Coordinated Biosynthesis of Essential Cell Envelope Components: Lipopolysaccharide and Fatty Acids Requires LapD, Acyl Carrier Protein, and Fully Hexaacylated Lipid A
Abstract
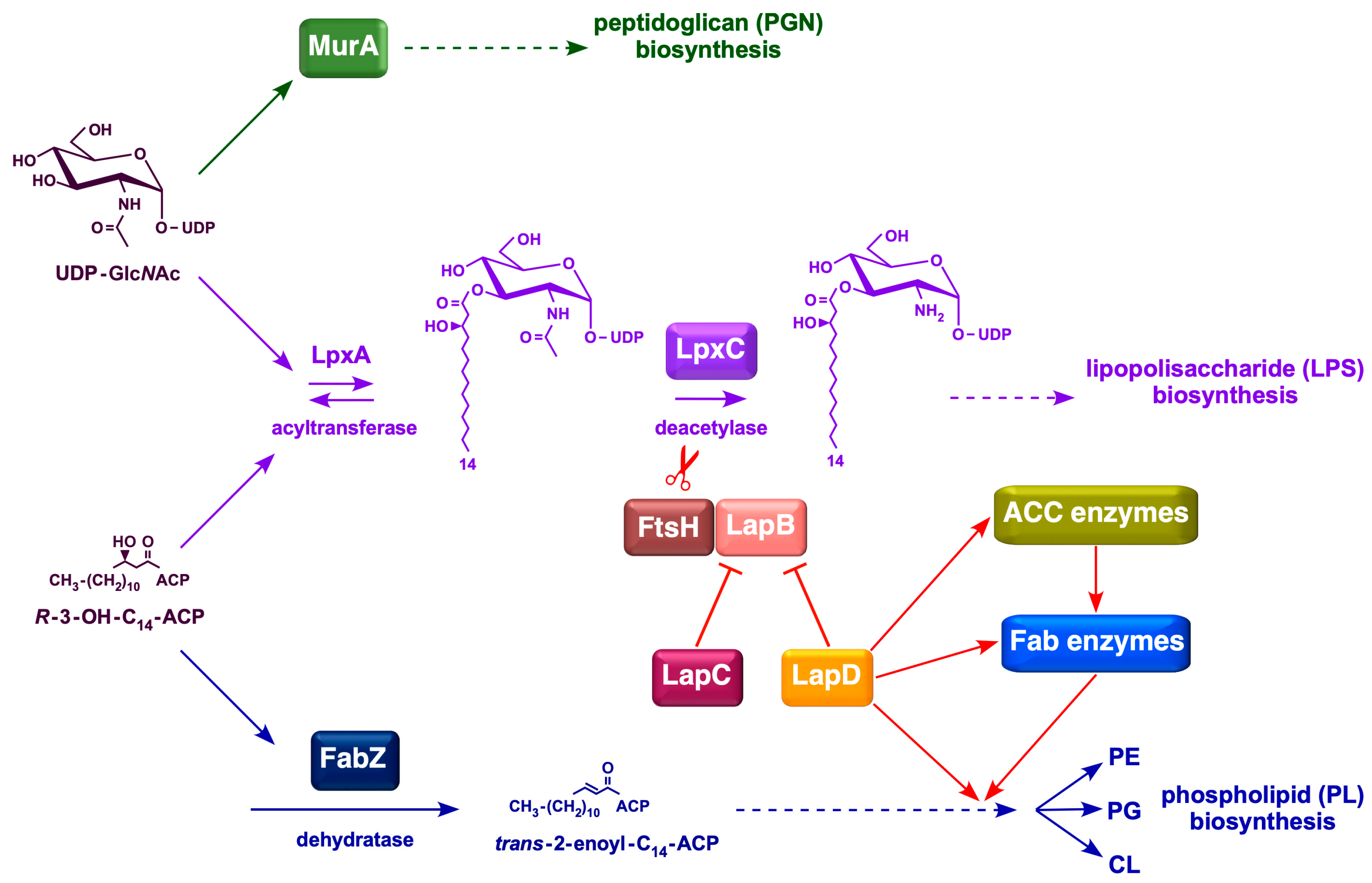
1. Introduction
2. Results
2.1. Suppressor-Free Δ(lapD lpxM) Bacteria Are Viable in Minimal Medium Under Slow Growth Conditions but Require Extragenic Suppressors for the Growth in Rich Medium
2.2. Suppressor Mutations Mapping to Genes Encoding Different Subunits of Acetyl Coenzyme A Carboxylase Can Bypass the Lethal Phenotype of Δ(lapD lpxM) Bacteria
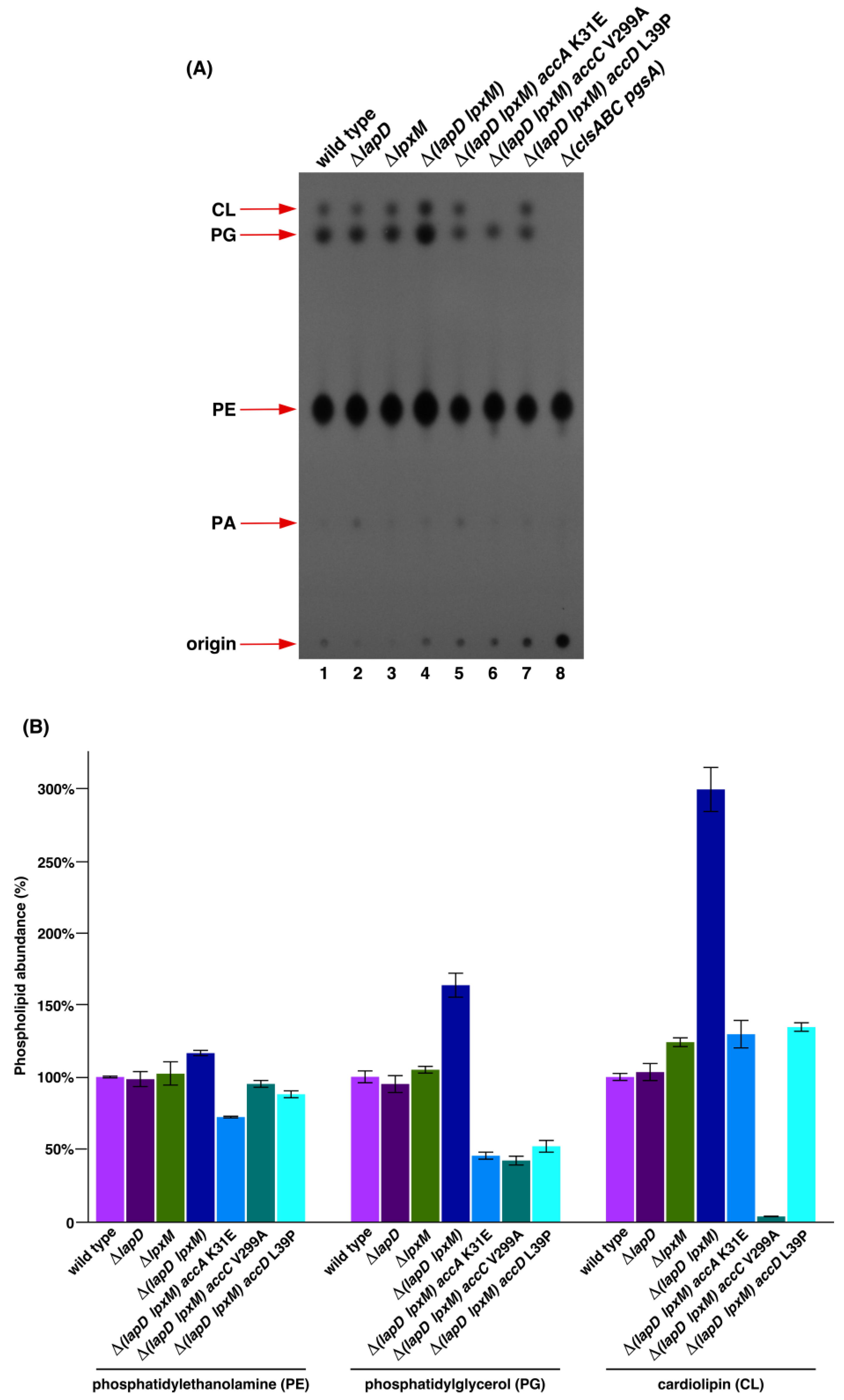
2.3. Suppressor Mutations in Acetyl-CoA Carboxylase Complex That Bypass the Lethal Phenotype of Δ(lapD lpxM) Bacteria Accumulate Reduced Amounts of Phospholipid Species
2.4. Suppressor Mutations in Acetyl-CoA Carboxylase Complex That Bypass the Lethal Phenotype of Δ(lapD lpxM) Bacteria Are Recessive
2.5. Suppressor Mutations in Acetyl-CoA Carboxylase Complex That Overcome the Lethal Phenotype of Δ(lapD lpxM) Bacteria Do Not Alter LpxC Levels
2.6. Lethality of Δ(lapD lpxM) Bacteria Due to Gross Changes in the Amounts of Free Fatty Acids That Can Be Bypassed by Mutations in the ACC Complex
2.7. Inhibition of Fatty Acid Biosynthesis by Exogenous Addition of Either Triclosan or Cerulenin Overcomes Lethality of Δ(lapD lpxM) Bacteria
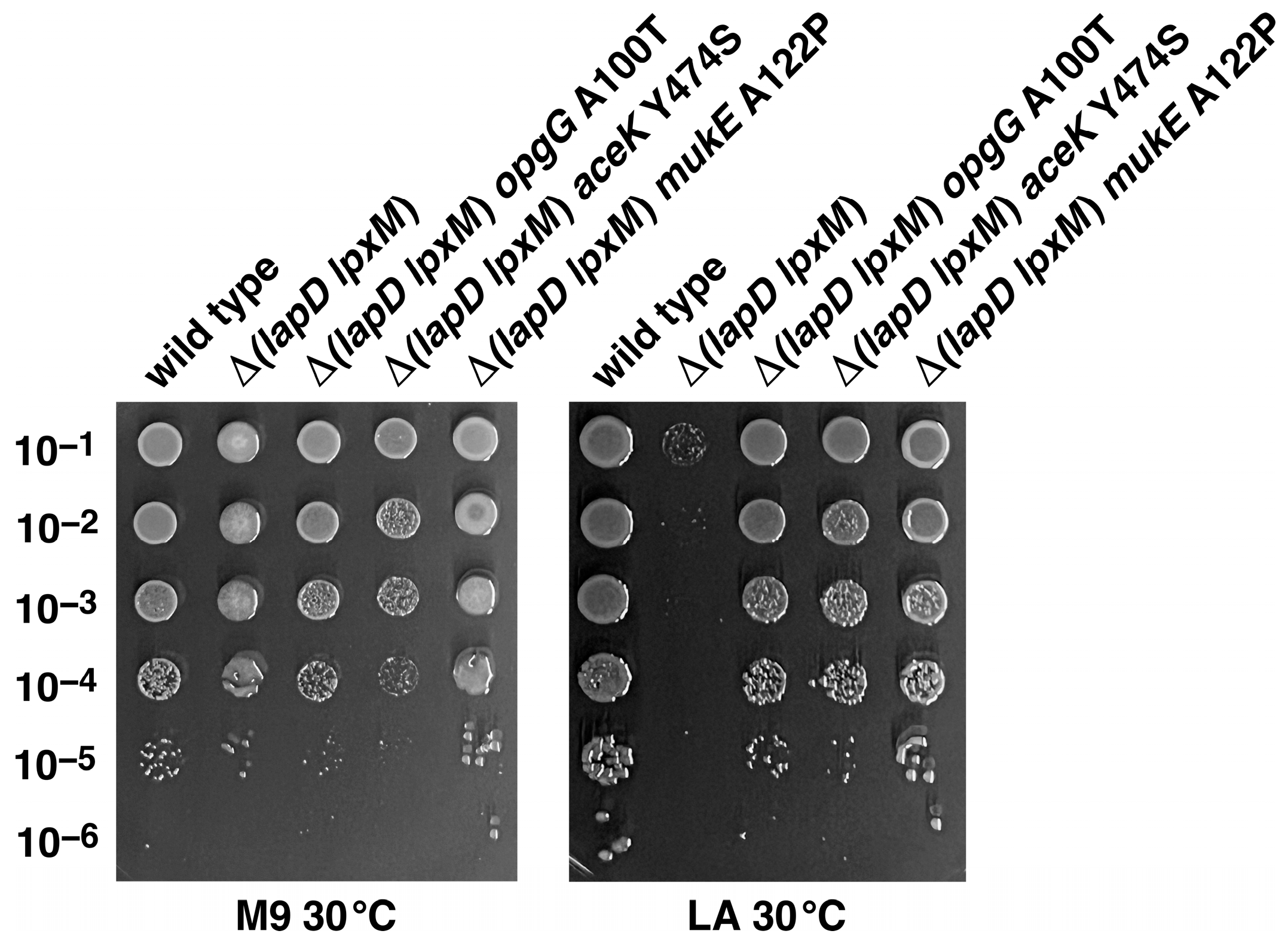
2.8. Suppressor Mutations Mapping to opgG, aceK and mukE Genes Can Bypass the Lethal Phenotype of Δ(lapD lpxM) Bacteria at 37 °C
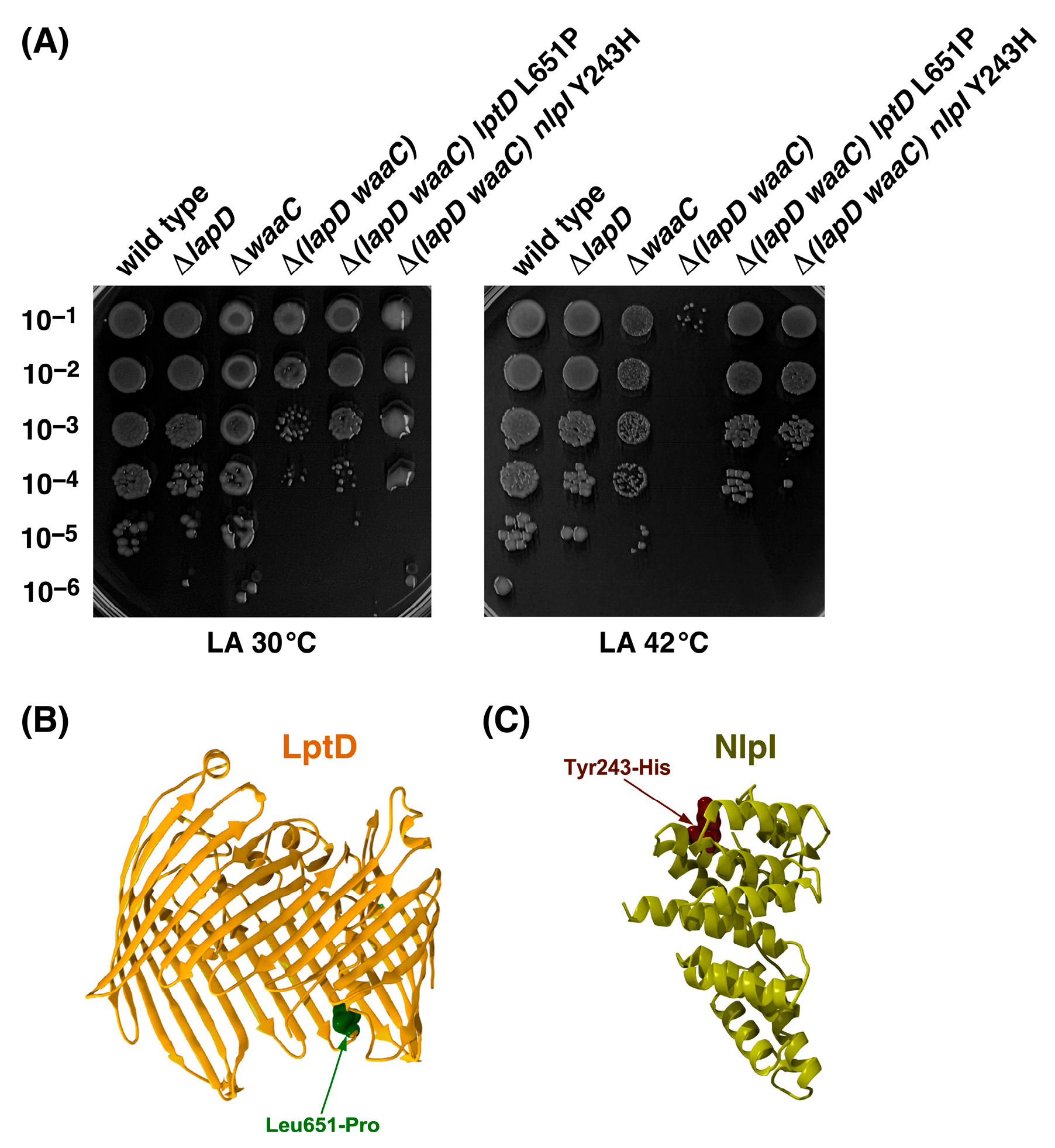
2.9. Suppressor Mutations Mapping to lptD and nlpI Genes Can Bypass the Synthetic Lethal Phenotype of Δ(lapD waaC) Bacteria at 42 °C
2.10. Overexpression of Either the acpP Gene or the accB Gene Overcome Cell Morphology Defects of ΔlapD Bacteria
2.11. Overexpression of Either acpP or menI Overcomes Vancomycin Sensitivity of ΔlapD Bacteria
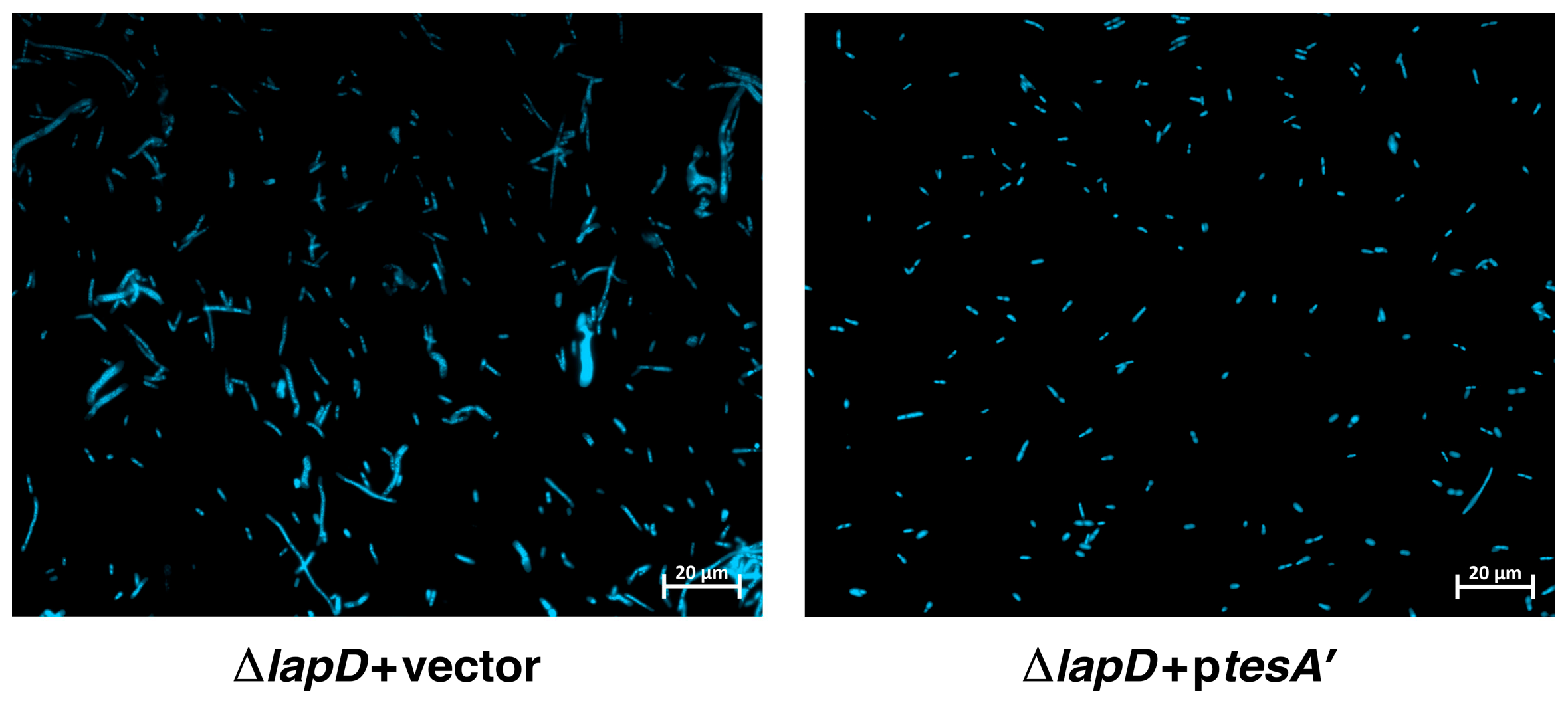
2.12. Overexpression Signal Sequence-Less TesA (TesA’) Overcome Cell Morphology Defects of ΔlapD Bacteria
2.13. Induction of Either the Acyl-Carrier Protein or AccB Subunit of the Acetyl-CoA Carboxylase Enzyme or TesA’ Represses Phospholipid Biosynthesis in ΔlapD Bacteria
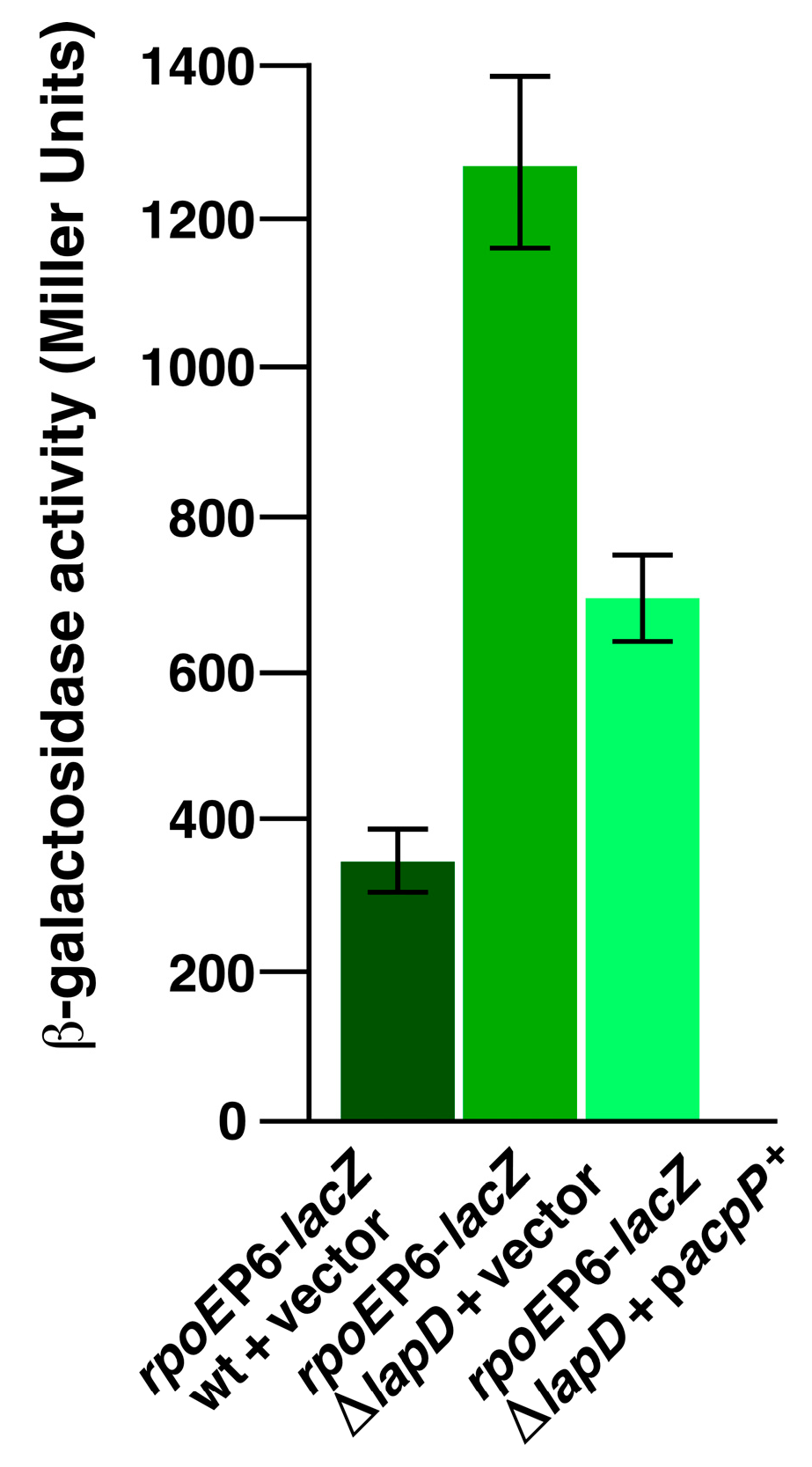
2.14. Overexpression of the acpP Gene Suppresses Elevated RpoE-Regulated Envelope Stress Response of ΔlapD Bacteria
2.15. Essentiality of the mepS Gene in ΔlapD Bacteria
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids and Media
4.2. Strain Construction and the Isolation of Extragenic Chromosomal Suppressors
4.3. The Identification of Multicopy Suppressors, Whose Overexpression Overcomes Vancomycin Sensitivity of ΔlapD Bacteria
4.4. The Construction of Plasmid Expressing Signal Sequence-Less tesA Gene
4.5. Isolation and Analysis of 32P-Labeled Phospholipids Species
4.6. Isolation of Fatty Acids and Their Analysis by Gas Chromatography
4.7. Examination of Cellular Morphology
4.8. Immunoblotting and Measurement of β-Galactosidase Activity
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Lindner, B.; Brade, H.; Raina, S. Molecular basis of lipopolysaccharide heterogeneity in Escherichia coli: Envelope stress-responsive regulators control the incorporation of glycoforms with a third 3-deoxy-α-D-manno-oct-2-ulosonic acid and rhamnose. J. Biol. Chem. 2011, 286, 42787–42807. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef]
- Clementz, T.; Bednarski, J.J.; Raetz, C.R. Function of the htrB high temperature requirement gene of Escherichia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J. Biol. Chem. 1996, 271, 12095–12102. [Google Scholar] [CrossRef] [PubMed]
- Clementz, T.; Zhou, Z.; Raetz, C.R. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. Acylation by MsbB follows laurate incorporation by HtrB. J. Biol. Chem. 1997, 272, 10353–10360. [Google Scholar] [CrossRef]
- Byers, D.M.; Gong, H. Acyl carrier protein: Structure-function relationships in a conserved multifunctional protein family. Biochem. Cell Biol. 2007, 85, 649–662. [Google Scholar] [CrossRef]
- Klein, G.; Wieczorek, A.; Szuster, M.; Raina, S. Checkpoints that regulate balanced biosynthesis of lipopolysaccharide and its essentiality in Escherichia coli. Int. J. Mol. Sci. 2022, 23, 189. [Google Scholar] [CrossRef]
- Ogura, T.; Inoue, K.; Tatsuta, T.; Suzaki, T.; Karata, K.; Young, K.; Su, L.H.; Fierke, C.A.; Jackman, J.E.; Raetz, C.R.H.; et al. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol. 1999, 31, 833–844. [Google Scholar] [CrossRef]
- Sorensen, P.G.; Lutkenhaus, J.; Young, K.; Eveland, S.S.; Anderson, M.S.; Raetz, C.R.H. Regulation of UDP-3-O-[R-3-hydroxymyristoyl]-N-acetylglucosamine deacetylase in Escherichia coli. The second enzymatic step of lipid A biosynthesis. J. Biol. Chem. 1996, 271, 25898–25905. [Google Scholar] [CrossRef]
- Schäkermann, M.; Langklotz, S.; Narberhaus, F. FtsH-mediated coordination of lipopolysaccharide biosynthesis in Escherichia coli correlates with the growth rate and the alarmone (p)ppGpp. J. Bacteriol. 2013, 195, 1912–1919. [Google Scholar] [CrossRef]
- Klein, G.; Kobylak, N.; Lindner, B.; Stupak, A.; Raina, S. Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J. Biol. Chem. 2014, 289, 14829–14853. [Google Scholar] [CrossRef]
- Emiola, A.; Andrews, S.S.; Heller, C.; George, J. Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2016, 113, 3108–3113. [Google Scholar] [CrossRef]
- Clairfeuille, T.; Buchholz, K.R.; Li, Q.; Verschueren, E.; Liu, P.; Sangaraju, D.; Park, S.; Noland, C.L.; Storek, K.M.; Nickerson, N.N.; et al. Structure of the essential inner membrane lipopolysaccharide-PbgA complex. Nature 2020, 584, 479–483. [Google Scholar] [CrossRef]
- Fivenson, E.M.; Bernhardt, T.G. An essential membrane protein modulates the proteolysis of LpxC to control lipopolysaccharide synthesis in Escherichia coli. mBio 2020, 11, e00939-20. [Google Scholar] [CrossRef]
- Biernacka, D.; Gorzelak, P.; Klein, G.; Raina, S. Regulation of the first committed step in lipopolysaccharide biosynthesis catalyzed by LpxC requires the essential protein LapC (YejM) and HslVU protease. Int. J. Mol. Sci. 2020, 21, 9088. [Google Scholar] [CrossRef] [PubMed]
- Guest, R.L.; Guerra, D.S.; Wissler, M.; Grimm, J.; Silhavy, T.J. YejM modulates activity of the YciM/FtsH protease complex to prevent lethal accumulation of lipopolysaccharide. mBio 2020, 11, e00598-20. [Google Scholar] [CrossRef] [PubMed]
- Cian, M.B.; Giordano, N.P.; Masilamani, R.; Minor, K.E.; Delabroux, Z.D. Salmonella enterica serovar Typhimurium uses PdgA/YejM to regulate lipopolysaccharide assembly during bacteremia. Infect. Immun. 2020, 88, e00758Ce19. [Google Scholar] [CrossRef]
- Nguyen, D.; Kelly, K.; Qiu, N.; Misra, R. YejM controls LpxC levels by regulating protease activity of the FtsH/YciM complex of Escherichia coli. J. Bacteriol. 2020, 202, e00303–e00320. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.; Mi, W. Regulatory mechanisms of lipopolysaccharide synthesis in Escherichia coli. Nat. Commun. 2022, 13, 4576. [Google Scholar] [CrossRef]
- Wieczorek, A.; Sendobra, A.; Maniyeri, A.; Sugalska, M.; Klein, G.; Raina, S. A new factor LapD is required for the regulation of LpxC amounts and lipopolysaccharide trafficking. Int. J. Mol. Sci. 2022, 23, 9706. [Google Scholar] [CrossRef]
- Magnuson, K.; Jackowski, S.; Rock, C.O.; Cronan, J.E., Jr. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 1993, 57, 522–542. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Rock, C.O. Exogenous fatty acid metabolism in bacteria. Biochimie 2017, 141, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Vadia, S.; Tse, J.L.; Lucena, R.; Yang, Z.; Kellogg, D.R.; Wang, J.D.; Levin, P.A. Fatty acid availability sets cell envelope capacity and dictates microbial cell size. Curr. Biol. 2017, 27, 1757–1767.e5. [Google Scholar] [CrossRef]
- Noga, M.J.; Büke, F.; van den Broek, N.J.F.; Imholz, N.C.E.; Scherer, N.; Yang, F.; Bokinsky, G. Posttranslational control of PlsB is sufficient to coordinate membrane synthesis with growth in Escherichia coli. mBio 2020, 11, e02703-19. [Google Scholar] [CrossRef]
- Mehla, J.; Liechti, G.; Morgenstein, R.M.; Caufield, J.H.; Hosseinnia, A.; Gagarinova, A.; Phanse, S.; Goodacre, N.; Brockett, M.; Sakhawalkar, N.; et al. ZapG (YhcB/DUF1043), a novel cell division protein in gamma-proteobacteria linking the Z-ring to septal peptidoglycan synthesis. J. Biol. Chem. 2021, 296, 100700. [Google Scholar] [CrossRef]
- Goodall, E.C.A.; Isom, G.L.; Rooke, J.L.; Pullela, K.; Icke, C.; Yang, Z.; Boelter, G.; Jones, A.; Warner, I.; Da Costa, R.; et al. Loss of YhcB results in dysregulation of coordinated peptidoglycan, LPS and phospholipid synthesis during Escherichia coli cell growth. PLoS Genet. 2021, 17, e1009586. [Google Scholar] [CrossRef]
- Murata, M.; Fujimoto, H.; Nishimura, K.; Charoensuk, K.; Nagamitsu, H.; Raina, S.; Kosaka, T.; Oshima, T.; Ogasawara, N.; Yamada, M. Molecular strategy for survival at a critical high temperature in Escherichia coli. PLoS ONE 2011, 6, e20063. [Google Scholar] [CrossRef]
- Cronan, J.E. The classical, yet controversial, first enzyme of lipid synthesis: Escherichia coli acetyl-CoA carboxylase. Microbiol. Mol. Biol. Rev. 2021, 85, e0003221. [Google Scholar] [CrossRef]
- Stanley, H.M.; Trent, M.S. Loss of YhcB results in overactive fatty acid biosynthesis. mBio 2024, 15, e0079024. [Google Scholar] [CrossRef]
- Zhu, K.; Zhang, Y.M.; Rock, C.O. Transcriptional regulation of membrane lipid homeostasis in Escherichia coli. J. Biol. Chem. 2009, 284, 34880–34888. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Lindner, B.; Brabetz, W.; Brade, H.; Raina, S. Escherichia coli K-12 suppressor-free mutants lacking early glycosyltransferases and late acyltransferases: Minimal lipopolysaccharide structure and induction of envelope stress response. J. Biol. Chem. 2009, 284, 15369–15389. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.J.; Rock, C.O. Regulation of fatty acid elongation and initiation by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 1996, 271, 1833–1836. [Google Scholar] [CrossRef]
- Davis, M.S.; Cronan, J.E., Jr. Inhibition of Escherichia coli acetyl coenzyme A carboxylase by acyl-acyl carrier protein. J. Bacteriol. 2001, 183, 1499–1503. [Google Scholar] [CrossRef]
- Heath, R.J.; Rubin, J.R.; Holland, D.R.; Zhang, E.; Snow, M.E.; Rock, C.O. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J. Biol. Chem. 1999, 274, 11110–11114. [Google Scholar] [CrossRef]
- Omura, S. The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol. Rev. 1976, 40, 681–697. [Google Scholar] [CrossRef]
- LaPorte, D.C.; Koshland, D.E. A protein with kinase and phosphatase activities involved in regulation of tricarboxylic acid cycle. Nature 1982, 300, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Jia, Z. Structure of the bifunctional isocitrate dehydrogenase kinase/phosphatase. Nature 2010, 465, 961–965. [Google Scholar] [CrossRef]
- Zheng, J.; Yates, S.P.; Jia, Z. Structural and mechanistic insights into the bifunctional enzyme isocitrate dehydrogenase kinase/phosphatase AceK. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 2656–2668. [Google Scholar] [CrossRef][Green Version]
- Bohin, J.P.; Kennedy, E.P. Regulation of the synthesis of membrane-derived oligosaccharides in Escherichia coli. Assay of phosphoglycerol transferase I in vivo. J. Biol. Chem. 1984, 259, 8388–8393. [Google Scholar] [CrossRef] [PubMed]
- Therisod, H.; Weissborn, A.C.; Kennedy, E.P. An essential function for acyl carrier protein in the biosynthesis of membrane-derived oligosaccharides of Escherichia coli. Proc. Natl. Acad. Sci. USA 1986, 83, 7236–7240. [Google Scholar] [CrossRef]
- Yamanaka, K.; Ogura, T.; Niki, H.; Hiraga, S. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol. Gen. Genet. 1996, 250, 241–251. [Google Scholar]
- Gloyd, M.; Ghirlando, R.; Guarné, A. The role of MukE in assembling a functional MukBEF complex. J. Mol. Biol. 2011, 412, 578–590. [Google Scholar] [CrossRef]
- She, W.; Mordukhova, E.; Zhao, H.; Petrushenko, Z.M.; Rybenkov, V.V. Mutational analysis of MukE reveals its role in focal subcellular localization of MukBEF. Mol. Microbiol. 2013, 87, 539–552. [Google Scholar] [CrossRef]
- Chng, S.S.; Ruiz, N.; Chimalakonda, G.; Silhavy, T.J.; Kahne, D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl. Acad. Sci. USA 2010, 107, 5363–5368. [Google Scholar] [CrossRef]
- Tadokoro, A.; Hayashi, H.; Kishimoto, T.; Makino, Y.; Fujisaki, S.; Nishimura, Y. Interaction of the Escherichia coli lipoprotein NlpI with periplasmic Prc (Tsp) protease. J. Biochem. 2004, 135, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Banzhaf, M.; Yau, H.C.; Verheul, J.; Lodge, A.; Kritikos, G.; Mateus, A.; Cordier, B.; Hov, A.K.; Stein, F.; Wartel, M.; et al. Outer membrane lipoprotein NlpI scaffolds peptidoglycan hydrolases within multi-enzyme complexes in Escherichia coli. EMBO J. 2020, 39, e102246. [Google Scholar] [CrossRef]
- Wang, S.; Huang, C.H.; Lin, T.S.; Yeh, Y.Q.; Fan, Y.S.; Wang, S.W.; Tseng, H.C.; Huang, S.J.; Chang, Y.Y.; Jeng, U.S.; et al. Structural basis for recruitment of peptidoglycan endopeptidase MepS by lipoprotein NlpI. Nat. Commun. 2024, 15, 5461. [Google Scholar] [CrossRef]
- Ohara, M.; Wu, H.C.; Sankaran, K.; Rick, P.D. Identification and characterization of a new lipoprotein, NlpI, in Escherichia coli K-12. J. Bacteriol. 1999, 181, 4318–4325. [Google Scholar] [CrossRef] [PubMed]
- Raina, S.; Georgopoulos, C. The htrM gene, whose product is essential for Escherichia coli viability only at elevated temperatures, is identical to the rfaD gene. Nucleic Acids Res. 1991, 19, 3811–3819. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Ma, X.; Chen, X.; Jiang, M.; Song, H.; Guo, Z. Identification of a hotdog fold thioesterase involved in the biosynthesis of menaquinone in Escherichia coli. J. Bacteriol. 2013, 195, 2768–2775. [Google Scholar] [CrossRef]
- Latham, J.A.; Chen, D.; Allen, K.N.; Dunaway-Mariano, D. Divergence of substrate specificity and function in the Escherichia coli hotdog-fold thioesterase paralogs YdiI and YbdB. Biochemistry 2014, 53, 4775–4787. [Google Scholar] [CrossRef]
- Cho, H.; Cronan, J.E., Jr. Defective export of a periplasmic enzyme disrupts regulation of fatty acid synthesis. J. Biol. Chem. 1995, 270, 4216–4219. [Google Scholar] [CrossRef] [PubMed]
- Rowlett, V.W.; Mallampalli, V.K.P.S.; Karlstaedt, A.; Dowhan, W.; Taegtmeyer, H.; Margolin, W.; Vitrac, H. Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 2017, 199, e00849-16. [Google Scholar] [CrossRef]
- Betton, J.M.; Boscus, D.; Missiakas, D.; Raina, S.; Hofnung, M. Probing the structural role of an α β loop of maltose-binding protein by mutagenesis: Heat-shock induction by loop variants of the maltose-binding protein that form periplasmic inclusion bodies. J. Mol. Biol. 1996, 262, 140–150. [Google Scholar] [CrossRef]
- Lima, S.; Guo, M.S.; Chaba, R.; Gross, C.A.; Sauer, R.T. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 2013, 340, 837–841. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Stupak, A.; Biernacka, D.; Wojtkiewicz, P.; Lindner, B.; Raina, S. Multiple transcriptional factors regulate transcription of the rpoE gene in Escherichia coli under different growth conditions and when the lipopolysaccharide biosynthesis is defective. J. Biol. Chem. 2016, 291, 22999–23019. [Google Scholar] [CrossRef]
- Mohan, S.; Kelly, T.M.; Eveland, S.S.; Raetz, C.R.; Anderson, M.S. An Escherichia coli gene (fabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. Relation to fabA and suppression of mutations in lipid A biosynthesis. J. Biol. Chem. 1994, 269, 32896–32903. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Hamamoto, K.; Kitakawa, M. Inner membrane protein YhcB interacts with RodZ involved in cell shape maintenance in Escherichia coli. ISRN Mol. Biol. 2012, 2012, 304021. [Google Scholar] [CrossRef]
- Li, S.J.; Cronan, J.E., Jr. Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J. Bacteriol. 1993, 175, 332–340. [Google Scholar] [CrossRef]
- Karow, M.; Fayet, O.; Georgopoulos, C. The lethal phenotype caused by null mutations in the Escherichia coli htrB gene is suppressed by mutations in the accBC operon, encoding two subunits of acetyl coenzyme A carboxylase. J. Bacteriol. 1992, 174, 7407–7418. [Google Scholar] [CrossRef]
- Ganong, B.R.; Raetz, C.R. Massive accumulation of phosphatidic acid in conditionally lethal CDP-diglyceride synthetase mutants and cytidine auxotrophs of Escherichia coli. J. Biol. Chem. 1982, 257, 389–394. [Google Scholar] [CrossRef]
- Dowhan, W. CDP-diacylglycerol synthase of microorganisms. Biochim. Biophys. Acta 1997, 1348, 157–165. [Google Scholar] [CrossRef]
- Moffatt, C.B.; Plaman, B.A.; Rowe, S.J.; Caruso, A.; Early, S.A.; Ruiz, N.; Kahne, D. Inhibiting lipopolysaccharide biogenesis: The more you know the further you go. Annu. Rev. Biochem. 2025, 94, 137–160. [Google Scholar] [CrossRef]
- Dexter, J.P.; Gunawardena, J. Dimerization and bifunctionality confer robustness to the isocitrate dehydrogenase regulatory system in Escherichia coli. J. Biol. Chem. 2013, 288, 5770–5778. [Google Scholar] [CrossRef]
- Pavoncello, V.; Barras, F.; Bouveret, E. Degradation of exogenous fatty acids in Escherichia coli. Biomolecules 2022, 12, 1019. [Google Scholar] [CrossRef]
- Weart, R.B.; Lee, A.H.; Chien, A.C.; Haeusser, D.P.; Hill, N.S.; Levin, P.A. A metabolic sensor governing cell size in bacteria. Cell 2007, 130, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Davis, R.M.; Kishony, R.; Kahne, D.; Ruiz, N. Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2012, 109, E2561–E2568. [Google Scholar] [CrossRef]
- Prince, J.P.; Bolla, J.R.; Fisher, G.L.M.; Mäkelä, J.; Fournier, M.; Robinson, C.V.; Arciszewska, L.K.; Sherratt, D.J. Acyl carrier protein promotes MukBEF action in Escherichia coli chromosome organization-segregation. Nat. Commun. 2012, 12, 6721. [Google Scholar] [CrossRef] [PubMed]
- Truong, T.T.; Vettiger, A.; Bernhardt, T.G. Cell division is antagonized by the activity of peptidoglycan endopeptidases that promote cell elongation. Mol. Microbiol. 2020, 114, 966–978. [Google Scholar] [CrossRef] [PubMed]
- Som, N.; Reddy, M. Cross-talk between phospholipid synthesis and peptidoglycan expansion by a cell wall hydrolase. Proc. Natl. Acad. Sci. USA 2023, 120, e2300784120. [Google Scholar] [CrossRef]
- Gorzelak, P.; Klein, G.; Raina, S. Molecular basis of essentiality of early critical steps in the lipopolysaccharide biogenesis in Escherichia coli K-12: Requirement of MsbA, cardiolipin, LpxL, LpxM and GcvB. Int. J. Mol. Sci. 2021, 22, 5099. [Google Scholar] [CrossRef]
- Douglass, M.V.; Cléon, F.; Trent, M.S. Cardiolipin aids in lipopolysaccharide transport to the gram-negative outer membrane. Proc. Natl. Acad. Sci. USA 2021, 118, e2018329118. [Google Scholar] [CrossRef]
- Raina, S.; Missiakas, D.; Baird, L.; Kumar, S.; Georgopoulos, C. Identification and transcriptional analysis of the Escherichia coli htrE operon which is homologous to pap and related pilin operons. J. Bacteriol. 1993, 175, 5009–50021. [Google Scholar] [CrossRef]
- Kawazura, T.; Matsumoto, K.; Kojima, K.; Kato, F.; Kanai, T.; Niki, H.; Shiomi, D. Exclusion of assembled MreB by anionic phospholipids at cell poles confers cell polarity for bidirectional growth. Mol. Microbiol. 2017, 104, 472–486. [Google Scholar] [CrossRef]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Stark, M.J. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene 1987, 51, 255–267. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, M.; Gesteland, R.F.; Atkins, J.F. tRNA hopping: Enhancement by an expanded anticodon. EMBO J. 1989, 8, 4315–4323. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]
- Dartigalongue, C.; Loferer, H.; Raina, S. EcfE, a new essential inner membrane protease: Its role in the regulation of heat shock response in Escherichia coli. EMBO J. 2001, 20, 5908–5918. [Google Scholar] [CrossRef]
- Kleckner, N.; Bender, J.; Gottesman, S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991, 204, 139–180. [Google Scholar]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Ames, G.F. Lipids of Salmonella typhimurium and Escherichia coli: Structure and metabolism. J. Bacteriol. 1968, 95, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Deranieh, R.M.; Joshi, A.S.; Greenberg, M.L. Thin-layer chromatography of phospholipids. Methods Mol. Biol. 2013, 1033, 21–27. [Google Scholar]
- Politz, M.; Lennen, R.; Pfleger, B. Quantification of bacterial fatty acids by extraction and methylation. Bio Protoc. 2013, 3, e950. [Google Scholar] [CrossRef] [PubMed]









| Genotype | Number of Transformants | ||
|---|---|---|---|
| P1 ΔlapD LA 30 °C | P1 ΔlapD M9 30 °C | P1 ΔlapD LA 42 °C | |
| Wt | 1020 | 923 | ND 1 |
| ΔlpxM | 8 | 875 | ND |
| ΔlpxM p(lpxM+) | 768 | 901 | ND |
| ΔwaaC | 676 | 810 | 17 |
| ΔwaaC p(waaC+) | 740 | 720 | 830 |
| Gene | Function | Mutation Position | Amounts of Isolates |
|---|---|---|---|
| Suppressors of Δ(lapD lpxM) | |||
| accA | acetyl-CoA carboxyltransferase subunit α | K31E (AAA ⟶ GAA) | 2 |
| R175L (CGT ⟶ CTT) | 2 | ||
| accC | biotin carboxylase | K40T (AAA ⟶ ACA) | 1 |
| V42I (GTA ⟶ ATA) | 1 | ||
| R253H (CGT ⟶ CAT) | 1 | ||
| V299A (GTT ⟶ GCT) | 2 | ||
| I309F (ATC ⟶ TTC) | 1 | ||
| accD | acetyl-CoA carboxyltransferase subunit β | L39P (CTG ⟶ CCG) | 1 |
| S173P (TCG ⟶ CCG) | 1 | ||
| opgG | osmoregulated periplasmic glucans biosynthesis protein G | A100T (GCC ⟶ ACC) | 2 |
| aceK | isocitrate dehydrogenase kinase | Y474S (TAT ⟶ TCT) | 1 |
| mukE | chromosome partitioning | A122P (GCA ⟶ CCA) | 1 |
| Suppressors of Δ(lapD waaC) | |||
| lptD | lipopolysaccharide assembly | L651P (CTG ⟶ CCG) | 1 |
| nlpI | lipoprotein | Y243H (TAC ⟶ CAC) | 1 |
| lptD and bamC | lptD L651P (CTG ⟶ CCG) | 1 | |
| outer membrane protein assembly factor | bamC F248L (TTC ⟶ CTC) | ||
| M9 30 °C | ||||||
|---|---|---|---|---|---|---|
| wt | ΔlapD | Δ(lapD lpxM) | Δ(lapD lpxM) accA K31E | Δ(lapD lpxM) accC V299A | Δ(lapD lpxM) accD L39P | |
| C14:0 | 1.49 ± 0.13 | 1.88 ± 0.05 | 4.07 ± 0.04 | 3.11 ± 0.07 | 1.03 ± 0.07 | 2.62 ± 0.04 |
| C16:0 | 45.01 ± 1.04 | 45.35 ± 0.24 | 41.86 ± 0.07 | 43.17 ± 0.02 | 41.35 ± 0.06 | 45.94 ± 0.04 |
| C18:0 | 0.72 ± 0.03 | 0.95 ± 0.10 | 0.39 ± 0.04 | 0.58 ± 0.01 | 2.01 ± 0.10 | 0.61 ± 0.08 |
| C16:1 | 15.91 ± 0.40 | 19.32 ± 0.04 | 30.13 ± 0.20 | 30.12 ± 0.06 | 19.68 ± 0.08 | 20.99 ± 0.01 |
| cyclopropane | 17.93 ± 0.02 | 14.14 ± 0.07 | 7.89 ± 0.11 | 6.25 ± 0.12 | 5.39 ± 0.15 | 15.26 ± 0.11 |
| C18:1 | 18.94 ± 1.66 | 18.36 ± 0.19 | 15.66 ± 0.06 | 16.78 ± 0.23 | 30.54 ± 0.16 | 14.58 ± 0.20 |
| LB 37 °C | ||||||
| C14:0 | 3.21 ± 0.03 | 4.03 ± 0.04 | 3.85 ± 0.10 | 3.43 ± 0.03 | 2.14 ± 0.01 | 3.71 ± 0.11 |
| C16:0 | 42.60 ± 1.11 | 45.17 ± 0.14 | 43.40 ± 0.16 | 45.29 ± 0.07 | 44.58 ± 0.21 | 44.11 ± 1.94 |
| C18:0 | 3.21 ± 0.09 | 3.31 ± 0.08 | 0.87 ± 0.20 | 0.77 ± 0.06 | 1.35 ± 0.04 | 0.74 ± 0.05 |
| C16:1 | 31.41 ± 0.73 | 31.51 ± 0.02 | 28.78 ± 0.05 | 31.01 ± 0.14 | 25.09 ± 0.17 | 31.63 ± 1.86 |
| cyclopropane | 2.18 ± 0.35 | 2.53 ± 0.08 | 4.79 ± 0.06 | 2.85 ± 0.01 | 2.00 ± 0.04 | 2.37 ± 0.21 |
| C18:1 | 17.37 ± 0.629 | 13.45 ± 0.05 | 18.31 ± 0.03 | 16.66 ± 0.17 | 24.83 ± 0.08 | 17.43 ± 0.75 |
| Strains | Genotype | Reference |
|---|---|---|
| W3110 | λ−, IN (rrnD-rrnE)1, rph-1 | CGSC, Yale |
| GK1275 | W3110 lpxM<>aph | [31] |
| SR7336 | GK1275 lpxM<>frt | This study |
| SR8035 | W3110 waaC<>aph | [31] |
| SR25196 | W3110 lapD<>aph | This study |
| SR25204 | SR25196 lapD<>frt | This study |
| GK6173 | SR25204 lapD<>frt lpxM<>aph | This study |
| SR25236 | SR7336 lpxM<>frt lapD<>aph | This study |
| SR25217 | SR25204 lapD<>frt lpxM<>aph accD L39P | This study |
| SR25218 | SR25204 lapD<>frt lpxM<>aph accA K31E | This study |
| SR25219 | SR25204 lapD<>frt lpxM<>aph accC V299A | This study |
| SR25220 | SR25204 lapD<>frt lpxM<>aph accD S173P | This study |
| SR25224 | SR25204 lapD<>frt lpxM<>aph accA R175L | This study |
| SR25225 | SR25204 lapD<>frt lpxM<>aph accC R253H | This study |
| SR25235 | SR25204 lapD<>frt lpxM<>aph accA K31E | This study |
| SR25240 | SR25204 lapD<>frt lpxM<>aph accC V299A | This study |
| SR25241 | SR7336 lpxM<>frt lapD<>aph accC K40T | This study |
| SR25307 | SR25204 lapD<>frt lpxM<>aph accA R175L | This study |
| SR25438 | SR7336 lpxM<>frt lapD<>aph accC I309F | This study |
| SR25437 | SR7336 lpxM<>frt lapD<>aph opgG A100T | This study |
| SR25443 | SR7336 lpxM<>frt lapD<>aph opgG A100T | This study |
| SR25454 | SR7336 lpxM<>frt lapD<>aph accC V42I | This study |
| SR25452 | SR7336 lpxM<>frt lapD<>aph aceK Y474S | This study |
| SR25458 | SR7336 lpxM<>frt lapD<>aph mukE A122P | This study |
| SR25651 | SR25204 lapD<>frt waaC<>aph | This study |
| SR25660 | SR25204 lapD<>frt waaC<>aph lptD L651P | This study |
| SR25661 | SR25204 lapD<>frt waaC<>aph nlpI Y243H | This study |
| SR25662 | SR25204 lapD<>frt waaC<>aph lptD L651P bamC F248L | This study |
| RU857 | ΔpgsA Δ(clsABC) lpp2 | [74] |
| SR25199 | SR25204 lapD<>frt + pacpP+ | This study |
| SR25203 | SR25204 lapD<>frt + paccB+ | This study |
| SR25870 | SR25204 lapD<>frt + ptesA′ | This study |
| SR25872 | SR25204 lapD<>frt + pmenI | This study |
| SR25467 | W3110 accA K31E Tn10 | This study |
| SR25468 | W3110 accC V299A Tn10 | This study |
| SR25874 | W3110 + pCA24N | This study |
| SR23799 | SR25204 lapD<>frt + pCA24N | This study |
| SR25878 | W3110 + pTTQ18 | This study |
| SR25880 | SR25204 lapD<>frt + pTTQ18 | This study |
| Plasmids | Genotype | Reference |
| pCA24N | IPTG-inducible expression vector cmR | [75] |
| pKD3 | oriR6Kg, bla(AmpR), kan, rgnB(Ter), cat | [76] |
| pKD13 | oriR6Kg, bla(AmpR), kan, rgnB(Ter) | [76] |
| pKD46 | araBp-gam-bet-exo, bla(AmpR), repA101(ts) | [76] |
| pCP20 | ts replicon with inducible FLP recombinase | [76] |
| pTTQ18 | expression vector with LacI represor | [77] |
| pSR25845 | tesA′ in pTTQ18 | This study |
| JW1080 | acpP+ in pCA24N cmR | [75] |
| JW3223 | accB+ in pCA24N cmR | [75] |
| JW1676 | menI+ in pCA24N cmR | [75] |
| JW5539 | lapD+ in pCA24N cmR | [75] |
| pREG | cosmid cloning vector | [78] |
| pSR25460 | accA+ in pREG ampR | This study |
| pSR25461 | accC+ in pREG ampR | This study |
| pSR25469 | accD+ in pREG ampR | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeschke, M.; Ayyolath, A.; Maniyeri, A.; Raina, S.; Klein, G. Coordinated Biosynthesis of Essential Cell Envelope Components: Lipopolysaccharide and Fatty Acids Requires LapD, Acyl Carrier Protein, and Fully Hexaacylated Lipid A. Int. J. Mol. Sci. 2025, 26, 10993. https://doi.org/10.3390/ijms262210993
Jeschke M, Ayyolath A, Maniyeri A, Raina S, Klein G. Coordinated Biosynthesis of Essential Cell Envelope Components: Lipopolysaccharide and Fatty Acids Requires LapD, Acyl Carrier Protein, and Fully Hexaacylated Lipid A. International Journal of Molecular Sciences. 2025; 26(22):10993. https://doi.org/10.3390/ijms262210993
Chicago/Turabian StyleJeschke, Marta, Aravind Ayyolath, Akshay Maniyeri, Satish Raina, and Gracjana Klein. 2025. "Coordinated Biosynthesis of Essential Cell Envelope Components: Lipopolysaccharide and Fatty Acids Requires LapD, Acyl Carrier Protein, and Fully Hexaacylated Lipid A" International Journal of Molecular Sciences 26, no. 22: 10993. https://doi.org/10.3390/ijms262210993
APA StyleJeschke, M., Ayyolath, A., Maniyeri, A., Raina, S., & Klein, G. (2025). Coordinated Biosynthesis of Essential Cell Envelope Components: Lipopolysaccharide and Fatty Acids Requires LapD, Acyl Carrier Protein, and Fully Hexaacylated Lipid A. International Journal of Molecular Sciences, 26(22), 10993. https://doi.org/10.3390/ijms262210993








