Residual Genetic Material in Mature Red Blood Cells
Abstract
1. Introduction
2. Residual Genetic Material in Red Blood Cells
2.1. mRNAs
2.2. miRNAs: Potential Physiological Roles of Erythrocyte miRNAs
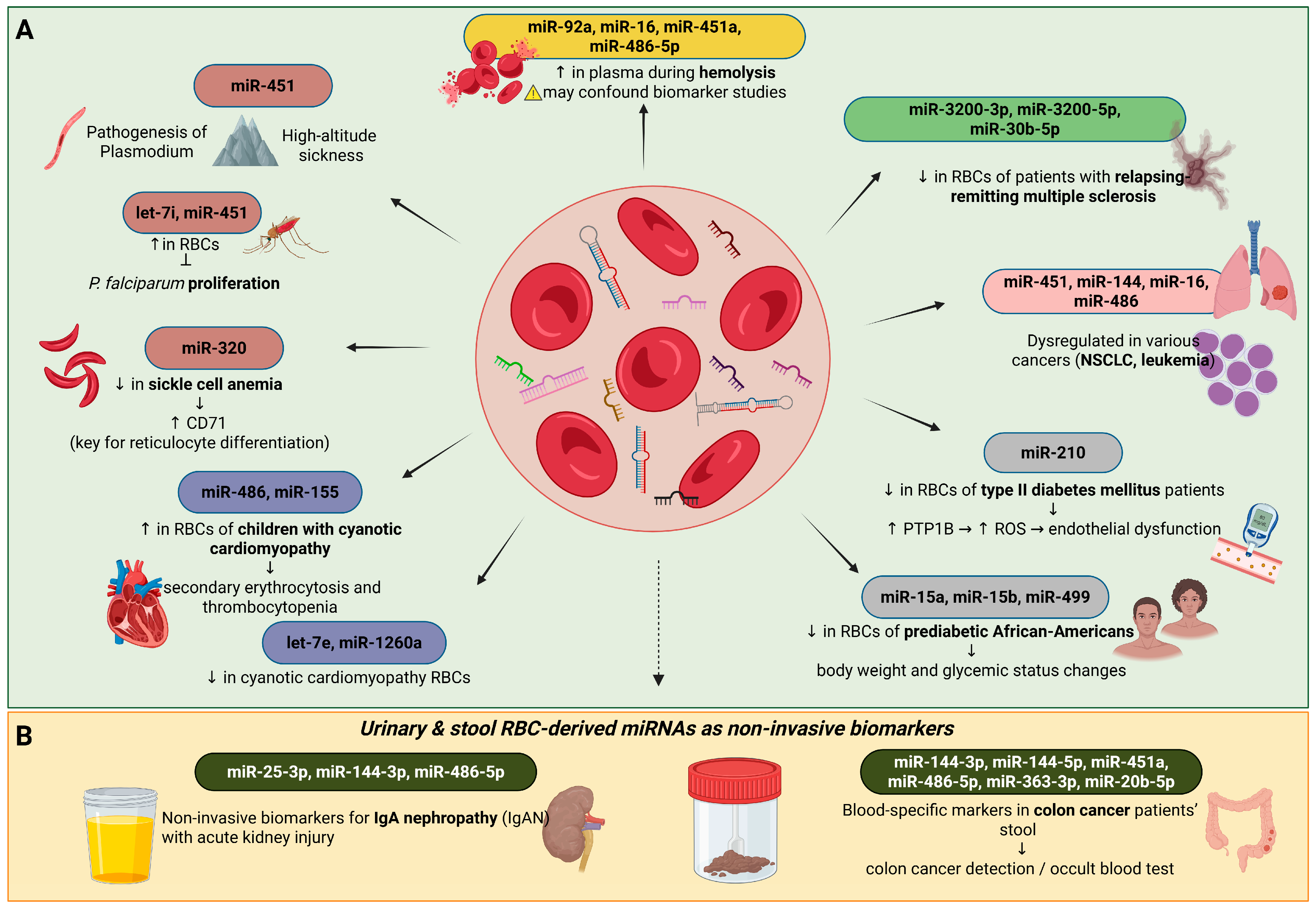
2.3. Possible Pathological Roles of Erythrocyte miRNAs in Various Diseases
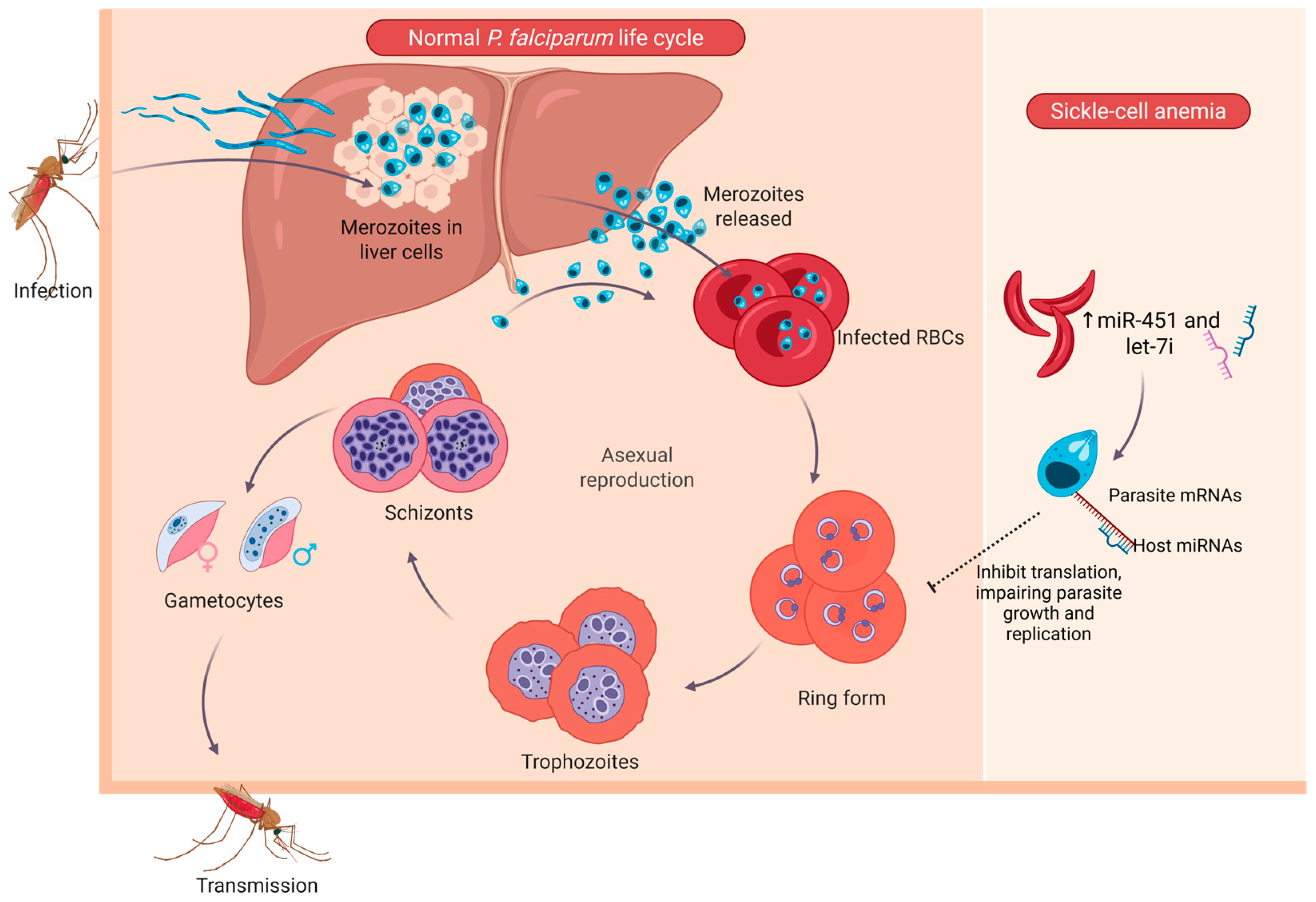
2.3.1. Erythrocyte miRNAs in Infectious Diseases
2.3.2. Erythrocyte miRNAs in Cancer
2.3.3. Erythrocyte miRNAs in Cardiovascular and Metabolic Regulation
2.3.4. Erythrocyte miRNAs as Biomarkers
2.4. lncRNAs
2.5. LincEPS and LncRNA-Saf in Erythroid Cell Survival
2.6. AlncRNA-EC7 and ShlncRNA-EC6 During Erythroid Differentiation
2.7. BGLT3 in the Regulation of Globin and UCA1 Gene Expression and Heme Biosynthesis
2.8. EVs (Extracellular Vesicles)
2.9. DNA
2.10. CircRNAs
3. Conclusions
4. Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AlncRNA | Antisense Long non-coding RNA |
| AlncRNA-EC7 | Erythroid-specific Long non-coding RNA |
| AS1 | Antisense RNA 1 |
| BFU-Es | Burst-Forming Unit–Erythroid |
| BGLT3 | Beta-Globin Locus Transcript 3 |
| CD71 | Transferrin Receptor 1 |
| CFU-Es | Colony-Forming Unit–Erythroid |
| csbDNA | Cell-surface-bound DNA |
| DAMPs | Damage-Associated Molecular Patterns |
| DNA | Deoxyribonucleic Acid |
| EGFR | Epidermal Growth Factor Receptor |
| ElncRNA | Enhancer Long non-coding RNA |
| GATA1 | GATA-binding factor 1 |
| IlncRNA | Intronic overlapping Long Non-Coding RNA |
| KLF1 | Krüppel-Like Factor 1 (Erythroid-specific transcription factor) |
| LincRNA | Long intergenic non-coding RNA |
| LncRNA | Long Non-coding RNA |
| miRNA | Micro-RNA |
| mRNA | Messenger RNA |
| mtDNA | Mitochondrial DNA |
| KRAS | Kirsten Rat Sarcoma Viral Oncogene Homolog |
| NGS | Next-Generation Sequencing |
| NSCLC | Non-Small Cell Lung Cancer |
| PlncRNA | Pseudogene Long non-coding RNA |
| PTP1B | Protein Tyrosine Phosphatase 1B |
| RBC | Red Blood Cell |
| RNA | Ribonucleic Acid |
| ROS | Reactive Oxygen Species |
| ShlncRNA | Small RNA host Long non-coding RNA |
| ShlncRNA-EC6 | Erythroid-specific long non-coding RNA |
| SLC4A1 | Solute Carrier Family 4 Member 1 |
| SMAD2 | Mothers Against Decapentaplegic Homolog 2 |
| SMAD4 | Mothers Against Decapentaplegic Homolog 4 |
| TAL1 | T-cell Acute Lymphocytic Leukemia protein 1 (also known as SCL, transcription factor regulating erythropoiesis) |
| TGF-β | Transforming Growth Factor β |
| TLR9 | Toll-Like Receptor 9 |
| UCA1 | Urothelial Cancer Associated 1 (lncRNA, regulator of heme metabolism in erythroblasts) |
References
- Liang, N.; Jiao, Z.; Zhang, C.; Wu, Y.; Wang, T.; Li, S.; Wang, Y.; Song, T.; Chen, J.-Q.; Liang, H.; et al. Mature Red Blood Cells Contain Long DNA Fragments and Could Acquire DNA from Lung Cancer Tissue. Adv. Sci. 2023, 10, 2206361. [Google Scholar] [CrossRef] [PubMed]
- Sears, D.A.; Udden, M.M. Howell-Jolly bodies: A brief historical review. Am. J. Med. Sci. 2012, 343, 407–409. [Google Scholar] [CrossRef]
- Felka, T.; Lemke, J.; Lemke, C.; Michel, S.; Liehr, T.; Claussen, U. DNA degradation during maturation of erythrocytes—Molecular cytogenetic characterization of Howell-Jolly bodies. Cytogenet. Genome Res. 2007, 119, 2–8. [Google Scholar] [CrossRef]
- Hotz, M.J.; Qing, D.; Shashaty, M.G.S.; Zhang, P.; Faust, H.; Sondheimer, N.; Rivella, S.; Worthen, G.S.; Mangalmurti, N.S. Red Blood Cells Homeostatically Bind Mitochondrial DNA through TLR9 to Maintain Quiescence and to Prevent Lung Injury. Am. J. Respir. Crit. Care Med. 2018, 197, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Tamkovich, S.; Laktionov, P. Cell-surface-bound circulating DNA in the blood: Biology and clinical application. IUBMB Life 2019, 71, 1201–1210. [Google Scholar] [CrossRef]
- Lam, L.K.M.; Murphy, S.; Kokkinaki, D.; Venosa, A.; Sherrill-Mix, S.; Casu, C.; Rivella, S.; Weiner, A.; Park, J.; Shin, S.; et al. DNA binding to TLR9 expressed by red blood cells promotes innate immune activation and anemia. Sci. Transl. Med. 2021, 13, eabj1008. [Google Scholar] [CrossRef]
- Usman, W.M.; Pham, T.C.; Kwok, Y.Y.; Vu, L.T.; Ma, V.; Peng, B.; Chan, Y.S.; Wei, L.; Chin, S.M.; Azad, A.; et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018, 9, 2359. [Google Scholar] [CrossRef]
- Biagiotti, S.; Canonico, B.; Tiboni, M.; Abbas, F.; Perla, E.; Montanari, M.; Battistelli, M.; Papa, S.; Casettari, L.; Rossi, L.; et al. Efficient and highly reproducible production of red blood cell-derived extracellular vesicle mimetics for the loading and delivery of RNA molecules. Sci. Rep. 2024, 14, 14610. [Google Scholar] [CrossRef]
- Liang, R.; Menon, V.; Qiu, J.; Arif, T.; Renuse, S.; Lin, M.; Nowak, R.; Hartmann, B.; Tzavaras, N.; Benson, D.L.; et al. Mitochondrial localization and moderated activity are key to murine erythroid enucleation. Blood Adv. 2021, 5, 2490–2504. [Google Scholar] [CrossRef]
- Wakabayashi, I.; Sotoda, Y.; Eguchi, R. Relationships among erythrocyte-derived microRNAs in serum of healthy donors. Clin. Chim. Acta 2020, 507, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Cao, X.; Berger, C.K.; Foote, P.H.; Mahoney, D.W.; Simonson, J.A.; Anderson, B.W.; Yab, T.C.; Taylor, W.R.; Boardman, L.A.; et al. Novel Approach to Fecal Occult Blood Testing by Assay of Erythrocyte-Specific microRNA Markers. Dig. Dis. Sci. 2017, 62, 1985–1994. [Google Scholar] [CrossRef]
- Ryan, P.; Atreya, C. Blood cell microRNAs: What are they and what future do they hold? Transfus. Med. Rev. 2011, 25, 247–251. [Google Scholar] [CrossRef]
- Joshi, U.; Jani, D.; George, L.B.; Highland, H. Human erythrocytes’ perplexing behaviour: Erythrocytic microRNAs. Mol. Cell. Biochem. 2025, 480, 923–935. [Google Scholar] [CrossRef]
- Fakoya, A.O.; Amraei, R. Histology, Howell Jolly Bodies. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Bastos, R.N.; Volloch, Z.; Aviv, H. Messenger RNA population analysis during erythroid differentiation: A kinetical approach. J. Mol. Biol. 1977, 110, 191–203. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Anastasiadi, A.T.; Tzounakas, V.L.; Nemkov, T.; Reisz, J.A.; Kriebardis, A.G.; Zimring, J.C.; Spitalnik, S.L.; Busch, M.P. Red Blood Cell Metabolism In Vivo and In Vitro. Metabolites 2023, 13, 793. [Google Scholar] [CrossRef]
- Goh, S.H.; Josleyn, M.; Lee, Y.T.; Danner, R.L.; Gherman, R.B.; Cam, M.C.; Miller, J.L. The human reticulocyte transcriptome. Physiol. Genom. 2007, 30, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Doss, J.F.; Corcoran, D.L.; Jima, D.D.; Telen, M.J.; Dave, S.S.; Chi, J.T. A comprehensive joint analysis of the long and short RNA transcriptomes of human erythrocytes. BMC Genom. 2015, 16, 952. [Google Scholar] [CrossRef]
- Schoenberg, D.R. Mechanisms of endonuclease-mediated mRNA decay. Wiley Interdiscip. Rev. RNA 2011, 2, 582–600. [Google Scholar] [CrossRef]
- Shi, L.; Lin, Y.-H.; Sierant, M.C.; Zhu, F.; Cui, S.; Guan, Y.; Sartor, M.A.; Tanabe, O.; Lim, K.-C.; Engel, J.D. Developmental transcriptome analysis of human erythropoiesis. Hum. Mol. Genet. 2014, 23, 4528–4542. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Y.; Tong, J.; Gao, J.; Guo, Q.; Zhang, L.; Wang, B.; Zhao, H.; Wang, H.; Jiang, E.; et al. Long non-coding RNA-dependent mechanism to regulate heme biosynthesis and erythrocyte development. Nat. Commun. 2018, 9, 4386. [Google Scholar] [CrossRef]
- Xu, P.; Palmer, L.E.; Lechauve, C.; Zhao, G.; Yao, Y.; Luan, J.; Vourekas, A.; Tan, H.; Peng, J.; Schuetz, J.D.; et al. Regulation of gene expression by miR-144/451 during mouse erythropoiesis. Blood 2019, 133, 2518–2528. [Google Scholar] [CrossRef] [PubMed]
- Randrianarison-Huetz, V.; Laurent, B.; Bardet, V.; Blobe, G.C.; Huetz, F.; Duménil, D. Gfi-1B controls human erythroid and megakaryocytic differentiation by regulating TGF-beta signaling at the bipotent erythro-megakaryocytic progenitor stage. Blood 2010, 115, 2784–2795. [Google Scholar] [CrossRef]
- Masaki, S.; Ohtsuka, R.; Abe, Y.; Muta, K.; Umemura, T. Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem. Biophys. Res. Commun. 2007, 364, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Shaham, L.; Vendramini, E.; Ge, Y.; Goren, Y.; Birger, Y.; Tijssen, M.R.; McNulty, M.; Geron, I.; Schwartzman, O.; Goldberg, L.; et al. MicroRNA-486-5p is an erythroid oncomiR of the myeloid leukemias of Down syndrome. Blood 2015, 125, 1292–1301. [Google Scholar] [CrossRef]
- Sun, L.; Fan, F.; Li, R.; Niu, B.; Zhu, L.; Yu, S.; Wang, S.; Li, C.; Wang, D. Different Erythrocyte MicroRNA Profiles in Low- and High-Altitude Individuals. Front. Physiol. 2018, 9, 1099. [Google Scholar] [CrossRef]
- Rivkin, N.; Chapnik, E.; Mildner, A.; Barshtein, G.; Porat, Z.; Kartvelishvily, E.; Dadosh, T.; Birger, Y.; Amir, G.; Yedgar, S.; et al. Erythrocyte survival is controlled by microRNA-142. Haematologica 2017, 102, 676–685. [Google Scholar] [CrossRef]
- Sarachana, T.; Kulkarni, S.; Atreya, C.D. Evaluation of small noncoding RNAs in ex vivo stored human mature red blood cells: Changes in noncoding RNA levels correlate with storage lesion events. Transfusion 2015, 55, 2672–2683. [Google Scholar] [CrossRef]
- Țichil, I.; Mitre, I.; Zdrenghea, M.T.; Bojan, A.S.; Tomuleasa, C.I.; Cenariu, D. A Review of Key Regulators of Steady-State and Ineffective Erythropoiesis. J. Clin. Med. 2024, 13, 2585. [Google Scholar] [CrossRef]
- Walzer, K.A.; Chi, J.T. Trans-kingdom small RNA transfer during host-pathogen interactions: The case of P. falciparum and erythrocytes. RNA Biol. 2017, 14, 442–449. [Google Scholar] [CrossRef] [PubMed]
- LaMonte, G.; Philip, N.; Reardon, J.; Lacsina, J.R.; Majoros, W.; Chapman, L.; Thornburg, C.D.; Telen, M.J.; Ohler, U.; Nicchitta, C.V.; et al. Translocation of Sickle Cell Erythrocyte MicroRNAs into Plasmodium falciparum Inhibits Parasite Translation and Contributes to Malaria Resistance. Cell Host Microbe 2012, 12, 187–199. [Google Scholar] [CrossRef]
- Meibalan, E.; Marti, M. Biology of Malaria Transmission. Cold Spring Harb. Perspect. Med. 2017, 7, a025452. [Google Scholar] [CrossRef]
- Gupta, H.; Wassmer, S.C. Harnessing the Potential of miRNAs in Malaria Diagnostic and Prevention. Front. Cell. Infect. Microbiol. 2021, 11, 793954. [Google Scholar] [CrossRef] [PubMed]
- de Castro, J.; Rodríguez, M.C.; Martínez-Zorzano, V.S.; Sánchez-Rodríguez, P.; Sánchez-Yagüe, J. Erythrocyte fatty acids as potential biomarkers in the diagnosis of advanced lung adenocarcinoma, lung squamous cell carcinoma, and small cell lung cancer. Am. J. Clin. Pathol. 2014, 142, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Liu, Q.; Hou, J.; Gu, Y.; Zhang, Y.; Chen, Z.; Fan, J.; Zhou, W.; Qiu, S.; Zhang, Y.; et al. Tumor-Induced Generation of Splenic Erythroblast-like Ter-Cells Promotes Tumor Progression. Cell 2018, 173, 634–648.e12. [Google Scholar] [CrossRef]
- Abraham, M.; Klein, S.; Bulvik, B.; Wald, H.; Weiss, I.D.; Olam, D.; Weiss, L.; Beider, K.; Eizenberg, O.; Wald, O.; et al. The CXCR4 inhibitor BL-8040 induces the apoptosis of AML blasts by downregulating ERK, BCL-2, MCL-1 and cyclin-D1 via altered miR-15a/16-1 expression. Leukemia 2017, 31, 2336–2346. [Google Scholar] [CrossRef]
- Fang, X.; Shen, F.; Lechauve, C.; Xu, P.; Zhao, G.; Itkow, J.; Wu, F.; Hou, Y.; Wu, X.; Yu, L.; et al. miR-144/451 represses the LKB1/AMPK/mTOR pathway to promote red cell precursor survival during recovery from acute anemia. Haematologica 2018, 103, 406–416. [Google Scholar] [CrossRef]
- Krakowsky, R.H.E.; Wurm, A.A.; Gerloff, D.; Katzerke, C.; Bräuer-Hartmann, D.; Hartmann, J.U.; Wilke, F.; Thiede, C.; Müller-Tidow, C.; Niederwieser, D.; et al. miR-451a abrogates treatment resistance in FLT3-ITD-positive acute myeloid leukemia. Blood Cancer J. 2018, 8, 36. [Google Scholar] [CrossRef]
- Bai, H.; Wu, S. miR-451: A Novel Biomarker and Potential Therapeutic Target for Cancer. OncoTargets Ther. 2019, 12, 11069–11082. [Google Scholar] [CrossRef]
- Leidinger, P.; Backes, C.; Dahmke, I.N.; Galata, V.; Huwer, H.; Stehle, I.; Bals, R.; Keller, A.; Meese, E. What makes a blood cell based miRNA expression pattern disease specific?—A miRNome analysis of blood cell subsets in lung cancer patients and healthy controls. Oncotarget 2014, 5, 9484–9497. [Google Scholar] [CrossRef]
- Kontidou, E.; Collado, A.; Pernow, J.; Zhou, Z. Erythrocyte-Derived microRNAs: Emerging Players in Cardiovascular and Metabolic Disease. Arterioscler. Thromb. Vasc. Biol. 2023, 43, 628–636. [Google Scholar] [CrossRef]
- Mukai, N.; Nakayama, Y.; Murakami, S.; Tanahashi, T.; Sessler, D.I.; Ishii, S.; Ogawa, S.; Tokuhira, N.; Mizobe, T.; Sawa, T.; et al. Potential contribution of erythrocyte microRNA to secondary erythrocytosis and thrombocytopenia in congenital heart disease. Pediatr. Res. 2018, 83, 866–873. [Google Scholar] [CrossRef]
- Fluitt, M.B.; Kumari, N.; Nunlee-Bland, G.; Nekhai, S.; Gambhir, K.K. miRNA-15a, miRNA-15b, and miRNA-499 are Reduced in Erythrocytes of Pre-Diabetic African-American Adults. Jacobs J. Diabetes Endocrinol. 2016, 2, 14. [Google Scholar]
- Zhou, Z.; Collado, A.; Sun, C.; Tratsiakovich, Y.; Mahdi, A.; Winter, H.; Chernogubova, E.; Seime, T.; Narayanan, S.; Jiao, T.; et al. Downregulation of Erythrocyte miR-210 Induces Endothelial Dysfunction in Type 2 Diabetes. Diabetes 2022, 71, 285–297. [Google Scholar] [CrossRef]
- Duan, Z.Y.; Cai, G.Y.; Li, J.J.; Bu, R.; Chen, X.M. Urinary Erythrocyte-Derived miRNAs: Emerging Role in IgA Nephropathy. Kidney Blood Press. Res. 2017, 42, 738–748. [Google Scholar] [CrossRef]
- Groen, K.; Maltby, V.E.; Lea, R.A.; Sanders, K.A.; Fink, J.L.; Scott, R.J.; Tajouri, L.; Lechner-Scott, J. Erythrocyte microRNA sequencing reveals differential expression in relapsing-remitting multiple sclerosis. BMC Med. Genom. 2018, 11, 48. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Wang, Y.; Telen, M.J.; Chi, J.T. The genomic analysis of erythrocyte microRNA expression in sickle cell diseases. PLoS ONE 2008, 3, e2360. [Google Scholar] [CrossRef]
- Sun, L.; Yu, Y.; Niu, B.; Wang, D. Red Blood Cells as Potential Repositories of MicroRNAs in the Circulatory System. Front. Genet. 2020, 11, 442. [Google Scholar] [CrossRef] [PubMed]
- Gladwin, M.T. Cardiovascular complications in patients with sickle cell disease. Hematol. Am. Soc. Hematol. Educ. Program 2017, 2017, 423–430. [Google Scholar] [CrossRef]
- Jarrick, S.; Lundberg, S.; Sundström, J.; Symreng, A.; Warnqvist, A.; Ludvigsson, J.F. Immunoglobulin A nephropathy and ischemic heart disease: A nationwide population-based cohort study. BMC Nephrol. 2021, 22, 165. [Google Scholar] [CrossRef]
- Anastasiadi, A.T.; Arvaniti, V.Z.; Hudson, K.E.; Kriebardis, A.G.; Stathopoulos, C.; D’Alessandro, A.; Spitalnik, S.L.; Tzounakas, V.L. Exploring unconventional attributes of red blood cells and their potential applications in biomedicine. Protein Cell 2024, 15, 315–330. [Google Scholar] [CrossRef]
- Kulczyńska, K.; Siatecka, M. A regulatory function of long non-coding RNAs in red blood cell development. Acta Biochim. Pol. 2016, 63, 675–680. [Google Scholar] [CrossRef]
- Li, F.; Xiao, Y.; Huang, F.; Deng, W.; Zhao, H.; Shi, X.; Wang, S.; Yu, X.; Zhang, L.; Han, Z.; et al. Spatiotemporal-specific lncRNAs in the brain, colon, liver and lung of macaque during development. Mol. Biosyst. 2015, 11, 3253–3263. [Google Scholar] [CrossRef]
- Hu, W.; Yuan, B.; Flygare, J.; Lodish, H.F. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011, 25, 2573–2578. [Google Scholar] [CrossRef] [PubMed]
- Villamizar, O.; Chambers, C.B.; Mo, Y.Y.; Torry, D.S.; Hofstrand, R.; Riberdy, J.M.; Persons, D.A.; Wilber, A. Fas-antisense long noncoding RNA is differentially expressed during maturation of human erythrocytes and confers resistance to Fas-mediated cell death. Blood Cells Mol. Dis. 2016, 58, 57–66. [Google Scholar] [CrossRef]
- Alvarez-Dominguez, J.R.; Hu, W.; Yuan, B.; Shi, J.; Park, S.S.; Gromatzky, A.A.; van Oudenaarden, A.; Lodish, H.F. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood 2014, 123, 570–581. [Google Scholar] [CrossRef]
- Xu, C.; Shi, L. Long non-coding RNAs during normal erythropoiesis. Blood Sci. 2019, 1, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.F.; Pegtel, D.M.; Lambertz, U.; Leonardi, T.; O’Driscoll, L.; Pluchino, S.; Ter-Ovanesyan, D.; Nolte-‘t Hoen, E.N. ISEV position paper: Extracellular vesicle RNA analysis and bioinformatics. J. Extracell. Vesicles 2013, 2, 22859. [Google Scholar] [CrossRef]
- Solaguren-Beascoa, M.; Gámez-Valero, A.; Escaramís, G.; Herrero-Lorenzo, M.; Ortiz, A.M.; Minguet, C.; Gonzalo, R.; Bravo, M.I.; Costa, M.; Martí, E. Phospho-RNA-Seq Highlights Specific Small RNA Profiles in Plasma Extracellular Vesicles. Int. J. Mol. Sci. 2023, 24, 11653. [Google Scholar] [CrossRef]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef]
- Matsumoto, J.; Stewart, T.; Sheng, L.; Li, N.; Bullock, K.; Song, N.; Shi, M.; Banks, W.A.; Zhang, J. Transmission of α-synuclein-containing erythrocyte-derived extracellular vesicles across the blood-brain barrier via adsorptive mediated transcytosis: Another mechanism for initiation and progression of Parkinson’s disease? Acta Neuropathol. Commun. 2017, 5, 71. [Google Scholar] [CrossRef]
- Mumtaz, P.T.; Taban, Q.; Dar, M.A.; Mir, S.; Haq, Z.u.; Zargar, S.M.; Shah, R.A.; Ahmad, S.M. Deep Insights in Circular RNAs: From biogenesis to therapeutics. Biol. Proced. Online 2020, 22, 10. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, J.; Cao, X.; Cai, Z.; Zhao, F. Exploring the cellular landscape of circular RNAs using full-length single-cell RNA sequencing. Nat. Commun. 2022, 13, 3242. [Google Scholar] [CrossRef]
- Misir, S.; Wu, N.; Yang, B.B. Specific expression and functions of circular RNAs. Cell Death Differ. 2022, 29, 481–491. [Google Scholar] [CrossRef]
- Habara, A. Comparative circRNA Profiling in Human Erythroblasts Derived from Fetal Liver and Bone Marrow Hematopoietic Stem Cells Using Public RNA-Seq Data. Int. J. Mol. Sci. 2025, 26, 8397. [Google Scholar] [CrossRef] [PubMed]
- Nicolet, B.P.; Engels, S.; Aglialoro, F.; van den Akker, E.; von Lindern, M.; Wolkers, M.C. Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res. 2018, 46, 8168–8180. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, H.; Fu, L.; Xu, T. Investigating the Underlying Mechanisms of Circular RNAs and Their Application in Clinical Research of Cervical Cancer. Front. Genet. 2021, 12, 653051. [Google Scholar] [CrossRef]
- Nassiri, S.M.; Ahmadi Afshar, N.; Almasi, P. Insight into microRNAs’ involvement in hematopoiesis: Current standing point of findings. Stem Cell Res. Ther. 2023, 14, 282. [Google Scholar] [CrossRef] [PubMed]
- Juzenas, S.; Lindqvist, C.M.; Ito, G.; Dolshanskaya, Y.; Halfvarson, J.; Franke, A.; Hemmrich-Stanisak, G. Depletion of erythropoietic miR-486-5p and miR-451a improves detectability of rare microRNAs in peripheral blood-derived small RNA sequencing libraries. NAR Genom. Bioinform. 2020, 2, lqaa008. [Google Scholar] [CrossRef]
- Yang, F.; Ruan, H.; Li, S.; Hou, W.; Qiu, Y.; Deng, L.; Su, S.; Chen, P.; Pang, L.; Lai, K. Analysis of circRNAs and circRNA-associated competing endogenous RNA networks in β-thalassemia. Sci. Rep. 2022, 12, 8071. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Lv, A.; Zhang, S.; Zheng, J.; Lin, N.; Xu, L.; Huang, H. Peripheral blood circular RNA circ-0008102 may serve as a novel clinical biomarker in beta-thalassemia patients. Eur. J. Pediatr. 2024, 183, 1367–1379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, G.; Yuan, Z.; Zhang, Y.; Chen, X.; Huang, J.; Li, N.; Liu, Z.; Zhong, W.; Huang, H.; et al. Profiling and Bioinformatics Analysis Revealing Differential Circular RNA Expression about Storage Lesion Regulatory in Stored Red Blood Cells. Transfus. Med. Hemother. 2021, 49, 76–87. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dryllis, G.; Fortis, S.P.; Kouroupaki, A.; Tsamesidis, I.; Birtsas, V.; Tsantes, A.G.; Valsami, S.; Konstantopoulos, K.; Papageorgiou, E.G.; Pessach, I.; et al. Residual Genetic Material in Mature Red Blood Cells. Int. J. Mol. Sci. 2025, 26, 10774. https://doi.org/10.3390/ijms262110774
Dryllis G, Fortis SP, Kouroupaki A, Tsamesidis I, Birtsas V, Tsantes AG, Valsami S, Konstantopoulos K, Papageorgiou EG, Pessach I, et al. Residual Genetic Material in Mature Red Blood Cells. International Journal of Molecular Sciences. 2025; 26(21):10774. https://doi.org/10.3390/ijms262110774
Chicago/Turabian StyleDryllis, Georgios, Sotirios P. Fortis, Aspasia Kouroupaki, Ioannis Tsamesidis, Vassilios Birtsas, Andreas G. Tsantes, Serena Valsami, Konstantinos Konstantopoulos, Effie G. Papageorgiou, Ilias Pessach, and et al. 2025. "Residual Genetic Material in Mature Red Blood Cells" International Journal of Molecular Sciences 26, no. 21: 10774. https://doi.org/10.3390/ijms262110774
APA StyleDryllis, G., Fortis, S. P., Kouroupaki, A., Tsamesidis, I., Birtsas, V., Tsantes, A. G., Valsami, S., Konstantopoulos, K., Papageorgiou, E. G., Pessach, I., & Kriebardis, A. G. (2025). Residual Genetic Material in Mature Red Blood Cells. International Journal of Molecular Sciences, 26(21), 10774. https://doi.org/10.3390/ijms262110774








