Population Genetic Structure of Historic Olives (Olea europaea subsp. europaea) from Jordan
Abstract
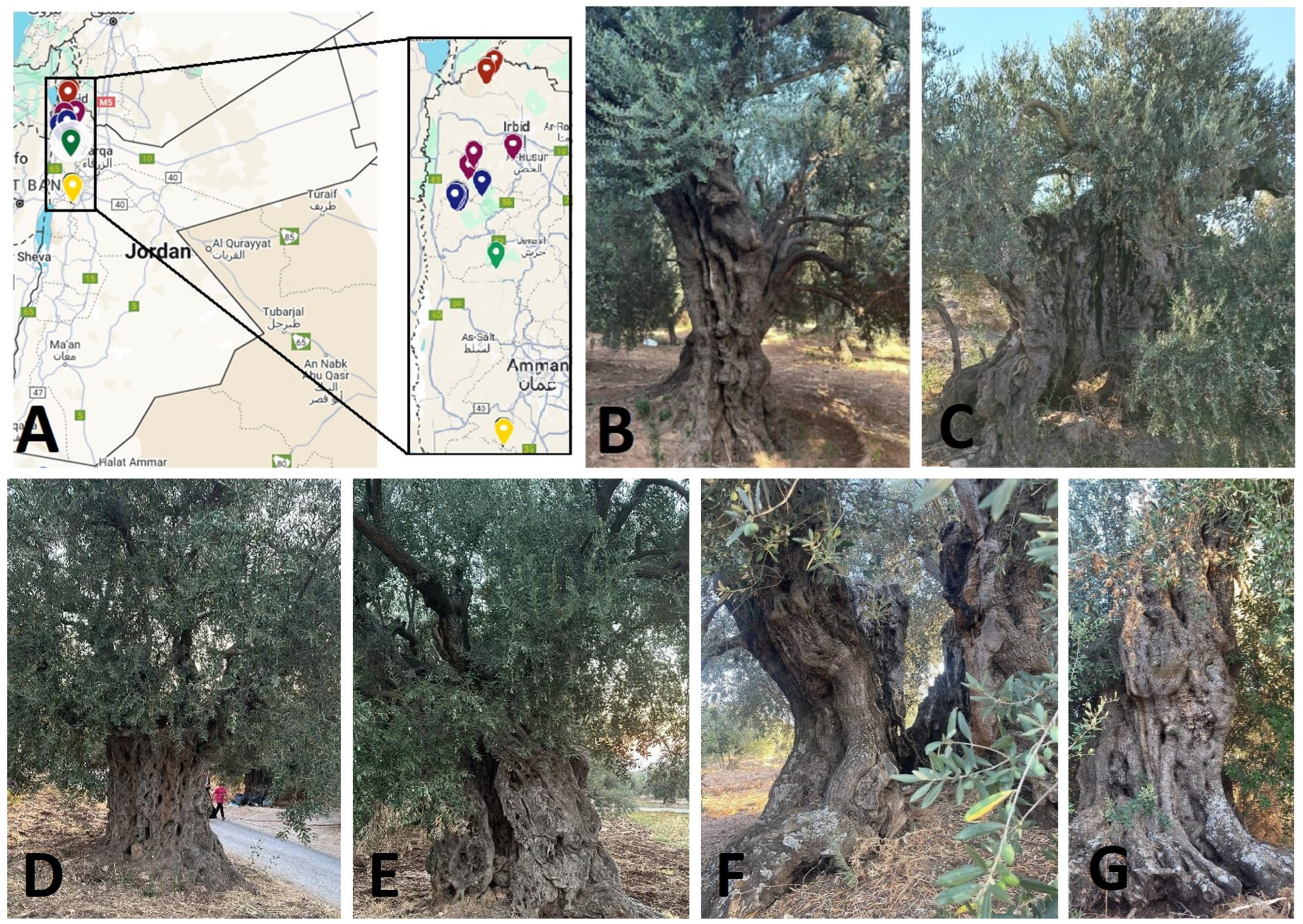
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. DNA Extraction
4.2. DNA Amplification
4.3. DNA Data Analysis and Interpretation
4.4. Phenotypic Data
4.5. Correlation Between Phenotypic and ISSR Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A | Ajloun |
| Cj | probability of confusion |
| CTAB | Cetyl Trimethyl Ammonium Bromide |
| D | discriminating power |
| EDTA | ethylenediaminetetraacetic acid |
| F | ‘Frantoia’ |
| i | Irbid |
| ISSRs | inter-simple sequence repeats |
| J | Jerash |
| M | ‘Mehras’ |
| N | ‘Nabali’ |
| NARC | National Agricultural Research Center |
| NGS | Next generattion sequencing |
| O | ‘Manzanillo’ |
| PCR | Polymerase Chain Reaction |
| PIC | Polymorphism Information Content |
| RAPD | random amplified polymorphic DNA |
| SNP | Single nucleiotide polymorphism |
| UPGMA | Unweighted Pair Group Method with Arithmetic Mean |
References
- Lavee, S.; Navero, D.B.; Bongi, G.; Jardak, T.; Loussert, R.; Martin, G.C.; Trigui, A. Biology and physiology of the olive. In World Olive Encyclopedia; The International Olive Council: Madrid, Spain, 1996. [Google Scholar]
- Department of Statistics, Jordan. 2025. Available online: https://dosweb.dos.gov.jo/ (accessed on 1 April 2025).
- Al-Khalifah, N.S.; Askari, E.; El-Kholy, M. Following Olive Footprints in Saudi Arabia. Following Olive Footprints (Olea europaea L.) Cultivation and Culture, Folklore and History, Traditions and Uses; (Scripta Horticulturae N. 13); CABI: Oxfordshire, UK, 2012. [Google Scholar]
- Dighton, A.; Fairbairn, A.; Bourke, S.; Faith, J.T.; Habgood, P. Bronze Age olive domestication in the north Jordan valley: New morphological evidence for regional complexity in early arboricultural practice from Pella in Jordan. Veg. Hist. Archaeobot. 2017, 26, 403–413. [Google Scholar] [CrossRef]
- Barazani, O.; Dag, A.; Dunseth, Z. The history of olive cultivation in the southern Levant. Front. Plant Sci. 2023, 14, 1131557. [Google Scholar] [CrossRef]
- Handziuk, N.M. Olive Oil and Urban Beginnings: A perspective from Early Bronze Age Northern Jordan. Doctoral Dissertation, University of Toronto, Toronto, ON, Canada, 2024. [Google Scholar]
- Al-Maaitah, M.; Al-Shdiefat, S.; Ayoub, S.; Al-Batsh, J.; Kattan, U.; Al-Julani, E. Jordan, where olive trees grew long before the advent of civilization. Olivae 2022, 129. Available online: https://www.internationaloliveoil.org/product/olivae-129-english-edition/ (accessed on 6 November 2025).
- Haddad, N.; Migdadi, H.; Brake, M.; Ayoub, S.; Obeidat, W.; Abusini, Y.; Aburumman, A.; Al-Shagour, B.; Al-Anasweh, E.; Sadder, M. Complete chloroplast genome sequence of historical olive (Olea europaea subsp. europaea) cultivar Mehras, in Jordan. Mitochondrial DNA Part B 2021, 6, 194–195. [Google Scholar] [CrossRef]
- Fabbri, A.; Lambardi, M.; Ozden-Tokatli, Y. Olive breeding. In Breeding Plantation Tree Crops: Tropical Species; Springer International Publishing: Cham, Switzerland, 2009; pp. 423–465. [Google Scholar]
- Aïachi Mezghani, M.; Laaribi, I.; Zouari, I.; Mguidich, A. Sustainability and plasticity of the olive tree cultivation in arid conditions. In Agriculture Productivity in Tunisia Under Stressed Environment; Springer International Publishing: Cham, Switzerland, 2021; pp. 27–56. [Google Scholar]
- Vollmann, J.; Rajcan, I. Handbook of Plant Breeding: Oil Crops; Springer International Publishing: Cham, Switzerland, 2009. [Google Scholar]
- Ateyyeh, A.F.; Sadder, M.T. Phylogenic clustering of three Jordanian olive (Olea europaea L.) cultivars within international collection. Adv. Hort. Sci. 2006, 20, 170–174. [Google Scholar]
- Ziar, C.; Amr, A.; Sadder, M.T. Genetic diversity and genome structure of historic Mediterranean olives. South-West. J. Hortic. Biol. Environ. 2024, 15, 39–51. [Google Scholar]
- Alkhatatbeh, H.; Sadder, M.; Haddad, N.; Al-Amad, I.; Brake, M.; Alsakarneh, N.; Alnajjar, A. Transcriptome analysis of historic olives reveals stress-specific biomarkers. Front. Plant Sci. 2025, 16, 1549305. [Google Scholar] [CrossRef]
- Sadder, M.T.; Ateyyeh, A.F.; Alswalmah, H.; Zakri, A.M.; Alsadon, A.A.; Al-Doss, A.A. Characterization of putative salinity-responsive biomarkers in olive (Olea europaea L.). Plant Genet. Resour. 2021, 19, 133–143. [Google Scholar] [CrossRef]
- Diez, C.M.; Trujillo, I.; Martinez-Urdiroz, N.; Barranco, D.; Rallo, L.; Marfil, P.; Gaut, B.S. Olive domestication and diversification in the Mediterranean Basin. New Phytol. 2015, 206, 436–447. [Google Scholar] [CrossRef]
- Bazakos, C.; Alexiou, K.G.; Ramos-Onsins, S.; Koubouris, G.; Tourvas, N.; Xanthopoulou, A.; Mellidou, I.; Moysiadis, T.; Vourlaki, I.-T.; Metzidakis, I.; et al. Whole genome scanning of a Mediterranean basin hotspot collection provides new insights into olive tree biodiversity and biology. Plant J. 2023, 116, 303–319. [Google Scholar] [CrossRef]
- Belaj, A.; Ninot, A.; Gómez-Gálvez, F.J.; El Riachy, M.; Gurbuz-Veral, M.; Torres, M.; Lazaj, A.; Klepo, T.; Paz, S.; Ugarte, J.; et al. Utility of EST-SNP markers for improving management and use of olive genetic resources: A case study at the Worldwide Olive Germplasm Bank of Córdoba. Plants 2022, 11, 921. [Google Scholar] [CrossRef]
- Gómez-Rodríguez, M.V.; Beuzon, C.; González-Plaza, J.J.; Fernández-Ocaña, A.M. Identification of an olive (Olea europaea L.) core collection with a new set of SSR markers. Genet. Resour. Crop Evol. 2021, 68, 117–133. [Google Scholar] [CrossRef]
- Sanz-Cortés, F.; Parfitt, D.E.; Romero, C.; Struss, D.; Llácer, G.; Badenes, M.L. Intraspecific olive diversity assessed with AFLP. Plant Breed. 2003, 122, 173–177. [Google Scholar] [CrossRef]
- Besnard, G.; Baradat, P.H.; Chevalier, D.; Tagmount, A.; Bervillé, A. Genetic differentiation in the olive complex (Olea europaea) revealed by RAPDs and RFLPs in the rRNA genes. Genet. Resour. Crop Evol. 2001, 48, 165–182. [Google Scholar] [CrossRef]
- Bracci, T.; Busconi, M.; Fogher, C.; Sebastiani, L. Molecular studies in olive (Olea europaea L.): Overview on DNA markers applications and recent advances in genome analysis. Plant Cell Rep. 2011, 30, 449–462. [Google Scholar] [CrossRef]
- Sadder, M.; Brake, M.; Ayoub, S.; Abusini, Y.; Al-Amad, I.; Haddad, N. Complete mitochondrial genome sequence of historical olive (Olea europaea Linnaeus 1753 subsp. europaea) cultivar Mehras in Jordan. Mitochondrial DNA Part B 2023, 8, 1205–1208. [Google Scholar] [CrossRef] [PubMed]
- Amiteye, S. Basic concepts and methodologies of DNA marker systems in plant molecular breeding. Heliyon 2021, 7, e08093. [Google Scholar] [CrossRef]
- Brake, M.; Migdadi, H.; Al-Gharaibeh, M.; Ayoub, S.; Haddad, N.; El Oqlah, A. Characterization of Jordanian olive cultivars (Olea europaea L.) using RAPD and ISSR molecular markers. Sci. Hortic. 2014, 176, 282–289. [Google Scholar] [CrossRef]
- Undal, V.S.; Ahir, M.R. The methods of molecular markers in genome analysis: A brief biotechnological approach. In Advances in Biotechnology and Molecular Biology; Weser Books: Zittau, Germany, 2023; p. 149. [Google Scholar]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef]
- Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. Soc. Vaud. Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Roldan-Ruiz, I.; Dendauw, J.; Van Bockstaele, E.; Depicker, A.; De Loose, M. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol. Breed. 2000, 6, 125–134. [Google Scholar] [CrossRef]
- Tessier, C.; David, J.; This, P.; Boursiquot, J.M.; Charrier, A. Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theor. Appl. Genet. 1999, 98, 171–177. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Wen, X.; Falush, D. Documentation for Structure Software: Version 2.3; University of Chicago: Chicago, IL, USA, 2010; Volume 1, p. 37. [Google Scholar]
- Cicatelli, A.; Fortunati, T.; De Feis, I.; Castiglione, S. Oil composition and genetic biodiversity of ancient and new olive (Olea europea L.) varieties and accessions of southern Italy. Plant Sci. 2013, 210, 82–92. [Google Scholar] [CrossRef]
- Ramanujan, S. Modular equations and approximations to π. Q. J. Math. 1914, 45, 350–372. [Google Scholar]
- Russ, J.C. The Image Processing Handbook; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Oksanen, J. Vegan: Community Ecology Package, version 2.3; R Studio: Boston, MA, USA, 2015.
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [PubMed]
- Galili, T. dendextend: An R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
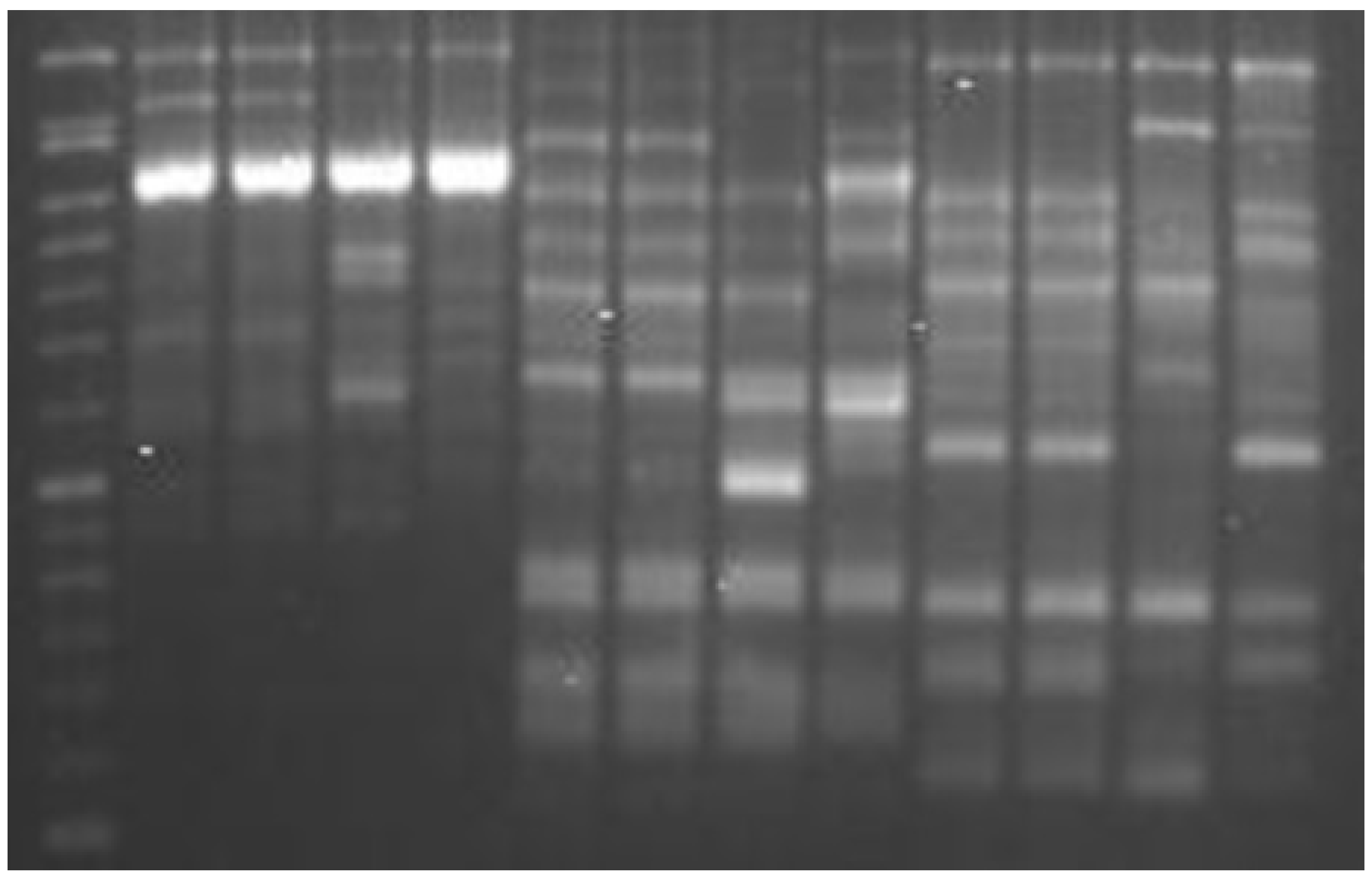
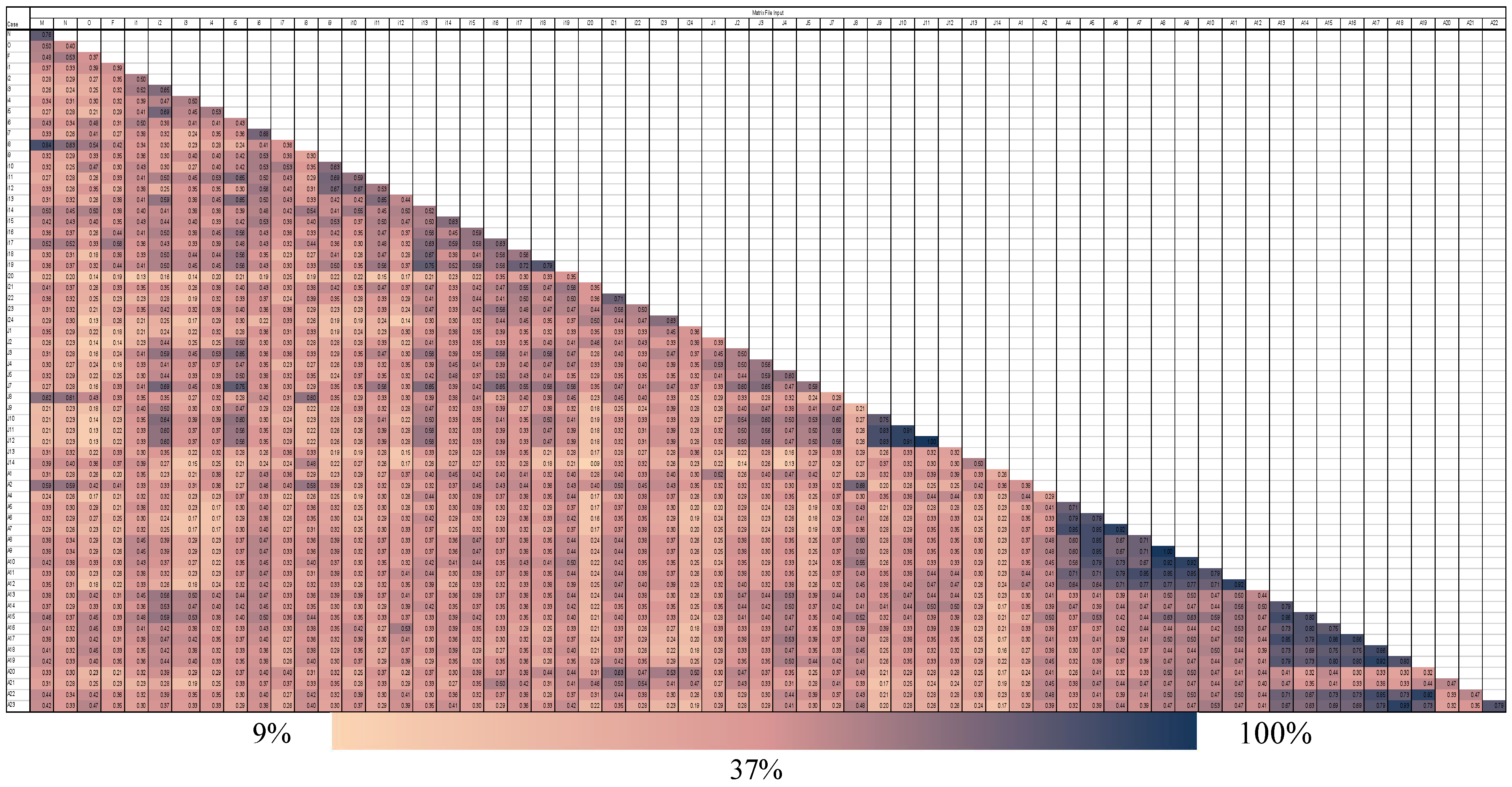
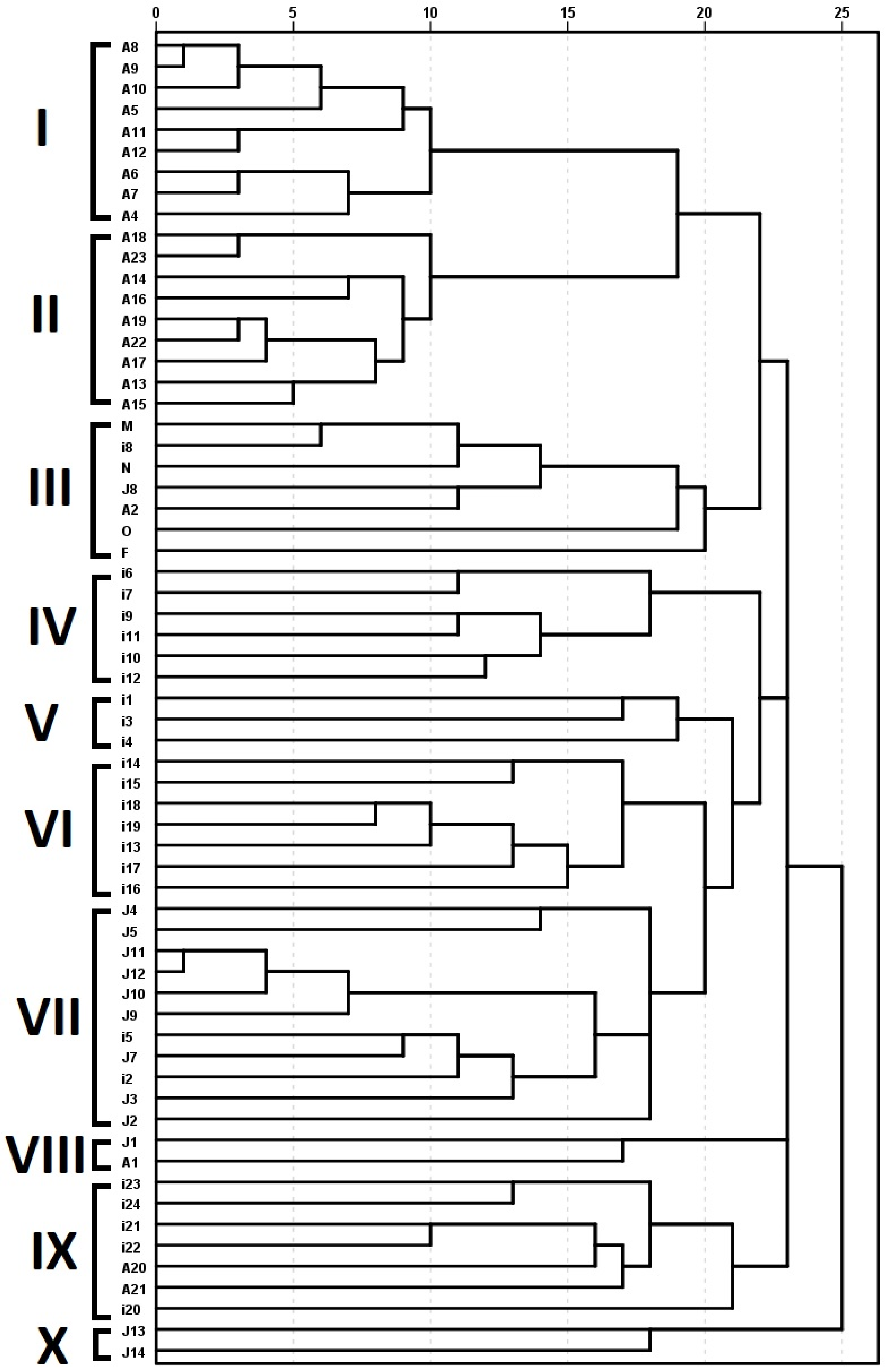
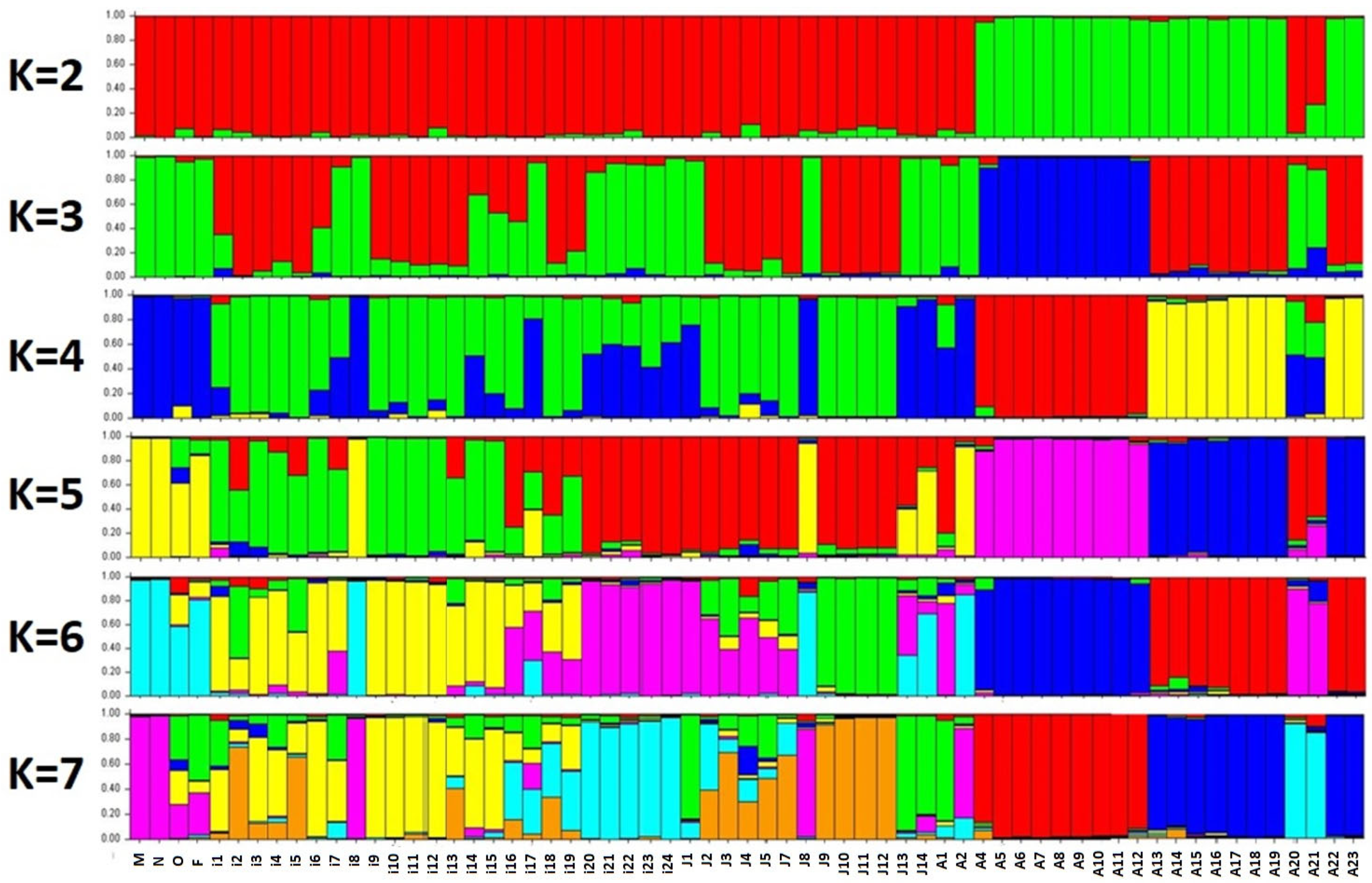




| Primer | Sequences (5′–3′) | Patterns * | D Value | Pic Value | Amplified Fragments | |
|---|---|---|---|---|---|---|
| Total | Polymorphic | |||||
| ISSR_807 | AGAGAGAGAGAGAGAGT | 63 | 0.70 | 0.27 | 17 | 16 |
| ISSR_810 | GAGAGAGAGAGAGAGAT | 63 | 0.61 | 0.28 | 17 | 17 |
| ISSR_825 | ACACACACACACACACT | 63 | 0.83 | 0.21 | 18 | 17 |
| Group | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||||||||
| M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | |
| Leaf length (cm) | 6.46 | 0.11 | 6.04 | 0.18 | 5.76 | 0.09 | 5.08 | 0.13 | 5.70 | 0.19 | 5.18 | 0.19 | 5.82 | 0.51 | 5.36 | 0.11 |
| Leaf width (cm) | 1.18 | 0.13 | 1.28 | 0.18 | 1.26 | 0.09 | 1.32 | 0.08 | 1.16 | 0.05 | 1.28 | 0.08 | 1.10 | 0.12 | 1.26 | 0.11 |
| Leaf area (cm2) | 3.66 | 0.37 | 4.90 | 0.54 | 4.45 | 0.26 | 3.43 | 0.21 | 2.58 | 0.28 | 3.58 | 0.59 | 4.60 | 0.55 | 3.82 | 1.04 |
| Leaf shape index | 5.52 | 0.54 | 4.78 | 0.56 | 4.59 | 0.24 | 3.86 | 0.16 | 4.92 | 0.16 | 4.05 | 0.14 | 5.30 | 0.19 | 4.28 | 0.31 |
| Leaf perimeter (cm) | 13.49 | 0.32 | 12.76 | 0.49 | 12.20 | 0.25 | 10.94 | 0.33 | 11.99 | 0.40 | 11.10 | 0.44 | 12.18 | 1.08 | 11.43 | 0.32 |
| Leaf roundness | 3.41 | 0.28 | 4.85 | 0.68 | 4.59 | 0.21 | 3.94 | 0.29 | 2.71 | 0.33 | 4.04 | 0.51 | 4.74 | 0.35 | 4.19 | 1.05 |
| Fruit length (mm) | 23.00 | 1.22 | 21.20 | 0.84 | 18.40 | 1.52 | 22.40 | 1.14 | 23.60 | 1.14 | 20.40 | 1.14 | 20.00 | 1.41 | 17.80 | 1.30 |
| Fruit width (mm) | 13.32 | 0.07 | 12.56 | 0.21 | 10.15 | 4.87 | 13.41 | 0.11 | 15.06 | 0.24 | 14.59 | 0.09 | 11.43 | 0.32 | 10.67 | 0.12 |
| Fruit shape index | 1.73 | 0.09 | 1.69 | 0.04 | 3.56 | 4.56 | 1.67 | 0.08 | 1.57 | 0.05 | 1.40 | 0.07 | 1.75 | 0.10 | 1.67 | 0.11 |
| Stone length (mm) | 16.80 | 0.84 | 15.80 | 1.92 | 17.00 | 1.58 | 15.40 | 1.14 | 14.40 | 1.82 | 14.40 | 1.14 | 20.00 | 1.00 | 14.80 | 0.84 |
| Stone width (mm) | 7.19 | 0.11 | 8.29 | 0.27 | 6.95 | 0.19 | 7.73 | 0.26 | 7.67 | 0.08 | 7.39 | 0.21 | 7.38 | 0.33 | 6.83 | 0.19 |
| Stone shape index | 2.34 | 0.08 | 1.90 | 0.18 | 2.44 | 0.17 | 1.99 | 0.08 | 1.88 | 0.22 | 1.95 | 0.10 | 2.71 | 0.09 | 2.17 | 0.07 |
| Flesh thickness (mm) | 3.07 | 0.03 | 2.14 | 0.06 | 1.60 | 2.38 | 2.84 | 0.10 | 3.70 | 0.09 | 3.60 | 0.07 | 2.03 | 0.05 | 1.92 | 0.07 |
| Fruit weight (g) | 1.65 | 0.17 | 1.66 | 0.15 | 1.83 | 0.24 | 2.11 | 0.16 | 2.21 | 0.37 | 1.99 | 0.19 | 3.66 | 0.19 | 1.25 | 0.08 |
| Stone weight (g) | 0.58 | 0.04 | 0.76 | 0.03 | 0.47 | 0.04 | 0.62 | 0.05 | 0.65 | 0.02 | 0.57 | 0.06 | 0.60 | 0.04 | 0.49 | 0.03 |
| Flesh weight (g) | 1.07 | 0.18 | 0.90 | 0.17 | 1.37 | 0.23 | 1.49 | 0.15 | 1.56 | 0.37 | 1.42 | 0.18 | 3.06 | 0.20 | 0.76 | 0.06 |
| Flesh % | 0.64 | 0.05 | 0.54 | 0.05 | 0.74 | 0.03 | 0.70 | 0.03 | 0.70 | 0.06 | 0.71 | 0.03 | 0.84 | 0.01 | 0.61 | 0.01 |
| Stone % | 0.36 | 0.05 | 0.46 | 0.05 | 0.26 | 0.03 | 0.30 | 0.03 | 0.30 | 0.06 | 0.29 | 0.03 | 0.16 | 0.01 | 0.39 | 0.01 |
| Flesh/stone ratio | 1.86 | 0.41 | 1.20 | 0.27 | 2.93 | 0.52 | 2.41 | 0.33 | 2.40 | 0.59 | 2.51 | 0.40 | 5.11 | 0.55 | 1.57 | 0.10 |
| Function | Eigenvalue | % of Variance | Cumulative % |
|---|---|---|---|
| 1 | 809.156 | 77.7 | 77.7 |
| 2 | 122.815 | 11.8 | 89.5 |
| 3 | 51.861 | 5.0 | 94.5 |
| 4 | 41.699 | 4.0 | 98.5 |
| Acc. | Region | Area | GPS | Acc. | Region | Area | GPS |
|---|---|---|---|---|---|---|---|
| M | Mshaqar | Mehras | 31.77394, 35.80324 | J5 | Jerash | Borma | 32.21637, 35.77961 |
| N | Mshaqar | Nabali | 31.77394, 35.80324 | J7 | Jerash | Borma | 32.21637, 35.77961 |
| O | Mshaqar | Manzanillo | 31.77394, 35.80324 | J8 | Jerash | Borma | 32.21637, 35.77961 |
| F | Mshaqar | Frantoio | 31.77394, 35.80324 | J9 | Jerash | Borma | 32.21637, 35.77961 |
| i1 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | J10 | Jerash | Borma | 32.21637, 35.77961 |
| i2 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | J11 | Jerash | Borma | 32.21637, 35.77961 |
| i3 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | J12 | Jerash | Borma | 32.21637, 35.77961 |
| i4 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | J13 | Jerash | Borma | 32.21637, 35.77961 |
| i5 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | J14 | Jerash | Borma | 32.21637, 35.77961 |
| i6 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A1 | Ajloun | Al Hashimiyya (Al mesier) | 32.36958, 35.65913 |
| i7 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A2 | Ajloun | Al Hashimiyya (Al mesier) | 32.36958, 35.65913 |
| i8 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A4 | Ajloun | Al Hashimiyya (Al mesier) | 32.36958, 35.65913 |
| i9 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A5 | Ajloun | Al Hashimiyya (Al mesier) | 32.36958, 35.65913 |
| i10 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A6 | Ajloun | Al Hashimiyya (Al mesier) | 32.36958, 35.65913 |
| i11 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A7 | Ajloun | Al Hashimiyya (Al mesier) | 32.36958, 35.65913 |
| i12 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A8 | Ajloun | Al Hashimiyya (Al mesier) | 32.36958, 35.65913 |
| i13 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A9 | Ajloun | Al Hashimiyya (Al mesier) | 32.36958, 35.65913 |
| i14 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A10 | Ajloun | Al Hashimiyya (Al mesier) | 32.36958, 35.65913 |
| i15 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A11 | Ajloun | Al Hashimiyya (Ashlaa) | 32.36147, 35.66961 |
| i16 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A12 | Ajloun | Al Hashimiyya (Ashlaa) | 32.36147, 35.66961 |
| i17 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A13 | Ajloun | Al Hashimiyya (Ashlaa) | 32.36147, 35.66961 |
| i18 | Irbid | Saham al Kaffarat | 32.70001, 35.77391 | A14 | Ajloun | Al Hashimiyya (Ashlaa) | 32.36147, 35.66961 |
| i19 | Irbid | Al-Korah | 32.44289, 35.70047 | A15 | Ajloun | Al Hashimiyya (Ashlaa) | 32.36147, 35.66961 |
| i20 | Irbid | Al-Korah | 32.43796, 35.69473 | A16 | Ajloun | Al Hashimiyya (Ashlaa) | 32.36147, 35.66961 |
| i21 | Irbid | Malka | 32.67751, 35.74889 | A17 | Ajloun | Orjan | 32.36291, 35.65904 |
| i22 | Irbid | Malka | 32.67589, 35.75027 | A18 | Ajloun | Al Hashimiyya (Amesier) | 32.36291, 35.65904 |
| i23 | Irbid | Der Abo Yousef | 32.48906, 35.83198 | A19 | Ajloun | Al Hashimiyya (Amesier) | 32.36207, 35.65798 |
| i24 | Irbid | Al-Korah | 32.46954, 35.71007 | A20 | Ajloun | Al Hashimiyya (Qarmash) | 32.36207, 35.65798 |
| J1 | Jerash | Borma | 32.21433, 35.78064 | A21 | Ajloun | Ajloun | 32.36432, 35.65283 |
| J2 | Jerash | Borma | 32.21433, 35.78064 | A22 | Ajloun | Wadi El Rayan | 32.39776, 35.73709 |
| J3 | Jerash | Borma | 32.21637, 35.77961 | A23 | Ajloun | Wadi El Rayan | 32.39776, 35.73709 |
| J4 | Jerash | Borma | 32.21637, 35.77961 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsakarneh, N.; Kayed, A.A.; Hammouh, F.; Alkhatatbeh, H.A.; Qutob, M.S.; Alkharabsheh, B.; Obeidat, W.M.; Ateyyeh, A.; Sadder, M.T. Population Genetic Structure of Historic Olives (Olea europaea subsp. europaea) from Jordan. Int. J. Mol. Sci. 2025, 26, 10863. https://doi.org/10.3390/ijms262210863
Alsakarneh N, Kayed AA, Hammouh F, Alkhatatbeh HA, Qutob MS, Alkharabsheh B, Obeidat WM, Ateyyeh A, Sadder MT. Population Genetic Structure of Historic Olives (Olea europaea subsp. europaea) from Jordan. International Journal of Molecular Sciences. 2025; 26(22):10863. https://doi.org/10.3390/ijms262210863
Chicago/Turabian StyleAlsakarneh, Nawal, Aseel Abu Kayed, Fadwa Hammouh, Hamad A. Alkhatatbeh, Maysoun S. Qutob, Bayan Alkharabsheh, Wisam M. Obeidat, Ahmad Ateyyeh, and Monther T. Sadder. 2025. "Population Genetic Structure of Historic Olives (Olea europaea subsp. europaea) from Jordan" International Journal of Molecular Sciences 26, no. 22: 10863. https://doi.org/10.3390/ijms262210863
APA StyleAlsakarneh, N., Kayed, A. A., Hammouh, F., Alkhatatbeh, H. A., Qutob, M. S., Alkharabsheh, B., Obeidat, W. M., Ateyyeh, A., & Sadder, M. T. (2025). Population Genetic Structure of Historic Olives (Olea europaea subsp. europaea) from Jordan. International Journal of Molecular Sciences, 26(22), 10863. https://doi.org/10.3390/ijms262210863








