H2A.Z and H3:K56Q Affect Transcription Through Chromatin and Yeast FACT-Dependent Nucleosome Unfolding
Abstract
1. Introduction
2. Results
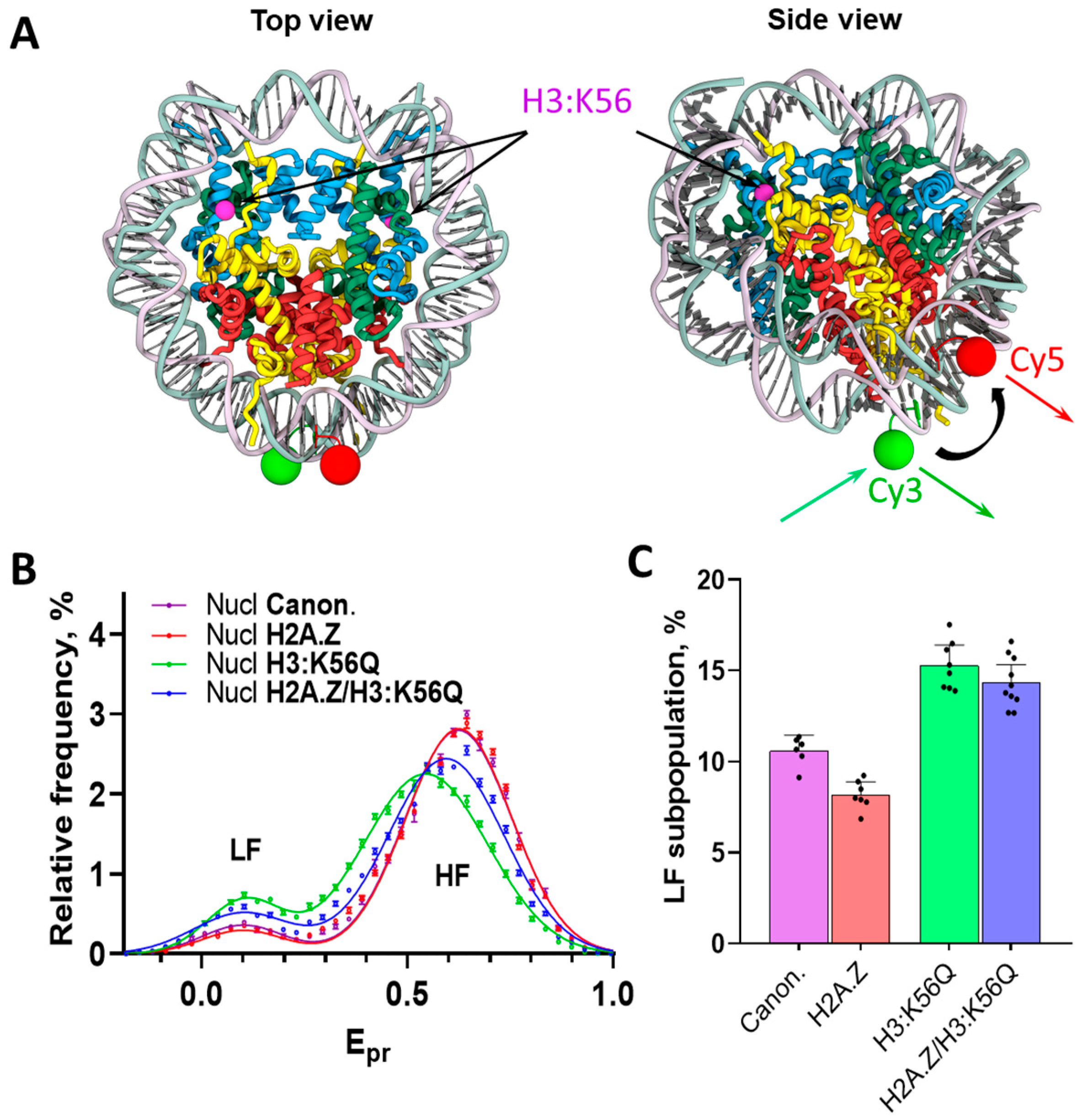
2.1. The Structure of Yeast Nucleosomes Is Minimally Affected by the Presence of Histones H2A.Z, H3:K56Q, or Their Combination
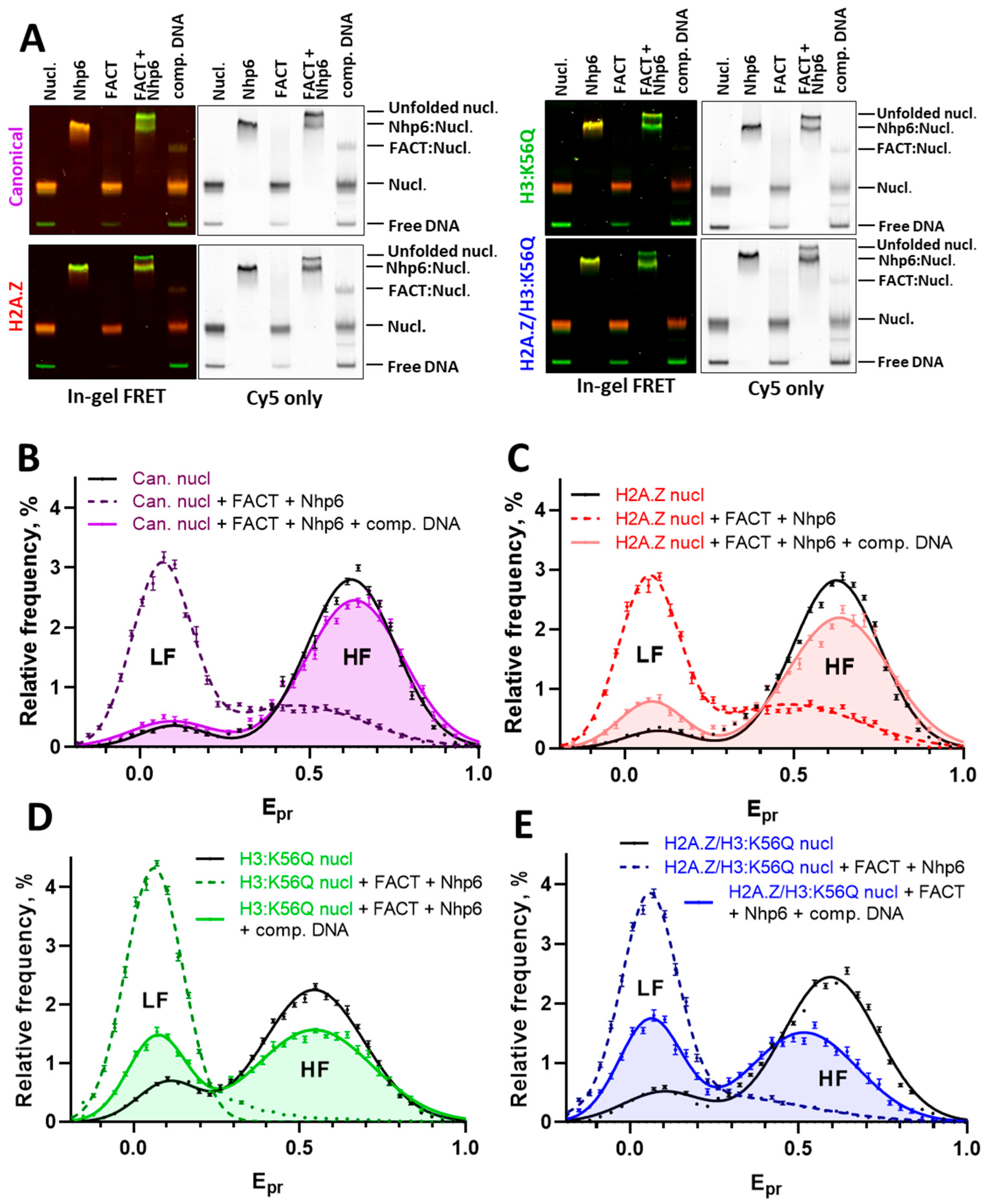
2.2. The Presence of Variant Histone H2A.Z Hampers Refolding of FACT-Unfolded Nucleosomes
2.3. The Mimic of Acetylation H3:K56Q Facilitates Nucleosome Unfolding by FACT and Inhibits Nucleosome Refolding
2.4. H2A.Z and H3:K56Q Cooperatively Affect FACT-Dependent Nucleosome Unfolding and Refolding
2.5. Histones Are Partially Evicted from Nhp6:FACT-Unfolded H2A.Z/H3:K56Q-Containing Nucleosomes
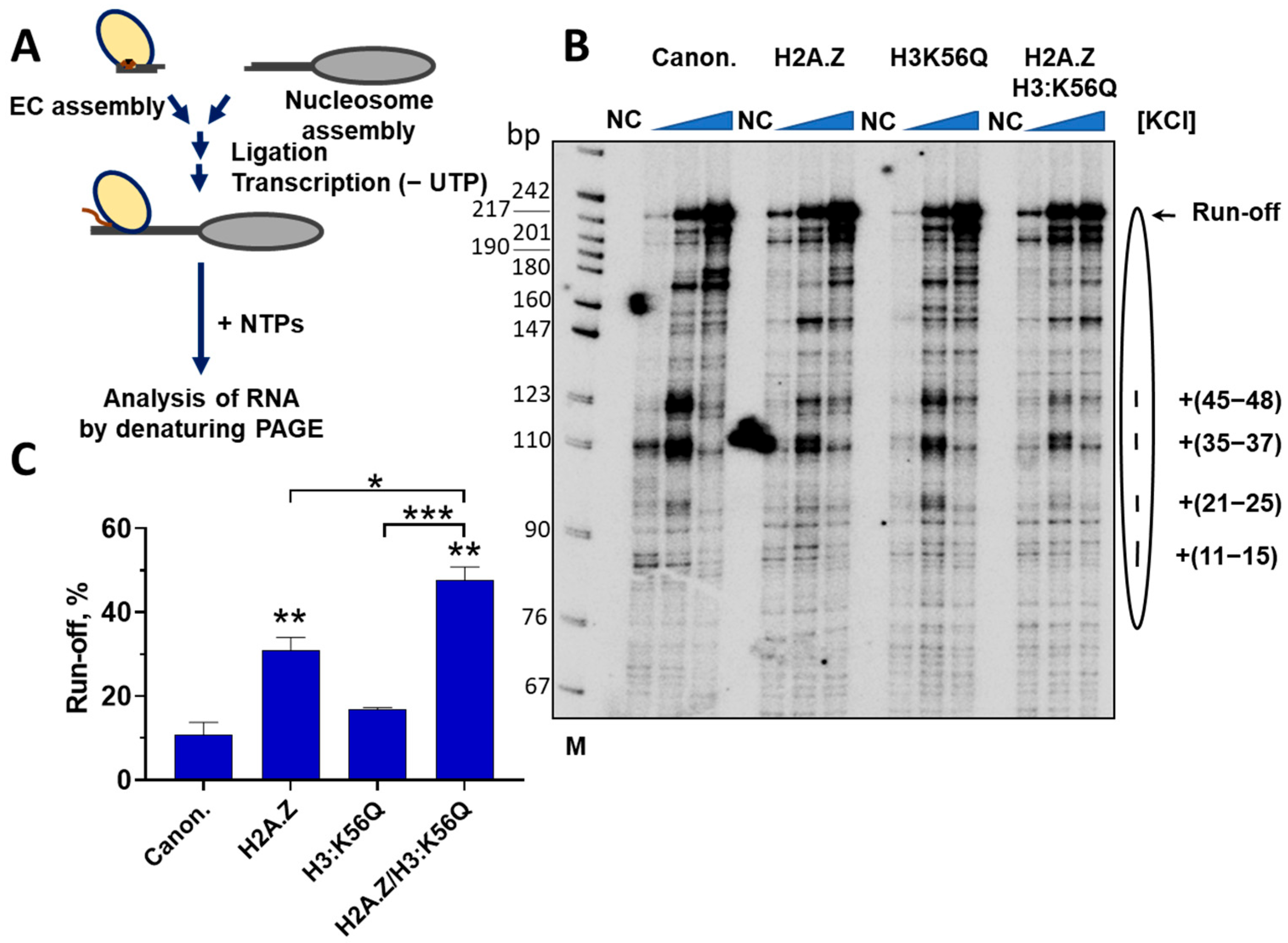
2.6. H3:K56Q and H2A.Z Additively Facilitate Transcription Through a Nucleosome
3. Discussion
3.1. Overview and Main Findings
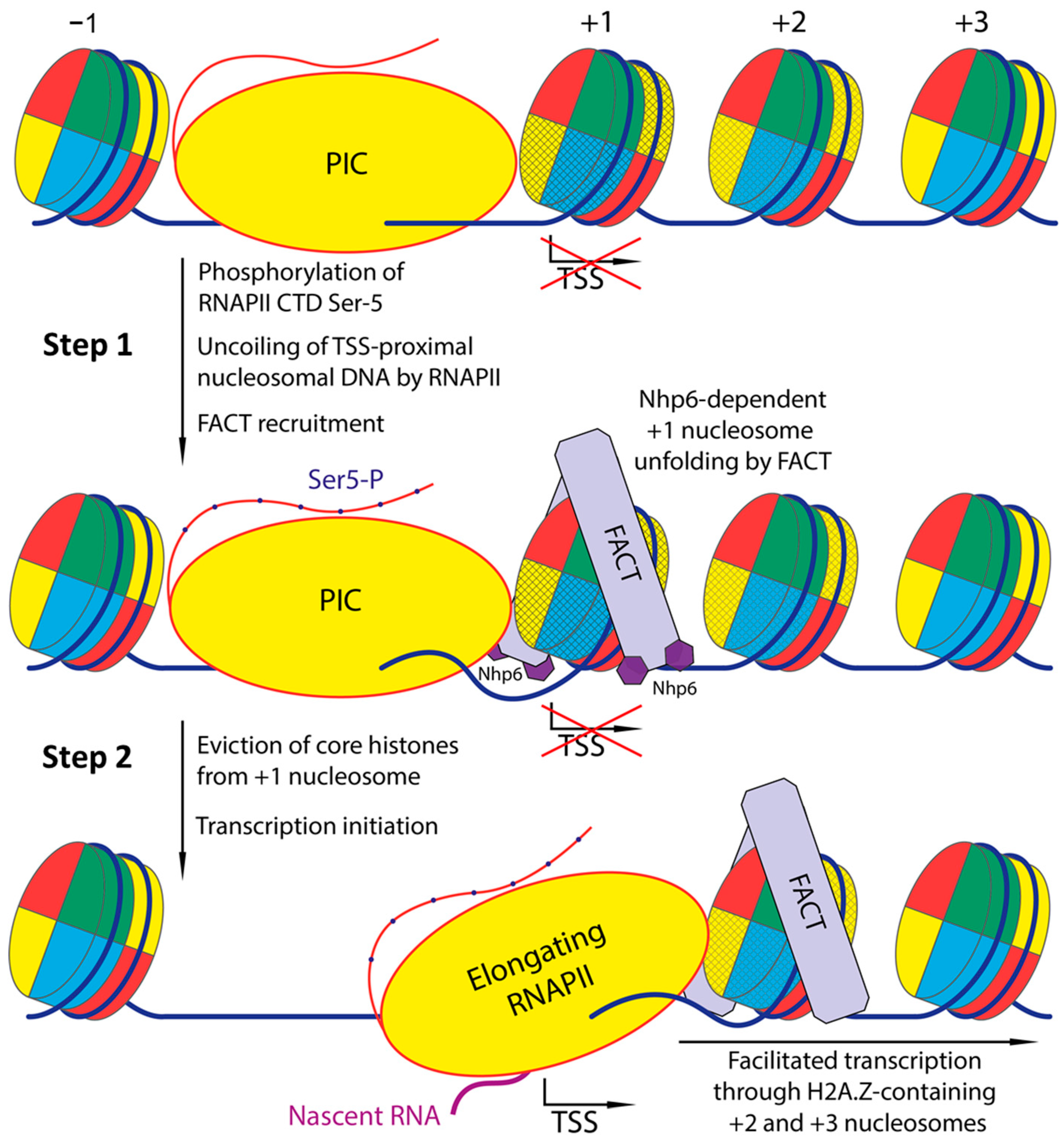
3.2. Current Models for +1 Nucleosome Turnover and the Role of FACT
3.3. FACT-Mediated H2A.Z Destabilization During Elongation of Transcription
3.4. Mechanistic Role of Nhp6 and Entry/Exit DNA Contacts
3.5. Initiation: Dislodging the +1 Nucleosome Before RNAPolII Transit
3.6. Early Elongation Through +2 and +3 Nucleosomes: Effects and Constraints
3.7. Integrated Model and Implications for Promoter Opening
4. Materials and Methods
4.1. Proteins
4.2. Nucleosome Assembly for spFRET Microscopy and Gel Purification
4.3. Nucleosome Unfolding by FACT, spFRET Microscopy, and Analysis of Electrophoretic Mobility of the Complexes
4.4. Transcription of Nucleosomes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CTD | C-terminal domain (of Rpb1) |
| EC | Elongation complex |
| EMSA | Electrophoretic mobility shift assay |
| FACT | Yeast FAcilitates Chromatin Transcription complex |
| H3:K56Ac | Histone H3 with acetylated K56 |
| H3:K56Q | Histone H3 with acetyl mimic substitution K56Q |
| NDR | Nucleosome-depleted region |
| PIC | Pre-initiation complex |
| RNAPII | RNA polymerase II |
| spFRET | Single-particle Förster resonance energy transfer |
| TFIIH | General transcription factor IIH |
| TSS | Transcription start site |
References
- Armeev, G.A.; Kniazeva, A.S.; Komarova, G.A.; Kirpichnikov, M.P.; Shaytan, A.K. Histone Dynamics Mediate DNA Unwrapping and Sliding in Nucleosomes. Nat. Commun. 2021, 12, 2387. [Google Scholar] [CrossRef] [PubMed]
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and Functions of ATP-Dependent Chromatin-Remodeling Enzymes. Cell 2013, 154, 490–503. [Google Scholar] [CrossRef]
- Zhou, C.Y.; Johnson, S.L.; Gamarra, N.I.; Narlikar, G.J. Mechanisms of ATP-Dependent Chromatin Remodeling Motors. Annu. Rev. Biophys. 2016, 45, 153–181. [Google Scholar] [CrossRef]
- Rhee, H.S.; Bataille, A.R.; Zhang, L.; Pugh, B.F. Subnucleosomal Structures and Nucleosome Asymmetry across a Genome. Cell 2014, 159, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- Bondarenko, V.A.; Steele, L.M.; Újvári, A.; Gaykalova, D.A.; Kulaeva, O.I.; Polikanov, Y.S.; Luse, D.S.; Studitsky, V.M. Nucleosomes Can Form a Polar Barrier to Transcript Elongation by RNA Polymerase II. Mol. Cell 2006, 24, 469–479. [Google Scholar] [CrossRef]
- Kireeva, M.L.; Hancock, B.; Cremona, G.H.; Walter, W.; Studitsky, V.M.; Kashlev, M. Nature of the Nucleosomal Barrier to RNA Polymerase II. Mol. Cell 2005, 18, 97–108. [Google Scholar] [CrossRef]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of Action and Regulation of ATP-Dependent Chromatin-Remodelling Complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Kaplan, N.; Moore, I.K.; Fondufe-Mittendorf, Y.; Gossett, A.J.; Tillo, D.; Field, Y.; LeProust, E.M.; Hughes, T.R.; Lieb, J.D.; Widom, J.; et al. The DNA-Encoded Nucleosome Organization of a Eukaryotic Genome. Nature 2009, 458, 362–366. [Google Scholar] [CrossRef]
- Zhang, Z.; Wippo, C.J.; Wal, M.; Ward, E.; Korber, P.; Pugh, B.F. A Packing Mechanism for Nucleosome Organization Reconstituted Across a Eukaryotic Genome. Science 2011, 332, 977–980. [Google Scholar] [CrossRef]
- Albert, I.; Mavrich, T.N.; Tomsho, L.P.; Qi, J.; Zanton, S.J.; Schuster, S.C.; Pugh, B.F. Translational and Rotational Settings of H2A.Z Nucleosomes across the Saccharomyces Cerevisiae Genome. Nature 2007, 446, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.M.; Ramachandran, S.; Henikoff, S. Nucleosomes Are Context-Specific, H2A.Z-Modulated Barriers to RNA Polymerase. Mol. Cell 2014, 53, 819–830. [Google Scholar] [CrossRef]
- Churchman, L.S.; Weissman, J.S. Nascent Transcript Sequencing Visualizes Transcription at Nucleotide Resolution. Nature 2011, 469, 368–373. [Google Scholar] [CrossRef]
- Wang, H.; Schilbach, S.; Ninov, M.; Urlaub, H.; Cramer, P. Structures of Transcription Preinitiation Complex Engaged with the +1 Nucleosome. Nat. Struct. Mol. Biol. 2023, 30, 226–232. [Google Scholar] [CrossRef]
- Raisner, R.M.; Hartley, P.D.; Meneghini, M.D.; Bao, M.Z.; Liu, C.L.; Schreiber, S.L.; Rando, O.J.; Madhani, H.D. Histone Variant H2A.Z Marks the 5′ Ends of Both Active and Inactive Genes in Euchromatin. Cell 2005, 123, 233–248, Erratum in Cell 2005, 134, 188. [Google Scholar] [CrossRef]
- Bagchi, D.N.; Battenhouse, A.M.; Park, D.; Iyer, V.R. The Histone Variant H2A.Z in Yeast Is Almost Exclusively Incorporated into the +1 Nucleosome in the Direction of Transcription. Nucleic Acids Res. 2020, 48, 157–170. [Google Scholar] [CrossRef]
- Santisteban, M.S.; Kalashnikova, T.; Smith, M.M. Histone H2A.Z Regulates Transcription and Is Partially Redundant with Nucleosome Remodeling Complexes. Cell 2000, 103, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Roberts, D.N.; Cairns, B.R. Genome-Wide Dynamics of Htz1, a Histone H2A Variant That Poises Repressed/Basal Promoters for Activation Through Histone Loss. Cell 2005, 123, 219–231. [Google Scholar] [CrossRef]
- Dhillon, N.; Oki, M.; Szyjka, S.J.; Aparicio, O.M.; Kamakaka, R.T. H2A.Z Functions to Regulate Progression Through the Cell Cycle. Mol. Cell Biol. 2006, 26, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Venters, B.J.; Wachi, S.; Mavrich, T.N.; Andersen, B.E.; Jena, P.; Sinnamon, A.J.; Jain, P.; Rolleri, N.S.; Jiang, C.; Hemeryck-Walsh, C.; et al. A Comprehensive Genomic Binding Map of Gene and Chromatin Regulatory Proteins in Saccharomyces. Mol. Cell 2011, 41, 480–492. [Google Scholar] [CrossRef]
- Li, S.; Wei, T.; Panchenko, A.R. Histone Variant H2A.Z Modulates Nucleosome Dynamics to Promote DNA Accessibility. Nat. Commun. 2023, 14, 769. [Google Scholar] [CrossRef] [PubMed]
- Kniazeva, A.S.; Armeev, G.A.; Shaytan, A.K. H2A-H2B Histone Dimer Plasticity and Its Functional Implications. Cells 2022, 11, 2837. [Google Scholar] [CrossRef]
- Dion, M.F.; Kaplan, T.; Kim, M.; Buratowski, S.; Friedman, N.; Rando, O.J. Dynamics of Replication-Independent Histone Turnover in Budding Yeast. Science 2007, 315, 1405–1408. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, T.; Liu, C.L.; Erkmann, J.A.; Holik, J.; Grunstein, M.; Kaufman, P.D.; Friedman, N.; Rando, O.J. Cell Cycle– and Chaperone-Mediated Regulation of H3K56ac Incorporation in Yeast. PLoS Genet. 2008, 4, e1000270. [Google Scholar] [CrossRef]
- Rufiange, A.; Jacques, P.-É.; Bhat, W.; Robert, F.; Nourani, A. Genome-Wide Replication-Independent Histone H3 Exchange Occurs Predominantly at Promoters and Implicates H3 K56 Acetylation and Asf1. Mol. Cell 2007, 27, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Resch, M.; Lilyestrom, W.; Clark, N.; Hansen, J.C.; Peterson, C.; Luger, K. Structural Characterization of H3K56Q Nucleosomes and Nucleosomal Arrays. Biochim. Et. Biophys. Acta (BBA)—Gene Regul. Mech. 2010, 1799, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Neumann, H.; Hancock, S.M.; Buning, R.; Routh, A.; Chapman, L.; Somers, J.; Owen-Hughes, T.; van Noort, J.; Rhodes, D.; Chin, J.W. A Method for Genetically Installing Site-Specific Acetylation in Recombinant Histones Defines the Effects of H3 K56 Acetylation. Mol. Cell 2009, 36, 153–163. [Google Scholar] [CrossRef]
- Ferreira, H.; Somers, J.; Webster, R.; Flaus, A.; Owen-Hughes, T. Histone Tails and the H3 AlphaN Helix Regulate Nucleosome Mobility and Stability. Mol. Cell Biol. 2007, 27, 4037–4048. [Google Scholar] [CrossRef]
- Masumoto, H.; Hawke, D.; Kobayashi, R.; Verreault, A. A Role for Cell-Cycle-Regulated Histone H3 Lysine 56 Acetylation in the DNA Damage Response. Nature 2005, 436, 294–298. [Google Scholar] [CrossRef]
- Topal, S.; Vasseur, P.; Radman-Livaja, M.; Peterson, C.L. Distinct Transcriptional Roles for Histone H3-K56 Acetylation During the Cell Cycle in Yeast. Nat. Commun. 2019, 10, 4372. [Google Scholar] [CrossRef]
- Tramantano, M.; Sun, L.; Au, C.; Labuz, D.; Liu, Z.; Chou, M.; Shen, C.; Luk, E. Constitutive Turnover of Histone H2A.Z at Yeast Promoters Requires the Preinitiation Complex. eLife 2016, 5, e14243. [Google Scholar] [CrossRef]
- Mohan, C.; Kim, L.M.; Hollar, N.; Li, T.; Paulissen, E.; Leung, C.T.; Luk, E. VivosX, a Disulfide Crosslinking Method to Capture Site-Specific, Protein-Protein Interactions in Yeast and Human Cells. eLife 2018, 7, e36654. [Google Scholar] [CrossRef]
- Ranjan, A.; Nguyen, V.Q.; Liu, S.; Wisniewski, J.; Kim, J.M.; Tang, X.; Mizuguchi, G.; Elalaoui, E.; Nickels, T.J.; Jou, V.; et al. Live-Cell Single Particle Imaging Reveals the Role of RNA Polymerase II in Histone H2A.Z Eviction. eLife 2020, 9, e55667. [Google Scholar] [CrossRef]
- Mason, P.B.; Struhl, K. The FACT Complex Travels with Elongating RNA Polymerase II and Is Important for the Fidelity of Transcriptional Initiation In Vivo. Mol. Cell Biol. 2003, 23, 8323–8333, Erratum in Mol. Cell Biol. 2004, 24, 6536. [Google Scholar] [CrossRef] [PubMed]
- Mylonas, C.; Tessarz, P. NET-Prism Enables RNA Polymerase-Dedicated Transcriptional Interrogation at Nucleotide Resolution. RNA Biol. 2019, 16, 1156–1165. [Google Scholar] [CrossRef] [PubMed]
- Jeronimo, C.; Angel, A.; Nguyen, V.Q.; Kim, J.M.; Poitras, C.; Lambert, E.; Collin, P.; Mellor, J.; Wu, C.; Robert, F. FACT Is Recruited to the +1 Nucleosome of Transcribed Genes and Spreads in a Chd1-Dependent Manner. Mol. Cell 2021, 81, 3542–3559.e11. [Google Scholar] [CrossRef] [PubMed]
- Formosa, T. The Role of FACT in Making and Breaking Nucleosomes. Biochim. Et Biophys. Acta (BBA)—Gene Regul. Mech. 2012, 1819, 247–255. [Google Scholar] [CrossRef]
- Grasser, K.D. The FACT Histone Chaperone: Tuning Gene Transcription in the Chromatin Context to Modulate Plant Growth and Development. Front. Plant Sci. 2020, 11, 85. [Google Scholar] [CrossRef]
- Gurova, K.; Chang, H.W.; Valieva, M.E.; Sandlesh, P.; Studitsky, V.M. Structure and Function of the Histone Chaperone FACT—Resolving FACTual Issues. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 892–904. [Google Scholar] [CrossRef]
- Xin, H.; Takahata, S.; Blanksma, M.; McCullough, L.; Stillman, D.J.; Formosa, T. YFACT Induces Global Accessibility of Nucleosomal DNA without H2A-H2B Displacement. Mol. Cell 2009, 35, 365–376. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, K.; Zhang, N.; Wei, H.; Tan, Y.Z.; Zhang, Z.; Carragher, B.; Potter, C.S.; D’Arcy, S.; Luger, K. FACT Caught in the Act of Manipulating the Nucleosome. Nature 2020, 577, 426–431. [Google Scholar] [CrossRef]
- Mayanagi, K.; Saikusa, K.; Miyazaki, N.; Akashi, S.; Iwasaki, K.; Nishimura, Y.; Morikawa, K.; Tsunaka, Y. Structural Visualization of Key Steps in Nucleosome Reorganization by Human FACT. Sci. Rep. 2019, 9, 10183. [Google Scholar] [CrossRef] [PubMed]
- Valieva, M.E.; Armeev, G.A.; Kudryashova, K.S.; Gerasimova, N.S.; Shaytan, A.K.; Kulaeva, O.I.; McCullough, L.L.; Formosa, T.; Georgiev, P.G.; Kirpichnikov, M.P.; et al. Large-Scale ATP-Independent Nucleosome Unfolding by a Histone Chaperone. Nat. Struct. Mol. Biol. 2016, 23, 1111–1116. [Google Scholar] [CrossRef]
- Sivkina, A.L.; Karlova, M.G.; Valieva, M.E.; McCullough, L.L.; Formosa, T.; Shaytan, A.K.; Feofanov, A.V.; Kirpichnikov, M.P.; Sokolova, O.S.; Studitsky, V.M. Electron Microscopy Analysis of ATP-Independent Nucleosome Unfolding by FACT. Commun. Biol. 2022, 5, 2. [Google Scholar] [CrossRef]
- Jeronimo, C.; Watanabe, S.; Kaplan, C.D.; Peterson, C.L.; Robert, F. The Histone Chaperones FACT and Spt6 Restrict H2A.Z from Intragenic Locations. Mol. Cell 2015, 58, 1113–1123. [Google Scholar] [CrossRef]
- Lee, K.P.; Baxter, H.J.; Guillemette, J.G.; Lawford, H.G.; Lewis, P.N. Structural Studies on Yeast Nucleosomes. Can. J. Biochem. 1982, 60, 379–388. [Google Scholar] [CrossRef]
- Leung, A.; Cheema, M.; González-Romero, R.; Eirin-Lopez, J.M.; Ausió, J.; Nelson, C.J. Unique Yeast Histone Sequences Influence Octamer and Nucleosome Stability. FEBS Lett. 2016, 590, 2629–2638. [Google Scholar] [CrossRef]
- Truong, D.M.; Boeke, J.D. Resetting the Yeast Epigenome with Human Nucleosomes. Cell 2017, 171, 1508–1519.e13. [Google Scholar] [CrossRef]
- Maluchenko, N.V.; Nilov, D.K.; Pushkarev, S.V.; Kotova, E.Y.; Gerasimova, N.S.; Kirpichnikov, M.P.; Langelier, M.-F.; Pascal, J.M.; Akhtar, M.d.S.; Feofanov, A.V.; et al. Mechanisms of Nucleosome Reorganization by PARP1. Int. J. Mol. Sci. 2021, 22, 12127. [Google Scholar] [CrossRef] [PubMed]
- Armeev, G.A.; Moiseenko, A.V.; Motorin, N.A.; Afonin, D.A.; Zhao, L.; Vasilev, V.A.; Oleinikov, P.D.; Glukhov, G.S.; Peters, G.S.; Studitsky, V.M.; et al. Structure and Dynamics of a Nucleosome Core Particle Based on Widom 603 DNA Sequence. Structure 2025, 33, 948–959.e5. [Google Scholar] [CrossRef]
- Winogradoff, D.; Aksimentiev, A. Molecular Mechanism of Spontaneous Nucleosome Unraveling. J. Mol. Biol. 2019, 431, 323–335. [Google Scholar] [CrossRef] [PubMed]
- McCullough, L.L.; Pham, T.H.; Parnell, T.J.; Connell, Z.; Chandrasekharan, M.B.; Stillman, D.J.; Formosa, T. Establishment and Maintenance of Chromatin Architecture Are Promoted Independently of Transcription by the Histone Chaperone FACT and H3-K56 Acetylation in Saccharomyces Cerevisiae. Genetics 2019, 211, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Heo, K.; Kim, H.; Choi, S.H.; Choi, J.; Kim, K.; Gu, J.; Lieber, M.R.; Yang, A.S.; An, W. FACT-Mediated Exchange of Histone Variant H2AX Regulated by Phosphorylation of H2AX and ADP-Ribosylation of Spt16. Mol. Cell 2008, 30, 86–97. [Google Scholar] [CrossRef]
- Gershon, L.; Kupiec, M. The Amazing Acrobat: Yeast’s Histone H3K56 Juggles Several Important Roles While Maintaining Perfect Balance. Genes 2021, 12, 342. [Google Scholar] [CrossRef] [PubMed]
- Kulaeva, O.I.; Gaykalova, D.A.; Pestov, N.A.; Golovastov, V.V.; Vassylyev, D.G.; Artsimovitch, I.; Studitsky, V.M. Mechanism of Chromatin Remodeling and Recovery During Passage of RNA Polymerase II. Nat. Struct. Mol. Biol. 2009, 16, 1272–1278. [Google Scholar] [CrossRef]
- Kulaeva, O.I.; Gaykalova, D.A.; Studitsky, V.M. Transcription Through Chromatin by RNA Polymerase II: Histone Displacement and Exchange. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2007, 618, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Kulaeva, O.I.; Studitsky, V.M. Mechanism of Histone Survival During Transcription by RNA Polymerase II. Transcription 2010, 1, 85–88. [Google Scholar] [CrossRef][Green Version]
- Kulaeva, O.I.; Hsieh, F.-K.; Studitsky, V.M. RNA Polymerase Complexes Cooperate to Relieve the Nucleosomal Barrier and Evict Histones. Proc. Natl. Acad. Sci. USA 2010, 107, 11325–11330. [Google Scholar] [CrossRef]
- Hsieh, F.-K.; Kulaeva, O.I.; Patel, S.S.; Dyer, P.N.; Luger, K.; Reinberg, D.; Studitsky, V.M. Histone Chaperone FACT Action During Transcription Through Chromatin by RNA Polymerase II. Proc. Natl. Acad. Sci. USA 2013, 110, 7654–7659. [Google Scholar] [CrossRef]
- Chang, H.-W.; Kulaeva, O.I.; Shaytan, A.K.; Kibanov, M.; Kuznedelov, K.; Severinov, K.V.; Kirpichnikov, M.P.; Clark, D.J.; Studitsky, V.M. Analysis of the Mechanism of Nucleosome Survival During Transcription. Nucleic Acids Res. 2014, 42, 1619–1627. [Google Scholar] [CrossRef]
- Chang, H.-W.; Valieva, M.E.; Safina, A.; Chereji, R.V.; Wang, J.; Kulaeva, O.I.; Morozov, A.V.; Kirpichnikov, M.P.; Feofanov, A.V.; Gurova, K.V.; et al. Mechanism of FACT Removal from Transcribed Genes by Anticancer Drugs Curaxins. Sci. Adv. 2018, 4, eaav2131. [Google Scholar] [CrossRef]
- Kotova, E.Y.; Hsieh, F.-K.; Chang, H.-W.; Maluchenko, N.V.; Langelier, M.-F.; Pascal, J.M.; Luse, D.S.; Feofanov, A.V.; Studitsky, V.M. Human PARP1 Facilitates Transcription Through a Nucleosome and Histone Displacement by Pol II In Vitro. Int. J. Mol. Sci. 2022, 23, 7107. [Google Scholar] [CrossRef]
- Han, J.; Zhou, H.; Li, Z.; Xu, R.-M.; Zhang, Z. The Rtt109-Vps75 Histone Acetyltransferase Complex Acetylates Non-Nucleosomal Histone H3. J. Biol. Chem. 2007, 282, 14158–14164. [Google Scholar] [CrossRef]
- Han, J.; Zhou, H.; Horazdovsky, B.; Zhang, K.; Xu, R.-M.; Zhang, Z. Rtt109 Acetylates Histone H3 Lysine 56 and Functions in DNA Replication. Science 2007, 315, 653–655. [Google Scholar] [CrossRef]
- Tsubota, T.; Berndsen, C.E.; Erkmann, J.A.; Smith, C.L.; Yang, L.; Freitas, M.A.; Denu, J.M.; Kaufman, P.D. Histone H3-K56 Acetylation Is Catalyzed by Histone Chaperone-Dependent Complexes. Mol. Cell 2007, 25, 703–712. [Google Scholar] [CrossRef]
- Fillingham, J.; Recht, J.; Silva, A.C.; Suter, B.; Emili, A.; Stagljar, I.; Krogan, N.J.; Allis, C.D.; Keogh, M.-C.; Greenblatt, J.F. Chaperone Control of the Activity and Specificity of the Histone H3 Acetyltransferase Rtt109. Mol. Cell Biol. 2008, 28, 4342–4353. [Google Scholar] [CrossRef] [PubMed]
- Berndsen, C.E.; Denu, J.M. Catalysis and Substrate Selection by Histone/Protein Lysine Acetyltransferases. Curr. Opin. Struct. Biol. 2008, 18, 682–689. [Google Scholar] [CrossRef]
- Rege, M.; Subramanian, V.; Zhu, C.; Hsieh, T.-H.S.; Weiner, A.; Friedman, N.; Clauder-Münster, S.; Steinmetz, L.M.; Rando, O.J.; Boyer, L.A.; et al. Chromatin Dynamics and the RNA Exosome Function in Concert to Regulate Transcriptional Homeostasis. Cell Rep. 2015, 13, 1610–1622. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhou, H.; Wurtele, H.; Davies, B.; Horazdovsky, B.; Verreault, A.; Zhang, Z. Acetylation of Histone H3 Lysine 56 Regulates Replication-Coupled Nucleosome Assembly. Cell 2008, 134, 244–255. [Google Scholar] [CrossRef]
- Su, L.; Xia, W.; Shen, T.; Liang, Q.; Wang, W.; Li, H.; Jiao, J. H2A.Z.1 Crosstalk with H3K56-Acetylation Controls Gliogenesis Through the Transcription of Folate Receptor. Nucleic Acids Res. 2018, 46, 8817–8831. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-H.; Alami, S.; Luk, E.; Wu, C.-H.; Sen, S.; Mizuguchi, G.; Wei, D.; Wu, C. Swc2 Is a Widely Conserved H2AZ-Binding Module Essential for ATP-Dependent Histone Exchange. Nat. Struct. Mol. Biol. 2005, 12, 1064–1071. [Google Scholar] [CrossRef]
- Luk, E.; Ranjan, A.; FitzGerald, P.C.; Mizuguchi, G.; Huang, Y.; Wei, D.; Wu, C. Stepwise Histone Replacement by SWR1 Requires Dual Activation with Histone H2A.Z and Canonical Nucleosome. Cell 2010, 143, 725–736. [Google Scholar] [CrossRef]
- Watanabe, S.; Radman-Livaja, M.; Rando, O.J.; Peterson, C.L. A Histone Acetylation Switch Regulates H2A.Z Deposition by the SWR-C Remodeling Enzyme. Science 2013, 340, 195–199. [Google Scholar] [CrossRef]
- Papamichos-Chronakis, M.; Watanabe, S.; Rando, O.J.; Peterson, C.L. Global Regulation of H2A.Z Localization by the INO80 Chromatin-Remodeling Enzyme Is Essential for Genome Integrity. Cell 2011, 144, 200–213. [Google Scholar] [CrossRef]
- Jeronimo, C.; Poitras, C.; Robert, F. Histone Recycling by FACT and Spt6 During Transcription Prevents the Scrambling of Histone Modifications. Cell Rep. 2019, 28, 1206–1218.e8. [Google Scholar] [CrossRef] [PubMed]
- Formosa, T.; Eriksson, P.; Wittmeyer, J.; Ginn, J.; Yu, Y.; Stillman, D.J. Spt16-Pob3 and the HMG Protein Nhp6 Combine to Form the Nucleosome-Binding Factor SPN. EMBO J. 2001, 20, 3506–3517. [Google Scholar] [CrossRef] [PubMed]
- Rhee, H.S.; Pugh, B.F. Genome-Wide Structure and Organization of Eukaryotic Pre-Initiation Complexes. Nature 2012, 483, 295–301, Erratum in Nature 2012, 487, 128. [Google Scholar] [CrossRef]
- True, J.D.; Muldoon, J.J.; Carver, M.N.; Poorey, K.; Shetty, S.J.; Bekiranov, S.; Auble, D.T. The Modifier of Transcription 1 (Mot1) ATPase and Spt16 Histone Chaperone Co-Regulate Transcription Through Preinitiation Complex Assembly and Nucleosome Organization. J. Biol. Chem. 2016, 291, 15307–15319. [Google Scholar] [CrossRef] [PubMed]
- Takahata, S.; Yu, Y.; Stillman, D.J. FACT and Asf1 Regulate Nucleosome Dynamics and Coactivator Binding at the HO Promoter. Mol. Cell 2009, 34, 405–415. [Google Scholar] [CrossRef]
- Ransom, M.; Williams, S.K.; Dechassa, M.L.; Das, C.; Linger, J.; Adkins, M.; Liu, C.; Bartholomew, B.; Tyler, J.K. FACT and the Proteasome Promote Promoter Chromatin Disassembly and Transcriptional Initiation. J. Biol. Chem. 2009, 284, 23461–23471. [Google Scholar] [CrossRef]
- Davey, C.A.; Sargent, D.F.; Luger, K.; Maeder, A.W.; Richmond, T.J. Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9Å Resolution. J. Mol. Biol. 2002, 319, 1097–1113. [Google Scholar] [CrossRef]
- Hall, M.A.; Shundrovsky, A.; Bai, L.; Fulbright, R.M.; Lis, J.T.; Wang, M.D. High-Resolution Dynamic Mapping of Histone-DNA Interactions in a Nucleosome. Nat. Struct. Mol. Biol. 2009, 16, 124–129. [Google Scholar] [CrossRef]
- Farnung, L.; Ochmann, M.; Engeholm, M.; Cramer, P. Structural Basis of Nucleosome Transcription Mediated by Chd1 and FACT. Nat. Struct. Mol. Biol. 2021, 28, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Bajwa, P.; Johnson, F.C.; Bhaumik, S.R.; Shilatifard, A. Rtt109 Is Required for Proper H3K56 Acetylation. J. Biol. Chem. 2006, 281, 37270–37274. [Google Scholar] [CrossRef]
- Mylonas, C.; Tessarz, P. Transcriptional Repression by FACT Is Linked to Regulation of Chromatin Accessibility at the Promoter of ES Cells. Life Sci. Alliance 2018, 1, e201800085. [Google Scholar] [CrossRef]
- Luger, K.; Rechsteiner, T.J.; Richmond, T.J. Preparation of Nucleosome Core Particle from Recombinant Histones. Methods Enzym. 1999, 304, 3–19. [Google Scholar] [CrossRef]
- Ruone, S.; Rhoades, A.R.; Formosa, T. Multiple Nhp6 Molecules Are Required to Recruit Spt16-Pob3 to Form YFACT Complexes and to Reorganize Nucleosomes. J. Biol. Chem. 2003, 278, 45288–45295. [Google Scholar] [CrossRef] [PubMed]
- Biswas, D.; Yu, Y.; Prall, M.; Formosa, T.; Stillman, D.J. The Yeast FACT Complex Has a Role in Transcriptional Initiation. Mol. Cell Biol. 2005, 25, 5812–5822. [Google Scholar] [CrossRef]
- Koleske, A.J.; Chao, D.M.; Young, R.A. Purification of Yeast RNA Polymerase II Holoenzymes. Methods Enzym. 1996, 273, 176–184. [Google Scholar] [CrossRef]
- Lowary, P.T.; Widom, J. New DNA Sequence Rules for High Affinity Binding to Histone Octamer and Sequence-Directed Nucleosome Positioning. J. Mol. Biol. 1998, 276, 19–42. [Google Scholar] [CrossRef] [PubMed]
- Yap, E.P.; McGee, J.O. Short PCR Product Yields Improved by Lower Denaturation Temperatures. Nucleic Acids Res. 1991, 19, 1713. [Google Scholar] [CrossRef]
- Kudryashova, K.S.; Chertkov, O.V.; Nikitin, D.V.; Pestov, N.A.; Kulaeva, O.I.; Efremenko, A.V.; Solonin, A.S.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A. V Preparation of Mononucleosomal Templates for Analysis of Transcription with RNA Polymerase Using SpFRET. Methods Mol. Biol. 2015, 1288, 395–412. [Google Scholar] [CrossRef] [PubMed]
- Thåström, A.; Bingham, L.M.; Widom, J. Nucleosomal Locations of Dominant DNA Sequence Motifs for Histone–DNA Interactions and Nucleosome Positioning. J. Mol. Biol. 2004, 338, 695–709. [Google Scholar] [CrossRef]
- Gaykalova, D.A.; Kulaeva, O.I.; Bondarenko, V.A.; Studitsky, V.M. Preparation and Analysis of Uniquely Positioned Mononucleosomes. Methods Mol. Biol. 2009, 523, 109–123. [Google Scholar] [CrossRef]
- Kireeva, M.L.; Komissarova, N.; Waugh, D.S.; Kashlev, M. The 8-Nucleotide-Long RNA:DNA Hybrid Is a Primary Stability Determinant of the RNA Polymerase II Elongation Complex. J. Biol. Chem. 2000, 275, 6530–6536. [Google Scholar] [CrossRef] [PubMed]
- Kireeva, M.L.; Walter, W.; Tchernajenko, V.; Bondarenko, V.; Kashlev, M.; Studitsky, V.M. Nucleosome Remodeling Induced by RNA Polymerase II: Loss of the H2A/H2B Dimer During Transcription. Mol. Cell 2002, 9, 541–552. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afonin, D.; Ukrainets, E.R.; Kotova, E.; Gerasimova, N.S.; Armeev, G.A.; Kirpichnikov, M.P.; Feofanov, A.V.; Studitsky, V.M. H2A.Z and H3:K56Q Affect Transcription Through Chromatin and Yeast FACT-Dependent Nucleosome Unfolding. Int. J. Mol. Sci. 2025, 26, 10887. https://doi.org/10.3390/ijms262210887
Afonin D, Ukrainets ER, Kotova E, Gerasimova NS, Armeev GA, Kirpichnikov MP, Feofanov AV, Studitsky VM. H2A.Z and H3:K56Q Affect Transcription Through Chromatin and Yeast FACT-Dependent Nucleosome Unfolding. International Journal of Molecular Sciences. 2025; 26(22):10887. https://doi.org/10.3390/ijms262210887
Chicago/Turabian StyleAfonin, Dmitrii, Elizaveta R. Ukrainets, Elena Kotova, Nadezhda S. Gerasimova, Grigoriy A. Armeev, Mikhail P. Kirpichnikov, Alexey V. Feofanov, and Vasily M. Studitsky. 2025. "H2A.Z and H3:K56Q Affect Transcription Through Chromatin and Yeast FACT-Dependent Nucleosome Unfolding" International Journal of Molecular Sciences 26, no. 22: 10887. https://doi.org/10.3390/ijms262210887
APA StyleAfonin, D., Ukrainets, E. R., Kotova, E., Gerasimova, N. S., Armeev, G. A., Kirpichnikov, M. P., Feofanov, A. V., & Studitsky, V. M. (2025). H2A.Z and H3:K56Q Affect Transcription Through Chromatin and Yeast FACT-Dependent Nucleosome Unfolding. International Journal of Molecular Sciences, 26(22), 10887. https://doi.org/10.3390/ijms262210887






