |
| RNASEH1 | ribonuclease H1 | −1.129 | 5.80 × 10−1 | −1.711 | 4.67 × 10−2 | |
| PPIF | peptidylprolyl isomerase F | 1.729 | 2.93 × 10−2 | 1.068 | 1.99 × 10−1 | |
| PPID | peptidylprolyl isomerase D | −1.532 | 1.02 × 10−2 | −1.532 | 2.33 × 10−1 | |
| Growth factor | PROK1 | prokineticin 1 | −1.288 | 1.64 × 10−1 | −1.952 | 1.21 × 10−3 |
| FGF6 | fibroblast growth factor 6 | −1.63 | 5.64 × 10−3 | −1.619 | 1.50 × 10−3 | |
| DKK1 | dickkopf WNT signaling pathway inhibitor 1 | −3.605 | 5.46 × 10−3 | −3.891 | 6.04 × 10−3 | |
| BMP6 | bone morphogenetic protein 6 | −1.564 | 3.23 × 10−2 | −1.575 | 7.89 × 10−3 | |
| GRN | granulin precursor | −2.085 | 4.60 × 10−3 | −1.765 | 1.05 × 10−2 | |
| NRG1 | neuregulin 1 | −1.613 | 4.73 × 10−2 | −1.49 | 2.68 × 10−2 | |
| ANGPT2 | angiopoietin 2 | −1.235 | 2.46 × 10−1 | −1.784 | 4.88 × 10−2 | |
| Kinase | PRKCG | protein kinase C gamma | −1.608 | 1.95 × 10−2 | −1.664 | 6.01 × 10−4 |
| CKM | creatine kinase, M-type | −1.169 | 1.10 × 10−1 | −1.521 | 8.87 × 10−4 | |
| MET | MET proto-oncogene, receptor tyrosine kinase | −1.5 | 9.39 × 10−2 | −2.848 | 9.85 × 10−4 | |
| CDK2 | cyclin dependent kinase 2 | −1.257 | 2.15 × 10−1 | −2.395 | 1.54 × 10−3 | |
| EPHB2 | EPH receptor B2 | −1.079 | 2.10 × 10−1 | −1.619 | 2.32 × 10−3 | |
| FGFR1 | fibroblast growth factor receptor 1 | −2.071 | 3.29 × 10−3 | −2.488 | 3.24 × 10−3 | |
| EFNA2 | ephrin A2 | −1.659 | 4.84 × 10−3 | −1.306 | 4.19 × 10−3 | |
| INSR | insulin receptor | −1.429 | 2.23 × 10−2 | −1.759 | 5.56 × 10−3 | |
| ACVR1B | activin A receptor type 1B | −1.324 | 1.73 × 10−1 | −2.056 | 7.77 × 10−3 | |
| MAPK3 | mitogen-activated protein kinase 3 | −1.38 | 7.49 × 10−2 | −1.619 | 8.63 × 10−3 | |
| CHEK2 | checkpoint kinase 2 | −1.352 | 5.81 × 10−2 | −1.873 | 1.27 × 10−2 | |
| STC1 | stanniocalcin 1 | 2.403 | 2.88 × 10−3 | −1.619 | 1.40 × 10−2 | |
| GRK2 | G protein-coupled receptor kinase 2 | −1.495 | 7.34 × 10−2 | −1.505 | 1.41 × 10−2 | |
| RPS6KA3 | ribosomal protein S6 kinase A3 | −1.532 | 6.31 × 10−2 | −1.688 | 1.66 × 10−2 | |
| MAPKAPK3 | MAPK activated protein kinase 3 | −1.67 | 6.14 × 10−2 | −1.711 | 1.72 × 10−2 | |
| AKT1 | AKT serine/threonine kinase 1 | −1.366 | 1.55 × 10−1 | −1.63 | 1.81 × 10−2 | |
| AKT3 | AKT serine/threonine kinase 3 | −1.366 | 1.55 × 10−1 | −1.63 | 1.81 × 10−2 | |
| CAMK2D | calcium/calmodulin dependent protein kinase II delta | −1.5 | 4.20 × 10−2 | −1.597 | 2.54 × 10−2 | |
| AKT2 | AKT serine/threonine kinase 2 | −1.495 | 5.46 × 10−2 | −1.516 | 2.63 × 10−2 | |
| EPHA2 | EPH receptor A2 | 3 | 4.71 × 10−3 | 1.39 | 4.25 × 10−2 | |
| PIK3CA | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha | −1.625 | 4.25 × 10−2 | −1.803 | 4.38 × 10−2 | |
| PIK3R1 | phosphoinositide-3-kinase regulatory subunit 1 | −1.625 | 4.25 × 10−2 | −1.803 | 4.38 × 10−2 | |
| ERBB3 | erb-b2 receptor tyrosine kinase 3 | −1.485 | 8.60 × 10−2 | −1.873 | 4.39 × 10−2 | |
| PDPK1 | 3-phosphoinositide dependent protein kinase 1 | −1.537 | 6.06 × 10−2 | −1.625 | 4.61 × 10−2 | |
| CAMK2B | calcium/calmodulin dependent protein kinase II beta | −1.553 | 5.96 × 10−3 | −1.537 | 4.87 × 10−2 | |
| EPHA3 | EPH receptor A3 | 1.602 | 2.03 × 10−2 | 1.046 | 2.68 × 10−1 | |
| Peptidase | PCSK9 | proprotein convertase subtilisin/kexin type 9 | −5.205 | 7.16 × 10−3 | −6.084 | 1.24 × 10−3 |
| CMA1 | chymase 1 | −1.177 | 1.42 × 10−1 | −1.532 | 1.91 × 10−3 | |
| CTSA | cathepsin A | −2.107 | 4.83 × 10−3 | −2.354 | 2.04 × 10−3 | |
| CPE | carboxypeptidase E | 1.169 | 3.73 × 10−1 | 1.516 | 1.59 × 10−2 | |
| CASP3 | caspase 3 | −1.141 | 5.07 × 10−1 | −1.63 | 2.33 × 10−2 | |
| Phosphatase | PTPN6 | protein tyrosine phosphatase non-receptor type 6 | −1.537 | 1.17 × 10−3 | −1.495 | 1.48 × 10−3 |
| PON1 | paraoxonase 1 | −1.181 | 1.72 × 10−1 | −1.664 | 1.19 × 10−2 | |
| Transcription regulator | TBP | TATA-box binding protein | −1.537 | 5.77 × 10−2 | −1.853 | 1.57 × 10−3 |
| NACA | nascent polypeptide associated complex subunit alpha | 2.107 | 1.80 × 10−2 | 1.625 | 1.73 × 10−3 | |
| STAT6 | signal transducer and activator of transcription 6 | −1.664 | 2.05 × 10−2 | −1.58 | 8.39 × 10−3 | |
| HMGN1 | high mobility group nucleosome binding domain 1 | −1.068 | 6.46 × 10−1 | 1.892 | 9.84 × 10−3 | |
| STAT3 | signal transducer and activator of transcription 3 | −2.092 | 7.86 × 10−3 | −1.485 | 3.42 × 10−2 | |
| SMAD2 | SMAD family member 2 | −1.619 | 3.48 × 10−2 | −1.5 | 5.83 × 10−2 | |
| HMGB1 | high mobility group box 1 | −1.505 | 1.38 × 10−2 | −1.202 | 8.93 × 10−2 | |
| AIP | aryl hydrocarbon receptor interacting protein | −1.553 | 1.13 × 10−2 | −1.516 | 2.00 × 10−1 | |
| EEF1B2 | eukaryotic translation elongation factor 1 beta 2 | 1.141 | 4.88 × 10−1 | 2.189 | 1.28 × 10−2 | |
| EIF4EBP2 | eukaryotic translation initiation factor 4E binding protein 2 | −1.439 | 3.52 × 10−2 | −1.608 | 3.46 × 10−2 | |
| Transmembrane receptor | MICB | MHC class I polypeptide-related sequence B | −1.735 | 3.03 × 10−2 | −2.428 | 2.75 × 10−4 |
| RTN4R | reticulon 4 receptor | −2.979 | 1.40 × 10−2 | −2.77 | 4.62 × 10−4 | |
| TNFRSF1A | TNF receptor superfamily member 1A | −3.918 | 5.43 × 10−3 | −2.742 | 9.55 × 10−4 | |
| PLXNB2 | plexin B2 | −1.329 | 3.82 × 10−2 | −1.729 | 1.28 × 10−3 | |
| GFRA1 | GDNF family receptor alpha 1 | −1.945 | 1.53 × 10−2 | −2.014 | 2.20 × 10−3 | |
| RELT | RELT TNF receptor | −1.49 | 2.12 × 10−2 | −1.602 | 2.69 × 10−3 | |
| TNFRSF10D | TNF receptor superfamily member 10d | 3.63 | 1.02 × 10−2 | 3 | 2.86 × 10−3 | |
| LIFR | LIF receptor subunit alpha | 1.149 | 3.27 × 10−1 | −1.676 | 2.92 × 10−3 | |
| NRP1 | neuropilin 1 | −2.151 | 1.80 × 10−2 | −2.969 | 3.49 × 10−3 | |
| TNFRSF21 | TNF receptor superfamily member 21 | −2.523 | 8.55 × 10−4 | −2.471 | 3.58 × 10−3 | |
| KIR2DL4 | killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 4 | −1.699 | 1.01 × 10−2 | −1.809 | 4.25 × 10−3 | |
| SFRP1 | secreted frizzled related protein 1 | −1.21 | 6.59 × 10−2 | −2 | 5.01 × 10−3 | |
| IL6ST | interleukin 6 cytokine family signal transducer | −1.206 | 5.44 × 10−3 | −1.532 | 6.61 × 10−3 | |
| B2M | beta-2-microglobulin | 2.488 | 1.30 × 10−4 | 2.166 | 8.44 × 10−3 | |
| PLAUR | plasminogen activator, urokinase receptor | 1.866 | 8.83 × 10−3 | 1.454 | 9.80 × 10−3 | |
| AGER | advanced glycosylation end-product specific receptor | −1.181 | 2.49 × 10−1 | −1.602 | 1.82 × 10−2 | |
| IGF1R | insulin like growth factor 1 receptor | −1.49 | 1.73 × 10−1 | −1.809 | 2.30 × 10−2 | |
| PGLYRP1 | peptidoglycan recognition protein 1 | −1.469 | 5.55 × 10−2 | −1.575 | 2.39 × 10−2 | |
| ITGB1 | integrin subunit beta 1 | 1.125 | 6.18 × 10−1 | 1.636 | 3.43 × 10−2 | |
| transporter | ATP5PO | ATP synthase peripheral stalk subunit OSCP | 1.591 | 2.67 × 10−2 | 2.403 | 3.19 × 10−3 |
| MCL1 | MCL1 apoptosis regulator, BCL2 family member | −1.395 | 4.72 × 10−2 | −1.608 | 3.45 × 10−3 | |
| BPI | bactericidal permeability increasing protein | −1.608 | 4.50 × 10−3 | −1.711 | 3.68 × 10−3 | |
| ALB | albumin | −1.338 | 1.27 × 10−1 | −1.886 | 3.71 × 10−3 | |
| AFM | afamin | 1.189 | 1.64 × 10−1 | 1.67 | 1.09 × 10−2 | |
| SNX4 | sorting nexin 4 | −1.613 | 2.04 × 10−2 | −1.879 | 1.06 × 10−1 | |
| Other | SPARC | secreted protein acidic and cysteine rich | 1.202 | 5.39 × 10−1 | 1.558 | 2.44 × 10−4 |
| MICA | MHC class I polypeptide-related sequence A | −1.772 | 4.92 × 10−2 | −2.196 | 4.85 × 10−4 | |
| AMIGO2 | adhesion molecule with Ig like domain 2 | −2.621 | 1.93 × 10−2 | −1.945 | 5.42 × 10−4 | |
| GREM1 | gremlin 1, DAN family BMP antagonist | −1.537 | 1.05 × 10−2 | −1.5 | 1.24 × 10−3 | |
| CCNA2 | cyclin A2 | −1.257 | 2.15 × 10−1 | −2.395 | 1.54 × 10−3 | |
| DYNLL1 | dynein light chain LC8-type 1 | −1.157 | 1.61 × 10−1 | −1.58 | 1.95 × 10−3 | |
| RSPO2 | R-spondin 2 | −1.653 | 1.75 × 10−2 | −1.735 | 2.09 × 10−3 | |
| ISG15 | ISG15 ubiquitin like modifier | 7.21 | 4.52 × 10−3 | 8.168 | 2.49 × 10−3 | |
| FGF12 | fibroblast growth factor 12 | −1.197 | 1.23 × 10−1 | −1.765 | 3.70 × 10−3 | |
| CRISP3 | cysteine rich secretory protein 3 | −1.185 | 2.18 × 10−1 | −1.84 | 3.72 × 10−3 | |
| LAMA1 | laminin subunit alpha 1 | −4.377 | 1.17 × 10−3 | −5.098 | 4.98 × 10−3 | |
| LAMB1 | laminin subunit beta 1 | −4.377 | 1.17 × 10−3 | −5.098 | 4.98 × 10−3 | |
| LAMC1 | laminin subunit gamma 1 | −4.377 | 1.17 × 10−3 | −5.098 | 4.98 × 10−3 | |
| TIMP2 | TIMP metallopeptidase inhibitor 2 | −1.032 | 6.93 × 10−1 | −1.664 | 5.19 × 10−3 | |
| DKK4 | dickkopf WNT signaling pathway inhibitor 4 | −3.618 | 8.22 × 10−3 | −3.482 | 5.93 × 10−3 | |
| MFGE8 | milk fat globule EGF and factor V/VIII domain containing | −2.014 | 4.16 × 10−2 | −1.959 | 6.42 × 10−3 | |
| KREMEN2 | kringle containing transmembrane protein 2 | 1.165 | 3.25 × 10−1 | 1.659 | 6.87 × 10−3 | |
| H2AZ1 | H2A.Z variant histone 1 | −2.078 | 1.21 × 10−2 | −1.796 | 7.65 × 10−3 | |
| SERPINE2 | serpin family E member 2 | −2.648 | 1.65 × 10−4 | −2.258 | 7.96 × 10−3 | |
| GSN | gelsolin | −1.613 | 1.79 × 10−1 | −2.37 | 8.05 × 10−3 | |
| CFH | complement factor H | −1.84 | 5.70 × 10−3 | −1.564 | 8.60 × 10−3 | |
| TNC | tenascin C | −1.161 | 1.23 × 10−1 | −1.521 | 1.06 × 10−2 | |
| UNC5D | unc-5 netrin receptor D | −4.317 | 9.53 × 10−4 | −3.434 | 1.12 × 10−2 | |
| RPS7 | ribosomal protein S7 | 1.248 | 4.44 × 10−1 | 2.242 | 1.39 × 10−2 | |
| APP | amyloid beta precursor protein | −1.879 | 1.78 × 10−2 | −2.242 | 1.58 × 10−2 | |
| LGALS8 | galectin 8 | −1.569 | 6.70 × 10−2 | −1.532 | 1.78 × 10−2 | |
| DSG1 | desmoglein 1 | 1.094 | 5.43 × 10−1 | 1.532 | 1.86 × 10−2 | |
| RPS3A | ribosomal protein S3A | 1.31 | 3.53 × 10−1 | 2.403 | 2.16 × 10−2 | |
| H1-2 | H1.2 linker histone, cluster member | 2.77 | 7.98 × 10−3 | 4.925 | 2.18 × 10−2 | |
| SLITRK5 | SLIT and NTRK like family member 5 | −1.558 | 1.40 × 10−2 | −1.591 | 2.67 × 10−2 | |
| ITGA1 | integrin subunit alpha 1 | 1.125 | 6.18 × 10−1 | 1.636 | 3.43 × 10−2 | |
| SERPINE1 | serpin family E member 1 | 5.979 | 3.82 × 10−3 | 1.366 | 3.47 × 10−2 | |
| KIF23 | kinesin family member 23 | −2.63 | 8.74 × 10−3 | −2.181 | 3.77 × 10−2 | |
| MEPE | matrix extracellular phosphoglycoprotein | −1.454 | 4.26 × 10−2 | −1.548 | 4.62 × 10−2 | |
| IGFBP2 | insulin like growth factor binding protein 2 | 1.809 | 2.12 × 10−2 | 1.206 | 5.13 × 10−2 | |
| TGFBI | transforming growth factor beta induced | 1.505 | 1.41 × 10−2 | −1.113 | 6.83 × 10−2 | |
| CD55 | CD55 molecule (Cromer blood group) | 1.591 | 5.56 × 10−4 | 1.206 | 7.88 × 10−2 | |
| IGFBP6 | insulin like growth factor binding protein 6 | 1.959 | 5.31 × 10−4 | 1.169 | 8.05 × 10−2 | |
| CST3 | cystatin C | 1.972 | 3.32 × 10−3 | −1.181 | 1.98 × 10−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashid, M.-u.; Yasmin, T.; Coombs, K.M. HSPA5, a Host Cellular Heat-Shock Protein Required for Influenza a Virus Replication. Int. J. Mol. Sci. 2025, 26, 10998. https://doi.org/10.3390/ijms262210998
Rashid M-u, Yasmin T, Coombs KM. HSPA5, a Host Cellular Heat-Shock Protein Required for Influenza a Virus Replication. International Journal of Molecular Sciences. 2025; 26(22):10998. https://doi.org/10.3390/ijms262210998
Chicago/Turabian StyleRashid, Mahamud-ur, Tamanna Yasmin, and Kevin M. Coombs. 2025. "HSPA5, a Host Cellular Heat-Shock Protein Required for Influenza a Virus Replication" International Journal of Molecular Sciences 26, no. 22: 10998. https://doi.org/10.3390/ijms262210998
APA StyleRashid, M.-u., Yasmin, T., & Coombs, K. M. (2025). HSPA5, a Host Cellular Heat-Shock Protein Required for Influenza a Virus Replication. International Journal of Molecular Sciences, 26(22), 10998. https://doi.org/10.3390/ijms262210998






