STAT3 Inhibition to Treat Ulcerative Colitis-Associated Colorectal Cancer
Abstract
1. Introduction
2. Results
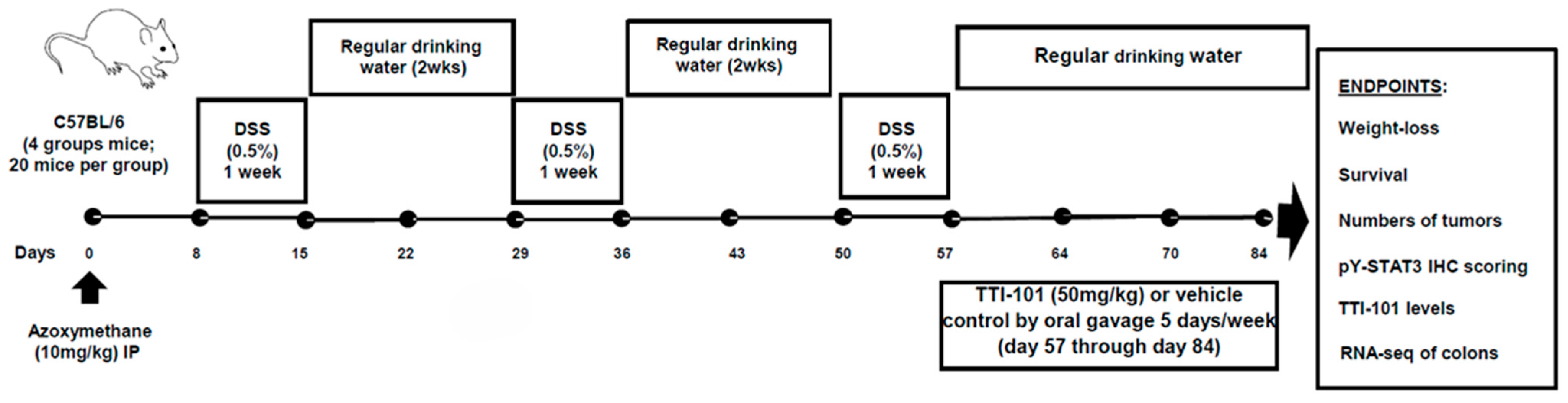
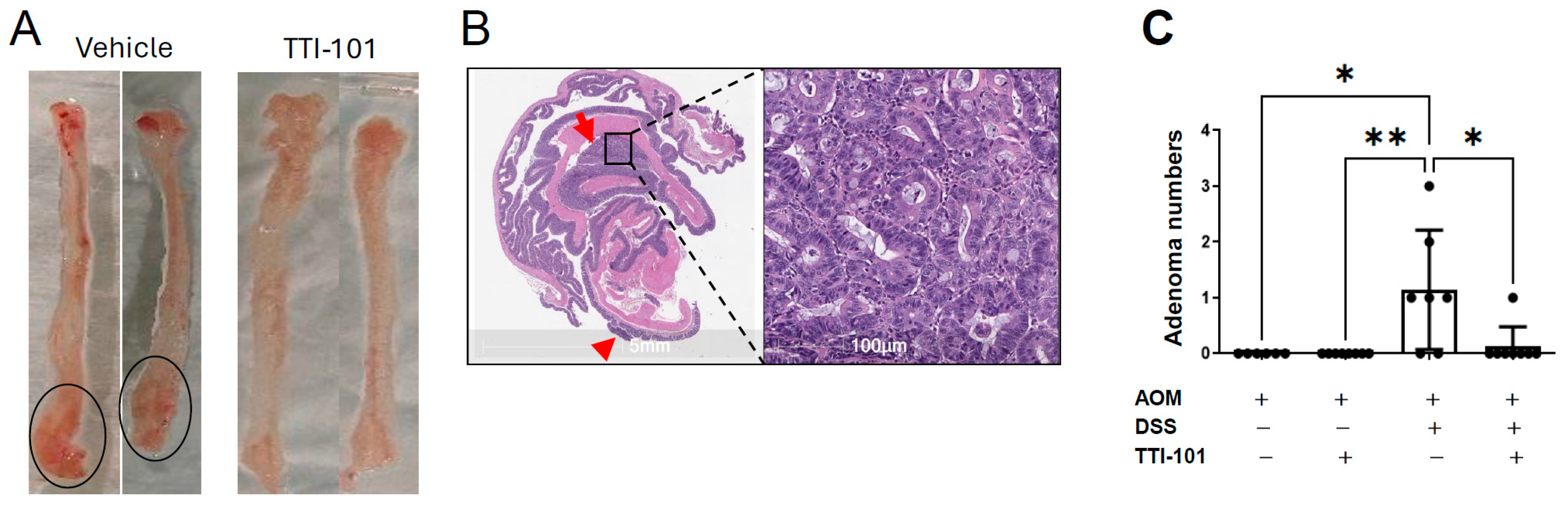
2.1. TTI-101 Reduced Adenoma Numbers in the AOM-DSS Mouse Model of Colitis-Induced CRC and Was Well Tolerated
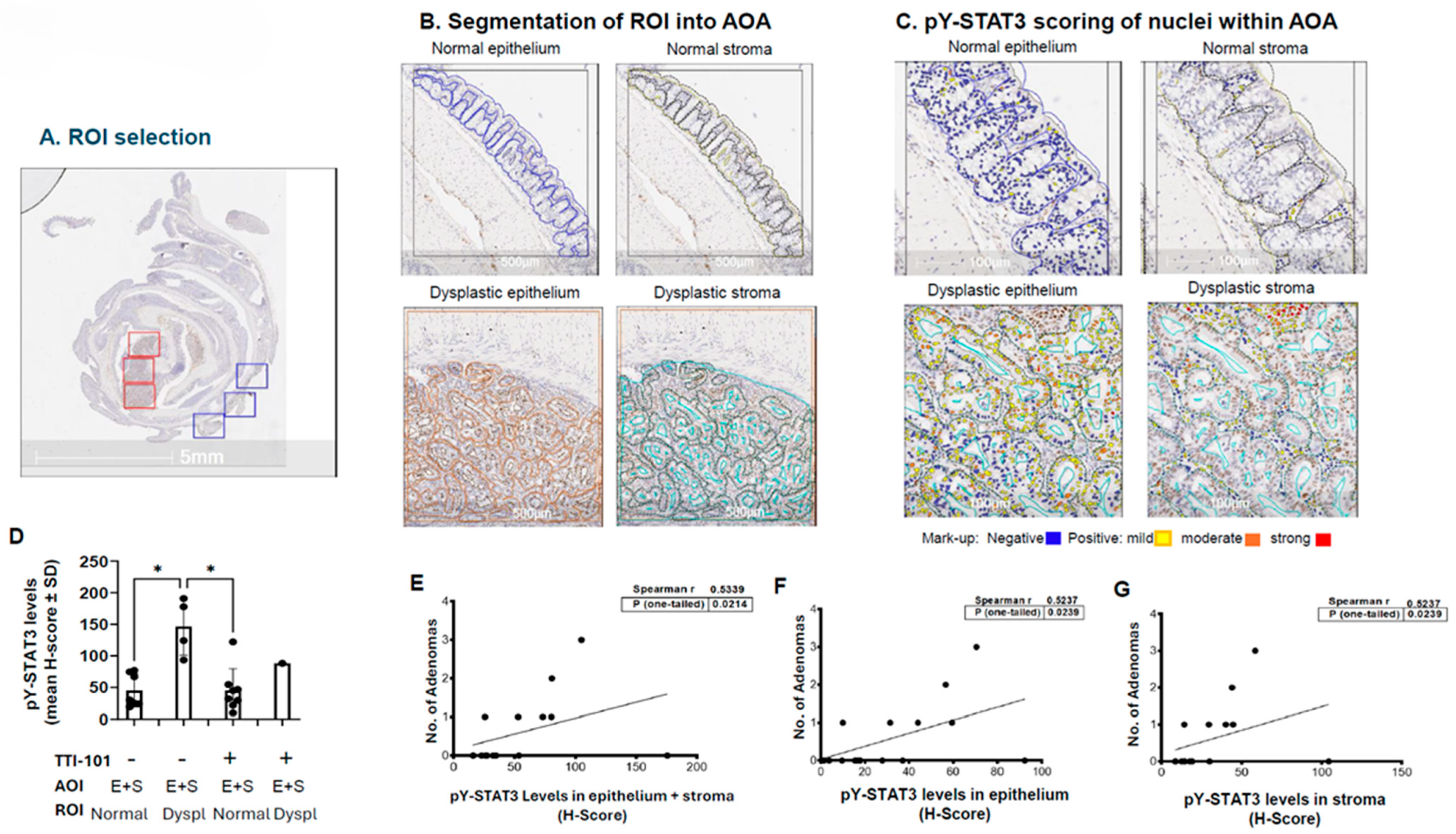
2.2. Levels of pY-STAT3 Were Increased in Dysplastic vs. Normal Colon Mucosa of AOM-DSS Mice and Correlated with Adenoma Number
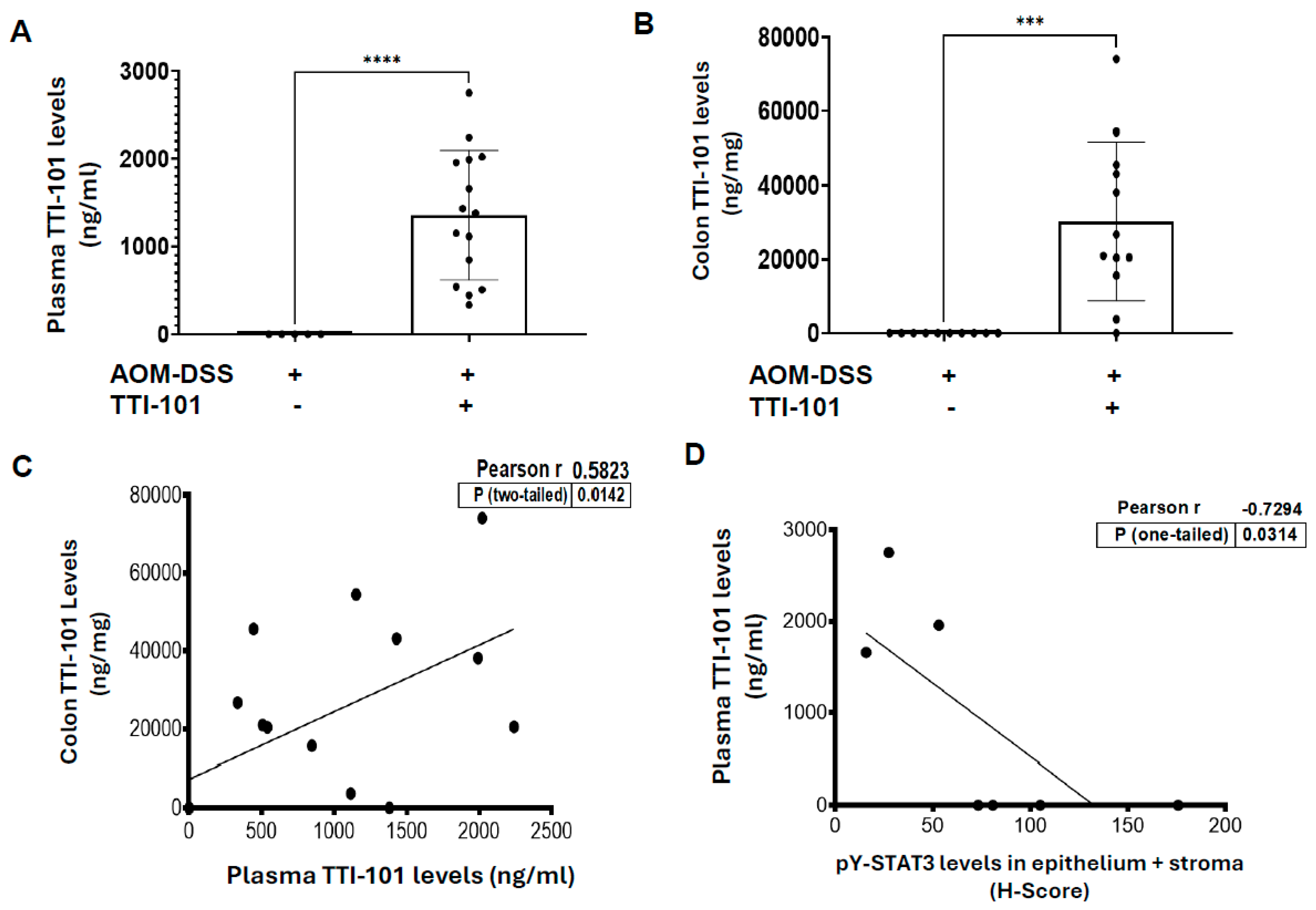
2.3. TTI-101 Was Detected at Pharmacologically Relevant Levels in Plasma and Colons of TTI-101-Treated AOM-DSS Mice; TTI-101 Levels Were Inversely Correlated to pY-STAT3 Levels
2.4. Reduction in Adenomas by TTI-101 Is Accompanied by Normalization of the Colon Transcriptome of AOM-DSS Mice
3. Discussion
4. Material and Methods
4.1. AOM-DSS Mouse Model of Colitis-CRC (Figure 1)
4.2. Measurement of TTI-101 in Plasma and Colons
4.3. IHC Staining and Digital Image Scoring for pY-STAT3
4.4. Total RNA Isolation, RNA Sequencing, and Analysis
4.5. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Cancer Society. Colorectal Cancer Facts & Figures 2023–2025; American Cancer Society: Atlanta, GA, USA, 2023. [Google Scholar]
- Castano-Milla, C.; Chaparro, M.; Gisbert, J.P. Systematic review with meta-analysis: The declining risk of colorectal cancer in ulcerative colitis. Aliment. Pharmacol. Ther. 2014, 39, 645–659. [Google Scholar] [CrossRef]
- Watanabe, T.; Konishi, T.; Kishimoto, J.; Kotake, K.; Muto, T.; Sugihara, K.; on behalf of the Japanese Society for Cancer of the Colon and Rectum. Ulcerative colitis-associated colorectal cancer shows a poorer survival than sporadic colorectal cancer: A nationwide Japanese study. Inflamm. Bowel Dis. 2011, 17, 802–808. [Google Scholar] [CrossRef]
- Ullman, T.A.; Itzkowitz, S.H. Intestinal inflammation and cancer. Gastroenterology 2011, 140, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.H.; Howard, J.K.; Lord, G.M. Transcription factor-driven regulation of ILC1 and ILC3. Trends Immunol. 2022, 43, 564–579. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Shi, S.; Ashworth, G.; Dong, C.; Liu, J.; Xing, F. ILC3 function as a double-edged sword in inflammatory bowel diseases. Cell Death Dis. 2019, 10, 315. [Google Scholar] [CrossRef] [PubMed]
- Kusaba, T.; Nakayama, T.; Yamazumi, K.; Yakata, Y.; Yoshizaki, A.; Nagayasu, T.; Sekine, I. Expression of p-STAT3 in human colorectal adenocarcinoma and adenoma; correlation with clinicopathological factors. J. Clin. Pathol. 2005, 58, 833–838. [Google Scholar] [CrossRef]
- Morikawa, T.; Baba, Y.; Yamauchi, M.; Kuchiba, A.; Nosho, K.; Shima, K.; Tanaka, N.; Huttenhower, C.; Frank, D.A.; Fuchs, C.S.; et al. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin. Cancer Res. 2011, 17, 1452–1462. [Google Scholar] [CrossRef]
- Marino, F.; Orecchia, V.; Regis, G.; Musteanu, M.; Tassone, B.; Jon, C.; Forni, M.; Calautti, E.; Chiarle, R.; Eferl, R.; et al. STAT3beta controls inflammatory responses and early tumor onset in skin and colon experimental cancer models. Am. J. Cancer Res. 2014, 4, 484–494. [Google Scholar]
- Bollrath, J.; Phesse, T.J.; von Burstin, V.A.; Putoczki, T.; Bennecke, M.; Bateman, T.; Nebelsiek, T.; Lundgren-May, T.; Canli, O.; Schwitalla, S.; et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell 2009, 15, 91–102. [Google Scholar] [CrossRef]
- Bharadwaj, U.; Eckols, T.K.; Xu, X.; Kasembeli, M.M.; Chen, Y.; Adachi, M.; Song, Y.; Mo, Q.; Lai, S.Y.; Tweardy, D.J. Small-molecule inhibition of STAT3 in radioresistant head and neck squamous cell carcinoma. Oncotarget 2016, 7, 26307–26330. [Google Scholar] [CrossRef]
- Kasembeli, M.M.; Kaparos, E.; Bharadwaj, U.; Allaw, A.; Khouri, A.; Acot, B.; Tweardy, D.J. Aberrant function of pathogenic STAT3 mutant proteins is linked to altered stability of monomers and homodimers. Blood 2023, 141, 1411–1424, Erratum in Blood 2023, 142, 2125. [Google Scholar] [CrossRef]
- Ziranu, P.; Pretta, A.; Aimola, V.; Cau, F.; Mariani, S.; D’Agata, A.P.; Codipietro, C.; Rizzo, D.; Dell’Utri, V.; Sanna, G.; et al. CD44: A New Prognostic Marker in Colorectal Cancer? Cancers 2024, 16, 1569. [Google Scholar] [CrossRef]
- Ding, Q.; Lu, P.; Xia, Y.; Ding, S.; Fan, Y.; Li, X.; Han, P.; Liu, J.; Tian, D.; Liu, M. CXCL9: Evidence and contradictions for its role in tumor progression. Cancer Med. 2016, 5, 3246–3259. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Wang, L.; Dong, Q.; Xu, K.; Ye, J.; Shao, X.; Yang, S.; Lu, C.; Chang, C.; Hou, Y.; et al. Aberrant LYZ expression in tumor cells serves as the potential biomarker and target for HCC and promotes tumor progression via csGRP78. Proc. Natl. Acad. Sci. USA 2023, 120, e2215744120. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Hu, Y.; Xiang, W.; Cai, Y.; Wang, Z.; Xiao, Q.; Liu, Y.; Li, Q.; Ding, K. Prognostic significance of matrix metalloproteinase 7 immunohistochemical expression in colorectal cancer: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 3281–3290. [Google Scholar] [PubMed]
- Duan, L.; Wu, R.; Ye, L.; Wang, H.; Yang, X.; Zhang, Y.; Chen, X.; Zuo, G.; Zhang, Y.; Weng, Y.; et al. S100A8 and S100A9 are associated with colorectal carcinoma progression and contribute to colorectal carcinoma cell survival and migration via Wnt/beta-catenin pathway. PLoS ONE 2013, 8, e62092. [Google Scholar] [CrossRef]
- Bockelman, C.; Beilmann-Lehtonen, I.; Kaprio, T.; Koskensalo, S.; Tervahartiala, T.; Mustonen, H.; Stenman, U.H.; Sorsa, T.; Haglund, C. Serum MMP-8 and TIMP-1 predict prognosis in colorectal cancer. BMC Cancer 2018, 18, 679. [Google Scholar] [CrossRef]
- Hsu, H.C.; Lee, Y.S.; Imbang, T.I.; Liu, T.C.; Hung, S.I. SLC11A1 predicts the overall survival of patients with colorectal cancer. Am. J. Cancer Res. 2024, 14, 2839–2851. [Google Scholar] [CrossRef]
- Zou, Q.; Lei, X.; Xu, A.; Li, Z.; He, Q.; Huang, X.; Xu, G.; Tian, F.; Ding, Y.; Zhu, W. Chemokines in progression, chemoresistance, diagnosis, and prognosis of colorectal cancer. Front. Immunol. 2022, 13, 724139. [Google Scholar] [CrossRef]
- Nimptsch, K.; Aleksandrova, K.; Boeing, H.; Janke, J.; Lee, Y.A.; Jenab, M.; Kong, S.Y.; Tsilidis, K.K.; Weiderpass, E.; Bueno-De-Mesquita, H.B.; et al. Plasma fetuin-A concentration, genetic variation in the AHSG gene and risk of colorectal cancer. Int. J. Cancer 2015, 137, 911–920. [Google Scholar] [CrossRef]
- Metzger, R.; Winter, L.; Bouznad, N.; Garzetti, D.; von Armansperg, B.; Rokavec, M.; Lutz, K.; Schafer, Y.; Krebs, S.; Winheim, E.; et al. CCL17 Promotes Colitis-Associated Tumorigenesis Dependent on the Microbiota. J. Immunol. 2022, 209, 2227–2238. [Google Scholar] [CrossRef]
- Chun, E.; Lavoie, S.; Michaud, M.; Gallini, C.A.; Kim, J.; Soucy, G.; Odze, R.; Glickman, J.N.; Garrett, W.S. CCL2 Promotes Colorectal Carcinogenesis by Enhancing Polymorphonuclear Myeloid-Derived Suppressor Cell Population and Function. Cell Rep. 2015, 12, 244–257. [Google Scholar] [CrossRef]
- Fahrer, J.; Wittmann, S.; Wolf, A.C.; Kostka, T. Heme Oxygenase-1 and Its Role in Colorectal Cancer. Antioxidants 2023, 12, 1989. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, K.; Sundararajan, V. IL-1beta and associated molecules as prognostic biomarkers linked with immune cell infiltration in colorectal cancer: An integrated statistical and machine learning approach. Discov. Oncol. 2025, 16, 252. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, H.; Ye, T.; Ye, Z. High expression of LEF1 correlates with poor prognosis in solid tumors, but not blood tumors: A meta-analysis. Biosci. Rep. 2020, 40, BSR20202520. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zheng, C.; Huang, L.; Lin, C.; Wang, J. USP18 directly regulates Snail1 protein through ubiquitination pathway in colorectal cancer. Cancer Cell Int. 2020, 20, 346. [Google Scholar] [CrossRef]
- Potievskaya, V.; Tyukanova, E.; Sekacheva, M.; Fashafsha, Z.; Fatyanova, A.; Potievskiy, M.; Kononova, E.; Kholstinina, A.; Polishchuk, E.; Shegai, P.; et al. Prognostic Significance of the Comprehensive Biomarker Analysis in Colorectal Cancer. Life 2025, 15, 1100. [Google Scholar] [CrossRef]
- Lohm, S.; Peduto-Eberl, L.; Lagadec, P.; Renggli-Zulliger, N.; Dudler, J.; Jeannin, J.F.; Juillerat-Jeanneret, L. Evaluation of the interaction between TGF beta and nitric oxide in the mechanisms of progression of colon carcinoma. Clin. Exp. Metastasis 2005, 22, 341–349. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Y.; Wu, Z.; Li, T.; Li, S.; Wang, C.; Ao, J.; Yang, C.; Zhou, Y. SOX11-dependent CATSPER1 expression controls colon cancer cell growth through regulation the PI3K/AKT signaling pathway. Genes. Genom. 2022, 44, 1415–1424. [Google Scholar] [CrossRef]
- Sanchez-Ramirez, D.; Mendoza-Rodriguez, M.G.; Aleman, O.R.; Candanedo-Gonzalez, F.A.; Rodriguez-Sosa, M.; Montesinos-Montesinos, J.J.; Salcedo, M.; Brito-Toledo, I.; Vaca-Paniagua, F.; Terrazas, L.I. Impact of STAT-signaling pathway on cancer-associated fibroblasts in colorectal cancer and its role in immunosuppression. World J. Gastrointest. Oncol. 2024, 16, 1705–1724. [Google Scholar] [CrossRef]
- Chen, J.; Ye, X.; Pitmon, E.; Lu, M.; Wan, J.; Jellison, E.R.; Adler, A.J.; Vella, A.T.; Wang, K. IL-17 inhibits CXCL9/10-mediated recruitment of CD8(+) cytotoxic T cells and regulatory T cells to colorectal tumors. J. Immunother. Cancer 2019, 7, 324. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, Z.; Deng, B.; Wu, D.; Quan, Y.; Min, Z. Interleukin 1beta/1RA axis in colorectal cancer regulates tumor invasion, proliferation and apoptosis via autophagy. Oncol. Rep. 2020, 43, 908–918. [Google Scholar] [CrossRef]
- Lang, Y.; Huang, H.; Jiang, H.; Wu, S.; Fang, Z.; Zhang, D.; Qian, H.; Liu, Y.; Yuan, M.; Xu, B.; et al. IL-1R2 promotes tumorigenesis and modulates the tumor immune microenvironment in colorectal cancer. Cancer Immunol. Immunother. 2025, 74, 284. [Google Scholar] [CrossRef]
- Chen, L.; Shu, P.; Zhang, X.; Ye, S.; Tian, L.; Shen, S.; Ma, J.; Ai, F.; Li, X. S100A8-Mediated Inflammatory Signaling Drives Colorectal Cancer Progression via the CXCL5/CXCR2 Axis. J. Cancer 2024, 15, 3452–3465. [Google Scholar] [CrossRef]
- Morley-Bunker, A.; Pearson, J.; Currie, M.J.; Morrin, H.; Whitehead, M.R.; Eglinton, T.; Walker, L.C. Assessment of intra-tumoural colorectal cancer prognostic biomarkers using RNA in situ hybridisation. Oncotarget 2019, 10, 1425–1439. [Google Scholar] [CrossRef]
- Jifu, C.; Lu, L.; Ding, J.; Lv, M.; Xia, J.; Wang, J.; Wang, P. USP18 Is Associated with PD-L1 Antitumor Immunity and Improved Prognosis in Colorectal Cancer. Biomolecules 2024, 14, 1191. [Google Scholar] [CrossRef] [PubMed]
- Schofield, J.H.; Longo, J.; Sheldon, R.D.; Albano, E.; Ellis, A.E.; Hawk, M.A.; Murphy, S.; Duong, L.; Rahmy, S.; Lu, X.; et al. Acod1 expression in cancer cells promotes immune evasion through the generation of inhibitory peptides. Cell Rep. 2024, 43, 113984. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Wang, Y.; Vallis, J.; Shariati, M.; Parfrey, P.S.; McLaughlin, J.R.; Wang, P.P.; Zhu, Y. Associations between polymorphisms in leptin and leptin receptor genes and colorectal cancer survival. Cancer Biol. Med. 2023, 20, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xu, Z.; Yang, B.; Zhou, X.; Kong, H. RASGRF1 Hypermethylation, a Putative Biomarker of Colorectal Cancer. Ann. Clin. Lab. Sci. 2018, 48, 3–10. [Google Scholar]
- Guo, S.; Sun, Y. OTOP2, Inversely Modulated by miR-3148, Inhibits CRC Cell Migration, Proliferation and Epithelial-Mesenchymal Transition: Evidence from Bioinformatics Data Mining and Experimental Verification. Cancer Manag. Res. 2022, 14, 1371–1384. [Google Scholar] [CrossRef]
- Li, M.; Tao, Z.; Zhao, Y.; Li, L.; Zheng, J.; Li, Z.; Chen, X. 5-methylcytosine RNA methyltransferases and their potential roles in cancer. J. Transl. Med. 2022, 20, 214. [Google Scholar] [CrossRef]
- Geerts, D.; Koster, J.; Albert, D.; Koomoa, D.L.; Feith, D.J.; Pegg, A.E.; Volckmann, R.; Caron, H.; Versteeg, R.; Bachmann, A.S. The polyamine metabolism genes ornithine decarboxylase and antizyme 2 predict aggressive behavior in neuroblastomas with and without MYCN amplification. Int. J. Cancer 2010, 126, 2012–2024. [Google Scholar] [CrossRef] [PubMed]
- Mullenger, J.L.; Zeidler, M.P.; Fragiadaki, M. Evaluating the Molecular Properties and Function of ANKHD1, and Its Role in Cancer. Int. J. Mol. Sci. 2023, 24, 12834. [Google Scholar] [CrossRef] [PubMed]
- Langford, D.J.; West, C.; Elboim, C.; Cooper, B.A.; Abrams, G.; Paul, S.M.; Schmidt, B.L.; Levine, J.D.; Merriman, J.D.; Dhruva, A.; et al. Variations in potassium channel genes are associated with breast pain in women prior to breast cancer surgery. J. Neurogenet. 2014, 28, 122–135. [Google Scholar] [CrossRef] [PubMed]
- Bakherad, M.; Salimi, M.; Angaji, S.A.; Mahjoubi, F.; Majidizadeh, T. LRIG1 expression and colorectal cancer prognosis. BMC Med. Genom. 2021, 14, 20. [Google Scholar] [CrossRef]
- Dariya, B.; Merchant, N.; Aliya, S.; Alam, A.; Nagaraju, G.P. EGFR and FGFR in Growth and Metastasis of Colorectal Cancer. In Role of Tyrosine Kinases in Gastrointestinal Malignancies; Nagaraju, G., Ed.; Springer: Singapore, 2018. [Google Scholar] [CrossRef]
- Xia, L.; Song, M.; Sun, M.; Wang, F.; Yang, C. Circular RNAs as Biomarkers for Cancer. In Circular RNAs; Advances in Experimental Medicine and Biology; Xiao, J., Ed.; Springer: Singapore, 2018; Volume 1087. [Google Scholar] [CrossRef]
- Thaker, A.I.; Shaker, A.; Rao, M.S.; Ciorba, M.A. Modeling colitis-associated cancer with azoxymethane (AOM) and dextran sulfate sodium (DSS). J. Vis. Exp. 2012, 4100. [Google Scholar] [CrossRef]
- Becker, C.; Fantini, M.C.; Wirtz, S.; Nikolaev, A.; Kiesslich, R.; Lehr, H.A.; Galle, P.R.; Neurath, M.F. In vivo imaging of colitis and colon cancer development in mice using high resolution chromoendoscopy. Gut 2005, 54, 950–954. [Google Scholar] [CrossRef]
- Meissl, K.; Macho-Maschler, S.; Muller, M.; Strobl, B. The good and the bad faces of STAT1 in solid tumours. Cytokine 2017, 89, 12–20. [Google Scholar] [CrossRef]
- Haricharan, S.; Dong, J.; Hein, S.; Reddy, J.P.; Du, Z.; Toneff, M.; Holloway, K.; Hilsenbeck, S.G.; Huang, S.; Atkinson, R.; et al. Mechanism and preclinical prevention of increased breast cancer risk caused by pregnancy. eLife 2013, 2, e00996. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Redell, M.S.; Ruiz, M.J.; Alonzo, T.A.; Gerbing, R.B.; Tweardy, D.J. Stat3 signaling in acute myeloid leukemia: Ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood 2011, 117, 5701–5709. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, J.; Dong, Y.; Tweardy, D.J.; Garibotto, G.; Mitch, W.E. Stat3 Activation Links a C/EBPdelta to Myostatin Pathway to Stimulate Loss of Muscle Mass. Cell Metab. 2013, 18, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Silva, K.A.; Dong, J.; Dong, Y.; Dong, Y.; Schor, N.; Tweardy, D.J.; Zhang, L.; Mitch, W.E. Inhibition of Stat3 suppresses caspase-3 and the ubiquitin-proteasome system leading to preservation of muscle mass in cancer cachexia. J. Biol. Chem. 2015, 290, 11177–11187. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.M.; Bharadwaj, U.; Eckols, T.K.; Kolosov, M.; Kasembeli, M.M.; Fridley, C.; Siller, R.; Tweardy, D.J. Small-molecule targeting of signal transducer and activator of transcription (STAT) 3 to treat non-small cell lung cancer. Lung Cancer 2015, 90, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Pedroza, M.; Le, T.T.; Lewis, K.; Karmouty-Quintana, H.; To, S.; George, A.T.; Blackburn, M.R.; Tweardy, D.J.; Agarwal, S.K. STAT-3 contributes to pulmonary fibrosis through epithelial injury and fibroblast-myofibroblast differentiation. FASEB J. 2015, 30, 129–140. [Google Scholar] [CrossRef]
- Xu, X.; Kasembeli, M.M.; Jiang, X.; Tweardy, B.J.; Tweardy, D.J. Chemical probes that competitively and selectively inhibit Stat3 activation. PLoS ONE 2009, 4, e4783. [Google Scholar] [CrossRef]
- Kasembeli, M.M.; Singhmar, P.; Ma, J.; Edralin, J.; Tang, Y.; Adams, C., 3rd; Heijnen, C.J.; Kavelaars, A.; Tweardy, D.J. TTI-101: A competitive inhibitor of STAT3 that spares oxidative phosphorylation and reverses mechanical allodynia in mouse models of neuropathic pain. Biochem. Pharmacol. 2021, 192, 114688, Erratum in Biochem. Pharmacol. 2022, 195, 114860. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Dong, Y.; Chen, Z.; Eckols, T.K.; Kasembeli, M.M.; Tweardy, D.J.; Mitch, W.E. Pharmacokinetics and pharmacodynamics of TTI-101, a STAT3 inhibitor that blocks muscle proteolysis in rats with chronic kidney disease. Am. J. Physiol. Ren. Physiol. 2020, 319, F84–F92, Erratum in Am. J. Physiol. Ren. Physiol. 2022, 322, F104. [Google Scholar] [CrossRef]
- Kettner, N.M.; Vijayaraghavan, S.; Durak, M.G.; Bui, T.; Kohansal, M.; Ha, M.J.; Liu, B.; Rao, X.; Wang, J.; Yi, M.; et al. Combined Inhibition of STAT3 and DNA Repair in Palbociclib-Resistant ER-Positive Breast Cancer. Clin. Cancer Res. 2019, 25, 3996–4013. [Google Scholar] [CrossRef]
- Robinson, P.; Magness, E.; Montoya, K.; Engineer, N.; Eckols, T.K.; Rodriguez, E.; Tweardy, D.J. Genetic and Small-Molecule Modulation of Stat3 in a Mouse Model of Crohn’s Disease. J. Clin. Med. 2022, 11, 7020. [Google Scholar] [CrossRef]
- Hoffman, K.A.; Villar, M.J.; Poveda, C.; Bottazzi, M.E.; Hotez, P.J.; Tweardy, D.J.; Jones, K.M. Signal Transducer and Activator of Transcription-3 Modulation of Cardiac Pathology in Chronic Chagasic Cardiomyopathy. Front. Cell. Infect. Microbiol. 2021, 11, 708325. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Z.; Wang, Y.; Tweardy, D.J.; Mitch, W.E. Stat3 activation induces insulin resistance via a muscle-specific E3 ubiquitin ligase Fbxo40. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E625–E635, Erratum in Am. J. Physiol. Endocrinol. Metab. 2022, 322, E330. [Google Scholar] [CrossRef]
- Jung, K.H.; Yoo, W.; Stevenson, H.L.; Deshpande, D.; Shen, H.; Gagea, M.; Yoo, S.Y.; Wang, J.; Eckols, T.K.; Bharadwaj, U.; et al. Multifunctional Effects of a Small-Molecule STAT3 Inhibitor on NASH and Hepatocellular Carcinoma in Mice. Clin. Cancer Res. 2017, 23, 5537–5546. [Google Scholar] [CrossRef]
- Gavino, A.C.; Nahmod, K.; Bharadwaj, U.; Makedonas, G.; Tweardy, D.J. STAT3 inhibition prevents lung inflammation, remodeling, and accumulation of Th2 and Th17 cells in a murine asthma model. Allergy 2016, 71, 1684–1692. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Vining, D.J.; Arora, S.P.; de Achaval, S.; Larson, J.; Kauh, J.; Cartwright, C.; Avritscher, R.; Alibhai, I.; Tweardy, D.J.; et al. Phase I Trial of TTI-101, a First-in-Class Oral Inhibitor of STAT3, in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2025, 31, 965–974. [Google Scholar] [CrossRef]

| Gene Symbol | Vehicle vs. Sham (Log 2-Fold Change) | TTI-101 vs. Vehicle (Log 2-Fold Change) | Role in CRC |
|---|---|---|---|
| Rps8-ps1 | 1.204 | −1.028 | No known role (NKR) |
| Camkv | 3.702 | −2.824 | NKR |
| Lypd8l | 2.378 | −1.572 | NKR |
| Pglyrp4 | 5.719 | −3.344 | NKR |
| Notum | 6.265 | −2.615 | NKR |
| Ccl2 | 1.629 | −1.124 | Progression/Metastasis |
| Tnfrsf11b | 2.706 | −1.606 | NKR |
| Abhd3 | 1.536 | −1.608 | NKR |
| Gvin1 | 1.518 | −1.100 | NKR |
| Cxcl3 | 6.815 | −3.389 | NKR |
| Disc1 | 1.258 | −1.248 | NKR |
| Acsbg1 | 2.345 | −1.431 | NKR |
| Plat | 2.306 | −1.851 | NKR |
| S100a9 | 6.150 | −3.654 | Progression/Metastasis |
| Tgm1 | 1.964 | −1.774 | NKR |
| Marcksl1 | 1.348 | −1.075 | NKR |
| Acod1 | 2.280 | −1.609 | NKR |
| Krt90 | 7.013 | −2.177 | NKR |
| Il1b | 2.833 | −2.191 | Progression/Metastasis |
| Trgc2 | 1.257 | −1.067 | NKR |
| Alb | 3.817 | −3.145 | NKR |
| Sbspon | 2.174 | −1.507 | NKR |
| Rpl27-ps1 | 1.788 | −1.222 | NKR |
| Gpr31a | 3.664 | −3.313 | NKR |
| Cers3 | 2.892 | −1.623 | NKR |
| Nkd1 | 1.888 | −1.182 | NKR |
| Dmgdh | 2.114 | −1.887 | NKR |
| R3hdml | 3.963 | −2.108 | NKR |
| Prox1os | 2.718 | −3.405 | NKR |
| Arhgdig | 1.196 | −1.010 | NKR |
| Ppbp | 3.807 | −2.576 | NKR |
| Fam89a | 1.682 | −1.255 | NKR |
| Pzp | 5.635 | −3.136 | NKR |
| Timp1 | 1.865 | −1.530 | Progression/Metastasis |
| S100a8 | 5.627 | −3.581 | NKR |
| Cbx3-ps8 | 2.339 | −2.428 | NKR |
| Ahsg | 2.228 | −2.551 | Progression/Metastasis |
| Spock2 | 2.297 | −1.976 | NKR |
| Alox12e | 2.745 | −2.427 | NKR |
| Rnf212 | 1.096 | −1.129 | NKR |
| Dsc3 | 1.850 | −1.413 | NKR |
| Lef1 | 1.605 | −1.221 | Progression/Metastases |
| Ascl4 | 5.100 | −1.815 | NKR |
| Ppp1r36 | 2.161 | −2.010 | NKR |
| Dusp2 | 1.076 | −1.084 | NKR |
| Asprv1 | 2.488 | −1.274 | NKR |
| Il1rl1 | 3.288 | −2.043 | NKR |
| Rpl19-ps3 | 3.993 | −1.718 | NKR |
| Halr1 | 2.994 | −2.124 | NKR |
| Col17a1 | 1.742 | −2.141 | NKR |
| Sox11 | 2.312 | −3.116 | Protection |
| Inhbb | 1.006 | −1.308 | NKR |
| Mogat2 | 1.495 | −1.056 | NKR |
| Chil1 | 1.165 | −1.754 | NKR |
| Lypd3 | 3.423 | −2.823 | NKR |
| Slc44a5 | 1.611 | −1.283 | NKR |
| Dnmt3c | 2.471 | −3.466 | NKR |
| Pdgfrl | 3.191 | −2.332 | NKR |
| Rab37 | 1.412 | −1.256 | NKR |
| Lyz1 | 4.044 | −2.348 | Progression/Metastases |
| Prr18 | 2.295 | −1.354 | NKR |
| Ccl17 | 3.056 | −1.452 | Progression/Metastases |
| Sp5 | 3.816 | −2.491 | NKR |
| Ncmap | 1.396 | −1.151 | NKR |
| Dsg3 | 2.043 | −1.326 | NKR |
| Crybb1 | 2.315 | −2.580 | NKR |
| Klrb1 | 1.837 | −1.168 | NKR |
| Alox12 | 2.685 | −1.417 | NKR |
| Slc26a9 | 4.146 | −2.985 | NKR |
| Il1r2 | 2.459 | −1.225 | NKR |
| Rpl3-ps2 | 1.597 | −1.247 | NKR |
| Gdf11 | 1.316 | −1.222 | NKR |
| Tbx3os2 | 2.779 | −2.150 | NKR |
| Prox1 | 1.818 | −1.335 | NKR |
| Rpl5-ps2 | 1.828 | −1.078 | NKR |
| Klra5 | 3.047 | −3.180 | NKR |
| Krt23 | 1.139 | −1.098 | NKR |
| Aif1l | 2.406 | −2.026 | NKR |
| Tspan32 | 1.684 | −1.850 | NKR |
| Fam162b | 1.123 | −1.687 | NKR |
| St3gal5 | 1.001 | −1.227 | NKR |
| Slc9a4 | 1.903 | −1.143 | NKR |
| Gata5 | 1.780 | −1.615 | NKR |
| Wfdc18 | 3.577 | −2.419 | NKR |
| Tmprss11d | 4.894 | −5.240 | NKR |
| Xcl1 | 1.802 | −2.270 | Progression/Metastases |
| Trim29 | 2.802 | −1.303 | NKR |
| Lect2 | 5.616 | −4.433 | NKR |
| Krt5 | 5.604 | −5.745 | NKR |
| Aox4 | 5.572 | −3.349 | NKR |
| Upk3a | 5.927 | −3.336 | NKR |
| Usp18 | 2.294 | −1.235 | Progression/Metastases |
| Tnfsf11 | 1.265 | −1.774 | Progression/Metastases |
| Trbc2 | 1.491 | −1.031 | NKR |
| Ly6d | 2.856 | −2.295 | NKR |
| Klrb1a | 1.845 | −2.020 | NKR |
| Ntf5 | 2.638 | −2.056 | NKR |
| Smim3 | 1.458 | −1.145 | NKR |
| Slc30a2 | 3.086 | −2.196 | NKR |
| Batf3 | 1.043 | −1.240 | NKR |
| Rpl7a-ps11 | 1.302 | −1.014 | NKR |
| Bpifb5 | 3.232 | −3.769 | NKR |
| Nkg7 | 1.515 | −1.697 | NKR |
| Wif1 | 3.845 | −2.663 | NKR |
| Cd3g | 1.157 | −1.223 | NKR |
| Prss56 | 11.002 | −6.618 | NKR |
| Prom2 | 1.500 | −1.907 | NKR |
| Reg3g | 3.918 | −4.756 | NKR |
| Cd3d | 1.484 | −1.002 | NKR |
| Ptpro | 1.279 | −1.183 | NKR |
| Rpl30-ps8 | 1.255 | −1.745 | NKR |
| Trbv2 | 3.593 | −4.271 | NKR |
| Cxcl9 | 2.395 | −1.727 | Progression/Metastases |
| Trgc1 | 1.715 | −1.434 | NKR |
| Ltf | 4.096 | −3.293 | Protection |
| Svopl | 1.613 | −1.845 | NKR |
| Ccdc146 | 2.126 | −2.022 | NKR |
| Rpl5-ps1 | 1.201 | −1.291 | NKR |
| Sema7a | 1.292 | −1.274 | NKR |
| Scgb2b20 | 3.004 | −3.324 | NKR |
| Erich2 | 1.527 | −1.446 | NKR |
| Psenen-ps | 1.069 | −1.482 | NKR |
| Cd244a | 1.594 | −1.089 | Immunosuppression |
| Cited1 | 3.753 | −3.005 | NKR |
| Adam8 | 1.110 | −1.273 | NKR |
| Reg3b | 4.126 | −6.043 | Protection |
| Scgb1b3 | 1.552 | −2.435 | NKR |
| Esyt3 | 2.903 | −2.166 | NKR |
| Col9a3 | 3.994 | −3.797 | NKR |
| Lgals2 | 1.817 | −1.878 | NKR |
| Ifitm1 | 1.988 | −1.670 | NKR |
| Adam28 | 2.704 | −4.919 | NKR |
| Adamts3 | 1.099 | −1.199 | NKR |
| Mab21l3 | 1.613 | −2.450 | NKR |
| Stx11 | 1.188 | −1.374 | NKR |
| Ceacam12 | 2.372 | −2.891 | NKR |
| Rpl27-ps3 | 1.092 | −1.204 | NKR |
| Morc1 | 2.447 | −3.211 | NKR |
| Bpifc | 2.966 | −5.168 | NKR |
| Muc6 | 2.216 | −1.503 | NKR |
| Scgb2b15 | 1.595 | −2.288 | NKR |
| Nanos1 | 1.309 | −1.468 | NKR |
| Zap70 | 1.656 | −1.609 | NKR |
| Gulo | 4.935 | −4.635 | NKR |
| Mmp7 | 5.399 | −4.059 | Progression/Metastases |
| Hmox1 | 1.292 | −1.175 | Progression/Metastases |
| Mt3 | 2.209 | −1.979 | NKR |
| Sprr2h | 2.384 | −4.243 | NKR |
| Rasl11a | 1.062 | −1.257 | NKR |
| Tesc | 1.592 | −1.848 | NKR |
| Rbm11 | 2.405 | −1.898 | NKR |
| Myl7 | 3.235 | −2.275 | NKR |
| Fzd10 | 1.186 | −1.961 | NKR |
| Akr1c18 | 2.300 | −3.300 | NKR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robinson, P.; Italia, Z.; Italia, Z.; Hoang, T.; Rodriguez, E.; Eckols, T.K.; Kasembeli, M.; Zorrilla, L.H.; Soto, L.M.S.; Mahalingam, R.; et al. STAT3 Inhibition to Treat Ulcerative Colitis-Associated Colorectal Cancer. Int. J. Mol. Sci. 2025, 26, 10808. https://doi.org/10.3390/ijms262110808
Robinson P, Italia Z, Italia Z, Hoang T, Rodriguez E, Eckols TK, Kasembeli M, Zorrilla LH, Soto LMS, Mahalingam R, et al. STAT3 Inhibition to Treat Ulcerative Colitis-Associated Colorectal Cancer. International Journal of Molecular Sciences. 2025; 26(21):10808. https://doi.org/10.3390/ijms262110808
Chicago/Turabian StyleRobinson, Prema, Zal Italia, Zara Italia, Tan Hoang, Emma Rodriguez, T. Kris Eckols, Moses Kasembeli, Leticia Hamana Zorrilla, Luisa Maren Solis Soto, Rajasekaran Mahalingam, and et al. 2025. "STAT3 Inhibition to Treat Ulcerative Colitis-Associated Colorectal Cancer" International Journal of Molecular Sciences 26, no. 21: 10808. https://doi.org/10.3390/ijms262110808
APA StyleRobinson, P., Italia, Z., Italia, Z., Hoang, T., Rodriguez, E., Eckols, T. K., Kasembeli, M., Zorrilla, L. H., Soto, L. M. S., Mahalingam, R., & Tweardy, D. J. (2025). STAT3 Inhibition to Treat Ulcerative Colitis-Associated Colorectal Cancer. International Journal of Molecular Sciences, 26(21), 10808. https://doi.org/10.3390/ijms262110808






