Hepatic Zonation in MASLD: Old Question, New Challenge in the Era of Spatial Omics
Abstract
1. Introduction
2. Zonation of Liver Architecture
3. Transcriptomics Signature of Liver Zonation
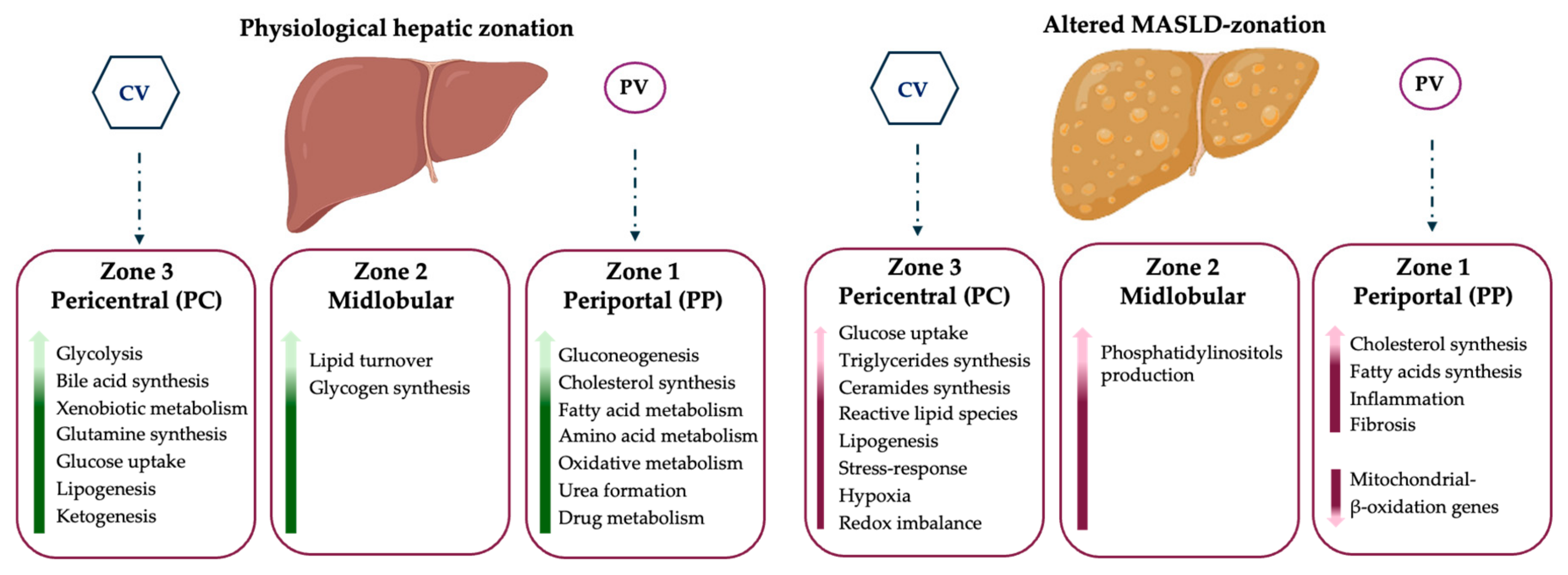
4. Metabolic Zonation: Physiological Status vs. MASLD
5. The Advent of Spatial-Omics and Advantages in Hepatology Research
5.1. Spatial Transcriptomics: Molecular Fundamentals and Applications in MASLD
5.2. Spatial Proteomics
5.3. Spatial Metabolomics and Lipidomics
5.4. Other Spatial Approaches (Spatial Genomics/Epigenomics)
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MASLD | Metabolic dysfunction-associated steatotic liver disease |
| MASH | Metabolic dysfunction-associated steatohepatitis |
| SLD | Steatotic liver disease |
| HCC | Hepatocellular carcinoma |
| IR | Insulin resistance |
| T2DM | Type 2 diabetes mellitus |
| ROS | Reactive oxygen species |
| FFA | Free fatty acids |
| mtDNA | Mitochondrial DNA |
| ER | Endoplasmic reticulum |
| LSECs | Liver Sinusoidal Endothelial cells |
| HSCs | Hepatic stellate cells |
| KCs | Kupffer cells |
| PC | Pericentral |
| PP | Periportal |
| Hh | Hedgehog |
| GSK3 | Glycogen synthase kinase-3 |
| APC | Adenoma polyposis coli |
| DVL | Dishevelled |
| LRP5/6 | LDL receptor–related protein 5/6 |
| KO | Knockout |
| Rspo | Rspondin |
| TCF | T-cell factor |
| NGS | Next-generation sequencing |
| scRNA-seq | Single-cell RNA sequencing |
| Drop-seq | Droplet-based scRNA-seq |
| HFD | High-fat diet |
| LDs | Lipid droplets |
| AMLN | Amylin |
| TREM2 | Triggering Receptor Expressed on Myeloid Cells 2 |
| TME | Tumor microenvironment |
| CAF | Cancer-associated fibroblasts |
| TIB | Tumor immune barrier |
| scDVP | Single-cell Deep Visual Proteomics |
| AI | Artificial intelligence |
| MS | Mass spectrometry |
| LAMs | Lipid-associated macrophages |
| LPCAT2 | Lysophosphatidylcholine Acyltransferase 2 |
| GRP35 | G-Protein-Coupled Receptor 35 |
| SEAM | Spatial single nuclear metabolomics |
| CNVs | Copy number variations |
References
- Younossi, Z.; Aggarwal, P.; Shrestha, I.; Fernandes, J.; Johansen, P.; Augusto, M.; Nair, S. The burden of non-alcoholic steatohepatitis: A systematic review of health-related quality of life and patient-reported outcomes. JHEP Rep. 2022, 4, 100525. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.K.; Chuah, K.H.; Rajaram, R.B.; Lim, L.L.; Ratnasingam, J.; Vethakkan, S.R. Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD): A State-of-the-Art Review. J. Obes. Metab. Syndr. 2023, 32, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Rumgay, H.; Arnold, M.; Ferlay, J.; Lesi, O.; Cabasag, C.J.; Vignat, J.; Laversanne, M.; McGlynn, K.A.; Soerjomataram, I. Global burden of primary liver cancer in 2020 and predictions to 2040. J. Hepatol. 2022, 77, 1598–1606. [Google Scholar] [CrossRef]
- Marchesini, G.; Brizi, M.; Bianchi, G.; Tomassetti, S.; Bugianesi, E.; Lenzi, M.; McCullough, A.J.; Natale, S.; Forlani, G.; Melchionda, N. Nonalcoholic fatty liver disease: A feature of the metabolic syndrome. Diabetes 2001, 50, 1844–1850. [Google Scholar] [CrossRef]
- Longo, M.; Meroni, M.; Paolini, E.; Macchi, C.; Dongiovanni, P. Mitochondrial dynamics and nonalcoholic fatty liver disease (NAFLD): New perspectives for a fairy-tale ending? Metab. Clin. Exp. 2021, 117, 154708. [Google Scholar] [CrossRef]
- Eskridge, W.; Cryer, D.R. Metabolic Dysfunction-Associated Steatotic Liver Disease and Metabolic Dysfunction-Associated Steatohepatitis: The Patient and Physician Perspective. J. Clin. Med. 2023, 12, 6216. [Google Scholar] [CrossRef]
- Han, H.S.; Kang, G.; Kim, J.S.; Choi, B.H.; Koo, S.H. Regulation of glucose metabolism from a liver-centric perspective. Exp. Mol. Med. 2016, 48, e218. [Google Scholar] [CrossRef]
- Godoy, P.; Hewitt, N.J.; Albrecht, U.; Andersen, M.E.; Ansari, N.; Bhattacharya, S.; Bode, J.G.; Bolleyn, J.; Borner, C.; Böttger, J.; et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch. Toxicol. 2013, 87, 1315–1530. [Google Scholar] [CrossRef]
- Jungermann, K.; Kietzmann, T. Zonation of parenchymal and nonparenchymal metabolism in liver. Annu. Rev. Nutr. 1996, 16, 179–203. [Google Scholar] [CrossRef]
- Gebhardt, R.; Matz-Soja, M. Liver zonation: Novel aspects of its regulation and its impact on homeostasis. World J. Gastroenterol. 2014, 20, 8491–8504. [Google Scholar] [CrossRef]
- Lambers, L.; Waschinsky, N.; Schleicher, J.; König, M.; Tautenhahn, H.-M.; Albadry, M.; Dahmen, U.; Ricken, T. Quantifying fat zonation in liver lobules: An integrated multiscale in silico model combining disturbed microperfusion and fat metabolism via a continuum biomechanical bi-scale, tri-phasic approach. Biomech. Model. Mechanobiol. 2024, 23, 631–653. [Google Scholar] [CrossRef]
- Rappaport, A.M. The structural and functional unit in the human liver (liver acinus). Anat. Rec. 1958, 130, 673–689. [Google Scholar] [CrossRef]
- Braeuning, A.; Ittrich, C.; Köhle, C.; Hailfinger, S.; Bonin, M.; Buchmann, A.; Schwarz, M. Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 2006, 273, 5051–5061. [Google Scholar] [CrossRef]
- Halpern, K.B.; Shenhav, R.; Matcovitch-Natan, O.; Toth, B.; Lemze, D.; Golan, M.; Massasa, E.E.; Baydatch, S.; Landen, S.; Moor, A.E.; et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 2017, 542, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Mowry, L.E.; Nejak-Bowen, K.N.; Okabe, H.; Diegel, C.R.; Lang, R.A.; Williams, B.O.; Monga, S.P. β-catenin signaling in murine liver zonation and regeneration: A Wnt-Wnt situation! Hepatology 2014, 60, 964–976. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, F.; Andersson, A.; Saarenpää, S.; Larsson, L.; Van Hul, N.; Kanatani, S.; Masek, J.; Ellis, E.; Barragan, A.; Mollbrink, A.; et al. Spatial Transcriptomics to define transcriptional patterns of zonation and structural components in the mouse liver. Nat. Commun. 2021, 12, 7046. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.A.; Delgado, T.C. Spatial metabolomics and its application in the liver. Hepatology 2024, 79, 1158–1179. [Google Scholar] [CrossRef]
- Sasse, D.; Spornitz, U.M.; Maly, I.P. Liver architecture. Enzyme 1992, 46, 8–32. [Google Scholar] [CrossRef]
- Saxena, R.; Theise, N.D.; Crawford, J.M. Microanatomy of the human liver-exploring the hidden interfaces. Hepatology 1999, 30, 1339–1346. [Google Scholar] [CrossRef]
- Kietzmann, T. Metabolic zonation of the liver: The oxygen gradient revisited. Redox Biol. 2017, 11, 622–630. [Google Scholar] [CrossRef]
- Droin, C.; Kholtei, J.E.; Bahar Halpern, K.; Hurni, C.; Rozenberg, M.; Muvkadi, S.; Itzkovitz, S.; Naef, F. Space-time logic of liver gene expression at sub-lobular scale. Nat. Metab. 2021, 3, 43–58. [Google Scholar] [CrossRef]
- Ben-Moshe, S.; Itzkovitz, S. Spatial heterogeneity in the mammalian liver. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 395–410. [Google Scholar] [CrossRef] [PubMed]
- Paris, J.; Henderson, N.C. Liver zonation, revisited. Hepatology 2022, 76, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Martini, T.; Naef, F.; Tchorz, J.S. Spatiotemporal Metabolic Liver Zonation and Consequences on Pathophysiology. Annu. Rev. Pathol. Mech. Dis. 2023, 18, 439–466. [Google Scholar] [CrossRef]
- Goel, C.; Monga, S.P.; Nejak-Bowen, K. Role and Regulation of Wnt/β-Catenin in Hepatic Perivenous Zonation and Physiological Homeostasis. Am. J. Pathol. 2022, 192, 4–17. [Google Scholar] [CrossRef]
- Aberle, H.; Bauer, A.; Stappert, J.; Kispert, A.; Kemler, R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997, 16, 3797–3804. [Google Scholar] [CrossRef]
- Liu, J.; Stevens, J.; Matsunami, N.; White, R.L. Targeted degradation of beta-catenin by chimeric F-box fusion proteins. Biochem. Biophys. Res. Commun. 2004, 313, 1023–1029. [Google Scholar] [CrossRef]
- Monga, S.P. Role of Wnt/β-catenin signaling in liver metabolism and cancer. Int. J. Biochem. Cell Biol. 2011, 43, 1021–1029. [Google Scholar] [CrossRef]
- Benhamouche, S.; Decaens, T.; Godard, C.; Chambrey, R.; Rickman, D.S.; Moinard, C.; Vasseur-Cognet, M.; Kuo, C.J.; Kahn, A.; Perret, C.; et al. Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Dev. Cell 2006, 10, 759–770. [Google Scholar] [CrossRef]
- Sekine, S.; Lan, B.Y.; Bedolli, M.; Feng, S.; Hebrok, M. Liver-specific loss of beta-catenin blocks glutamine synthesis pathway activity and cytochrome p450 expression in mice. Hepatology 2006, 43, 817–825. [Google Scholar] [CrossRef]
- Gougelet, A.; Torre, C.; Veber, P.; Sartor, C.; Bachelot, L.; Denechaud, P.D.; Godard, C.; Moldes, M.; Burnol, A.F.; Dubuquoy, C.; et al. T-cell factor 4 and β-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology 2014, 59, 2344–2357. [Google Scholar] [CrossRef]
- Colletti, M.; Cicchini, C.; Conigliaro, A.; Santangelo, L.; Alonzi, T.; Pasquini, E.; Tripodi, M.; Amicone, L. Convergence of Wnt signaling on the HNF4alpha-driven transcription in controlling liver zonation. Gastroenterology 2009, 137, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Burke, Z.D.; Reed, K.R.; Phesse, T.J.; Sansom, O.J.; Clarke, A.R.; Tosh, D. Liver zonation occurs through a beta-catenin-dependent, c-Myc-independent mechanism. Gastroenterology 2009, 136, 2316–2324.e2311. [Google Scholar] [CrossRef] [PubMed]
- Gougelet, A.; Sartor, C.; Senni, N.; Calderaro, J.; Fartoux, L.; Lequoy, M.; Wendum, D.; Talbot, J.N.; Prignon, A.; Chalaye, J.; et al. Hepatocellular Carcinomas with Mutational Activation of Beta-Catenin Require Choline and Can Be Detected by Positron Emission Tomography. Gastroenterology 2019, 157, 807–822. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Shen, J.; Yan, J.; Bott, A.J.; Maimouni, S.; Daguplo, H.Q.; Wang, Y.; Khayati, K.; Guo, J.Y.; Zhang, L.; et al. Glutamine synthetase limits β-catenin-mutated liver cancer growth by maintaining nitrogen homeostasis and suppressing mTORC1. J. Clin. Investig. 2022, 132, e161408. [Google Scholar] [CrossRef]
- Rocha, A.S.; Vidal, V.; Mertz, M.; Kendall, T.J.; Charlet, A.; Okamoto, H.; Schedl, A. The Angiocrine Factor Rspondin3 Is a Key Determinant of Liver Zonation. Cell Rep. 2015, 13, 1757–1764. [Google Scholar] [CrossRef]
- Huch, M.; Dorrell, C.; Boj, S.F.; van Es, J.H.; Li, V.S.; van de Wetering, M.; Sato, T.; Hamer, K.; Sasaki, N.; Finegold, M.J.; et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013, 494, 247–250. [Google Scholar] [CrossRef]
- Planas-Paz, L.; Orsini, V.; Boulter, L. The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat. Cell Biol. 2016, 18, 467–479. [Google Scholar] [CrossRef]
- Kietzmann, T. Liver Zonation in Health and Disease: Hypoxia and Hypoxia-Inducible Transcription Factors as Concert Masters. Int. J. Mol. Sci. 2019, 20, 2347. [Google Scholar] [CrossRef]
- Kietzmann, T.; Cornesse, Y.; Brechtel, K.; Modaressi, S.; Jungermann, K. Perivenous expression of the mRNA of the three hypoxia-inducible factor alpha-subunits, HIF1alpha, HIF2alpha and HIF3alpha, in rat liver. Biochem. J. 2001, 354, 531–537. [Google Scholar] [CrossRef]
- Rappaport, A.M.; Borowy, Z.J.; Lougheed, W.M.; Lotto, W.N. Subdivision of hexagonal liver lobules into a structural and functional unit; role in hepatic physiology and pathology. Anat. Rec. 1954, 119, 11–33. [Google Scholar] [CrossRef]
- Jungermann, K.; Kietzmann, T. Oxygen: Modulator of metabolic zonation and disease of the liver. Hepatology 2000, 31, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, J.; O’Brien, W.T.; Johnson, R.S.; LaManna, J.C.; Chavez, J.C.; Klein, P.S.; Simon, M.C. O2 regulates stem cells through Wnt/β-catenin signalling. Nat. Cell Biol. 2010, 12, 1007–1013. [Google Scholar] [CrossRef]
- Newton, I.P.; Kenneth, N.S.; Appleton, P.L.; Näthke, I.; Rocha, S. Adenomatous polyposis coli and hypoxia-inducible factor-1α have an antagonistic connection. Mol. Biol. Cell 2010, 21, 3630–3638. [Google Scholar] [CrossRef]
- Schmucker, D.L.; Mooney, J.S.; Jones, A.L. Stereological analysis of hepatic fine structure in the Fischer 344 rat. Influence of sublobular location and animal age. J. Cell Biol. 1978, 78, 319–337. [Google Scholar] [CrossRef]
- Novikoff, A.B. Cell heterogeneity within the hepatic lobule of the rat: Staining reactions. J. Histochem. Cytochem. Off. J. Histochem. Soc. 1959, 7, 240–244. [Google Scholar] [CrossRef]
- Loud, A.V. A quantitative stereological description of the ultrastructure of normal rat liver parenchymal cells. J. Cell Biol. 1968, 37, 27–46. [Google Scholar] [CrossRef]
- Jungermann, K. Metabolic zonation of liver parenchyma. Regulation of the glucostat of the liver. Die Naturwissenschaften 1985, 72, 76–84. [Google Scholar] [CrossRef]
- Jungermann, K.; Kietzmann, T. Role of oxygen in the zonation of carbohydrate metabolism and gene expression in liver. Kidney Int. 1997, 51, 402–412. [Google Scholar] [CrossRef]
- Sato, B.; Tanaka, A.; Mori, S.; Yanabu, N.; Kitai, T.; Tokuka, A.; Inomoto, T.; Iwata, S.; Yamaoka, Y.; Chance, B. Quantitative analysis of redox gradient within the rat liver acini by fluorescence images: Effects of glucagon perfusion. Biochim. Biophys. Acta 1995, 1268, 20–26. [Google Scholar] [CrossRef]
- Ballé, C.; Jungermann, K. Control of urea production, glutamine release and ammonia uptake in the perfused rat liver by the sympathetic innervation. Eur. J. Biochem. 1986, 158, 13–18. [Google Scholar] [CrossRef]
- Bartels, H.; Vogt, B.; Jungermann, K. Glycogen synthesis from pyruvate in the periportal and from glucose in the perivenous zone in perfused livers from fasted rats. FEBS Lett. 1987, 221, 277–283. [Google Scholar] [CrossRef]
- Nauck, M.; Wölfle, D.; Katz, N.; Jungermann, K. Modulation of the glucagon-dependent induction of phosphoenolpyruvate carboxykinase and tyrosine aminotransferase by arterial and venous oxygen concentrations in hepatocyte cultures. Eur. J. Biochem. 1981, 119, 657–661. [Google Scholar] [CrossRef]
- Schleicher, J.; Guthke, R.; Dahmen, U.; Dirsch, O.; Holzhuetter, H.G.; Schuster, S. A theoretical study of lipid accumulation in the liver-implications for nonalcoholic fatty liver disease. Biochim. Biophys. Acta 2014, 1841, 62–69. [Google Scholar] [CrossRef]
- Cheng, X.; Kim, S.Y.; Okamoto, H.; Xin, Y.; Yancopoulos, G.D.; Murphy, A.J.; Gromada, J. Glucagon contributes to liver zonation. Proc. Natl. Acad. Sci. USA 2018, 115, E4111–E4119. [Google Scholar] [CrossRef] [PubMed]
- Matz-Soja, M.; Hovhannisyan, A.; Gebhardt, R. Hedgehog signalling pathway in adult liver: A major new player in hepatocyte metabolism and zonation? Med. Hypotheses 2013, 80, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Sicklick, J.K.; Li, Y.X.; Choi, S.S.; Qi, Y.; Chen, W.; Bustamante, M.; Huang, J.; Zdanowicz, M.; Camp, T.; Torbenson, M.S.; et al. Role for hedgehog signaling in hepatic stellate cell activation and viability. Lab. Investig. J. Tech. Methods Pathol. 2005, 85, 1368–1380. [Google Scholar] [CrossRef] [PubMed]
- Omenetti, A.; Bass, L.M.; Anders, R.A.; Clemente, M.G.; Francis, H.; Guy, C.D.; McCall, S.; Choi, S.S.; Alpini, G.; Schwarz, K.B.; et al. Hedgehog activity, epithelial-mesenchymal transitions, and biliary dysmorphogenesis in biliary atresia. Hepatology 2011, 53, 1246–1258. [Google Scholar] [CrossRef]
- Diehl, A.M.; Chute, J. Underlying potential: Cellular and molecular determinants of adult liver repair. J. Clin. Investig. 2013, 123, 1858–1860. [Google Scholar] [CrossRef]
- Michelotti, G.A.; Machado, M.V.; Diehl, A.M. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 656–665. [Google Scholar] [CrossRef]
- Salaritabar, A.; Berindan-Neagoe, I.; Darvish, B.; Hadjiakhoondi, F.; Manayi, A.; Devi, K.P.; Barreca, D.; Orhan, I.E.; Süntar, I.; Farooqi, A.A.; et al. Targeting Hedgehog signaling pathway: Paving the road for cancer therapy. Pharmacol. Res. 2019, 141, 466–480. [Google Scholar] [CrossRef]
- Kolbe, E.; Aleithe, S.; Rennert, C.; Spormann, L.; Ott, F.; Meierhofer, D.; Gajowski, R.; Stöpel, C.; Hoehme, S.; Kücken, M.; et al. Mutual Zonated Interactions of Wnt and Hh Signaling Are Orchestrating the Metabolism of the Adult Liver in Mice and Human. Cell Rep. 2019, 29, 4553–4567.e4557. [Google Scholar] [CrossRef]
- Matz-Soja, M.; Aleithe, S.; Marbach, E.; Böttger, J.; Arnold, K.; Schmidt-Heck, W.; Kratzsch, J.; Gebhardt, R. Hepatic Hedgehog signaling contributes to the regulation of IGF1 and IGFBP1 serum levels. Cell Commun. Signal. CCS 2014, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Braeuning, A.; Menzel, M.; Kleinschnitz, E.M.; Harada, N.; Tamai, Y.; Köhle, C.; Buchmann, A.; Schwarz, M. Serum components and activated Ha-ras antagonize expression of perivenous marker genes stimulated by beta-catenin signaling in mouse hepatocytes. FEBS J. 2007, 274, 4766–4777. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Lefebvre, A.T.; Selzner, N.; Wrana, J.L.; Bhat, M. The hippo pathway: A master regulator of liver metabolism, regeneration, and disease. FASEB J. 2021, 35, e21570. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Schönberger, K.; Tchorz, J.S. Distinct hepatocyte identities in liver homeostasis and regeneration. JHEP Rep. 2023, 5, 100779. [Google Scholar] [CrossRef]
- Cunningham, R.P.; Porat-Shliom, N. Liver Zonation—Revisiting Old Questions with New Technologies. Front. Physiol. 2021, 12, 732929. [Google Scholar] [CrossRef]
- Brunt, E.M.; Kleiner, D.E.; Wilson, L.A.; Unalp, A.; Behling, C.E.; Lavine, J.E.; Neuschwander-Tetri, B.A. Portal chronic inflammation in nonalcoholic fatty liver disease (NAFLD): A histologic marker of advanced NAFLD-Clinicopathologic correlations from the nonalcoholic steatohepatitis clinical research network. Hepatology 2009, 49, 809–820. [Google Scholar] [CrossRef]
- Ekstedt, M.; Franzén, L.E.; Mathiesen, U.L.; Thorelius, L.; Holmqvist, M.; Bodemar, G.; Kechagias, S. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology 2006, 44, 865–873. [Google Scholar] [CrossRef]
- Xiong, X.; Kuang, H.; Ansari, S.; Liu, T.; Gong, J.; Wang, S.; Zhao, X.Y.; Ji, Y.; Li, C.; Guo, L.; et al. Landscape of Intercellular Crosstalk in Healthy and NASH Liver Revealed by Single-Cell Secretome Gene Analysis. Mol. Cell. 2019, 75, 644–660.e645. [Google Scholar] [CrossRef]
- Hendrikx, T.; Porsch, F.; Kiss, M.G.; Rajcic, D.; Papac-Miličević, N.; Hoebinger, C.; Goederle, L.; Hladik, A.; Shaw, L.E.; Horstmann, H.; et al. Soluble TREM2 levels reflect the recruitment and expansion of TREM2(+) macrophages that localize to fibrotic areas and limit NASH. J. Hepatol. 2022, 77, 1373–1385. [Google Scholar] [CrossRef]
- Brosch, M.; Kattler, K.; Herrmann, A.; von Schönfels, W.; Nordström, K.; Seehofer, D.; Damm, G.; Becker, T.; Zeissig, S.; Nehring, S.; et al. Epigenomic map of human liver reveals principles of zonated morphogenic and metabolic control. Nat. Commun. 2018, 9, 4150. [Google Scholar] [CrossRef]
- Steinman, J.B.; Salomao, M.A.; Pajvani, U.B. Zonation in NASH—A key paradigm for understanding pathophysiology and clinical outcomes. Liver Int. 2021, 41, 2534–2546. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Park, S.R.; Cho, C.S.; Xi, J.; Kang, H.M.; Lee, J.H. Holistic characterization of single-hepatocyte transcriptome responses to high-fat diet. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E244–E258. [Google Scholar] [CrossRef] [PubMed]
- Sveinbjornsson, G.; Ulfarsson, M.O.; Thorolfsdottir, R.B.; Jonsson, B.A.; Einarsson, E.; Gunnlaugsson, G.; Rognvaldsson, S.; Arnar, D.O.; Baldvinsson, M.; Bjarnason, R.G.; et al. Multiomics study of nonalcoholic fatty liver disease. Nat. Genet. 2022, 54, 1652–1663. [Google Scholar] [CrossRef]
- Longo, S.K.; Guo, M.G.; Ji, A.L.; Khavari, P.A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet. 2021, 22, 627–644. [Google Scholar] [CrossRef]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef]
- Katzenelenbogen, Y.; Sheban, F.; Yalin, A.; Yofe, I.; Svetlichnyy, D.; Jaitin, D.A.; Bornstein, C.; Moshe, A.; Keren-Shaul, H.; Cohen, M.; et al. Coupled scRNA-Seq and Intracellular Protein Activity Reveal an Immunosuppressive Role of TREM2 in Cancer. Cell 2020, 182, 872–885.e819. [Google Scholar] [CrossRef]
- Molgora, M.; Esaulova, E.; Vermi, W.; Hou, J.; Chen, Y.; Luo, J.; Brioschi, S.; Bugatti, M.; Omodei, A.S.; Ricci, B.; et al. TREM2 Modulation Remodels the Tumor Myeloid Landscape Enhancing Anti-PD-1 Immunotherapy. Cell 2020, 182, 886–900.e817. [Google Scholar] [CrossRef]
- Chung, B.K.; Øgaard, J.; Reims, H.M.; Karlsen, T.H.; Melum, E. Spatial transcriptomics identifies enriched gene expression and cell types in human liver fibrosis. Hepatol. Commun. 2022, 6, 2538–2550. [Google Scholar] [CrossRef] [PubMed]
- Khanal, S.; Liu, Y.; Bamidele, A.O.; Wixom, A.Q.; Washington, A.M.; Jalan-Sakrikar, N.; Cooper, S.A.; Vuckovic, I.; Zhang, S.; Zhong, J.; et al. Glycolysis in hepatic stellate cells coordinates fibrogenic extracellular vesicle release spatially to amplify liver fibrosis. Sci. Adv. 2024, 10, eadn5228. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xun, Z.; Ma, K.; Liang, S.; Li, X.; Zhou, S.; Sun, L.; Liu, Y.; Du, Y.; Guo, X.; et al. Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J. Hepatol. 2023, 78, 770–782. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, E.; Borner, G.H.H. Spatial proteomics: A powerful discovery tool for cell biology. Nat. Rev. Mol. Cell Biol. 2019, 20, 285–302. [Google Scholar] [CrossRef]
- Inverso, D.; Shi, J.; Lee, K.H.; Jakab, M.; Ben-Moshe, S.; Kulkarni, S.R.; Schneider, M.; Wang, G.; Komeili, M.; Vélez, P.A.; et al. A spatial vascular transcriptomic, proteomic, and phosphoproteomic atlas unveils an angiocrine Tie-Wnt signaling axis in the liver. Dev. Cell 2021, 56, 1677–1693.e1610. [Google Scholar] [CrossRef]
- Weiss, C.A.M.; Brown, L.A.; Miranda, L.; Pellizzoni, P.; Ben-Moshe, S.; Steigerwald, S.; Remmert, K.; Hernandez, J.; Borgwardt, K.; Rosenberger, F.A.; et al. Single cell spatial proteomics maps human liver zonation patterns and their vulnerability to fibrosis. bioRxiv 2025, 2025-04. [Google Scholar] [CrossRef]
- Guilliams, M.; Bonnardel, J.; Haest, B.; Vanderborght, B.; Wagner, C.; Remmerie, A.; Bujko, A.; Martens, L.; Thoné, T.; Browaeys, R.; et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 2022, 185, 379–396.e338. [Google Scholar] [CrossRef]
- Kang, S.W.S.; Cunningham, R.P.; Miller, C.B.; Brown, L.A.; Cultraro, C.M.; Harned, A.; Narayan, K.; Hernandez, J.; Jenkins, L.M.; Lobanov, A.; et al. A spatial map of hepatic mitochondria uncovers functional heterogeneity shaped by nutrient-sensing signaling. Nat. Commun. 2024, 15, 1799. [Google Scholar] [CrossRef]
- Seubnooch, P.; Montani, M.; Dufour, J.F.; Masoodi, M. Spatial lipidomics reveals zone-specific hepatic lipid alteration and remodeling in metabolic dysfunction-associated steatohepatitis. J. Lipid Res. 2024, 65, 100599. [Google Scholar] [CrossRef]
- Seubnooch, P.; Montani, M.; Tsouka, S.; Claude, E.; Rafiqi, U.; Perren, A.; Dufour, J.F.; Masoodi, M. Characterisation of hepatic lipid signature distributed across the liver zonation using mass spectrometry imaging. JHEP Rep. 2023, 5, 100725. [Google Scholar] [CrossRef]
- Hall, Z.; Bond, N.J.; Ashmore, T.; Sanders, F.; Ament, Z.; Wang, X.; Murray, A.J.; Bellafante, E.; Virtue, S.; Vidal-Puig, A.; et al. Lipid zonation and phospholipid remodeling in nonalcoholic fatty liver disease. Hepatology 2017, 65, 1165–1180. [Google Scholar] [CrossRef] [PubMed]
- Wattacheril, J.; Seeley, E.H.; Angel, P.; Chen, H.; Bowen, B.P.; Lanciault, C.; Caprioli, R.M.; Abumrad, N.; Flynn, C.R. Differential intrahepatic phospholipid zonation in simple steatosis and nonalcoholic steatohepatitis. PLoS ONE 2013, 8, e57165. [Google Scholar] [CrossRef] [PubMed]
- Otkur, W.; Zhang, Y.; Li, Y.; Bao, W.; Feng, T.; Wu, B.; Ma, Y.; Shi, J.; Wang, L.; Pei, S.; et al. Spatial multi-omics characterizes GPR35-relevant lipid metabolism signatures across liver zonation in MASLD. Life Metab. 2024, 3, loae021. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Rios, S.; O’Connor, I.P.; Kent, L.N.; Clouse, J.M.; Hadjiyannis, Y.; Koivisto, C.; Pecot, T.; Angel, P.M.; Drake, R.R.; Leone, G.; et al. Imaging Mass Spectrometry Reveals Alterations in N-Linked Glycosylation That Are Associated With Histopathological Changes in Nonalcoholic Steatohepatitis in Mouse and Human. Mol. Cell. Proteom. 2022, 21, 100225. [Google Scholar] [CrossRef]
- Alamri, H.; Patterson, N.H.; Yang, E.; Zoroquiain, P.; Lazaris, A.; Chaurand, P.; Metrakos, P. Mapping the triglyceride distribution in NAFLD human liver by MALDI imaging mass spectrometry reveals molecular differences in micro and macro steatosis. Anal. Bioanal. Chem. 2019, 411, 885–894. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhou, Q.; Cai, L.; Pan, L.; Sun, W.; Qumu, S.; Yu, S.; Feng, J.; Zhao, H. SEAM is a spatial single nuclear metabolomics method for dissecting tissue microenvironment. Nat. Methods 2021, 18, 1223–1232. [Google Scholar] [CrossRef]
- Matchett, K.P.; Paris, J.; Teichmann, S.A. Spatial genomics: Mapping human steatotic liver disease. Nat. Rev. Gastroenterol. Hepatol. 2024, 21, 646–660. [Google Scholar] [CrossRef]
- Llorens-Bobadilla, E.; Zamboni, M. Solid-phase capture and profiling of open chromatin by spatial ATAC. Nat. Biotechnol. 2023, 41, 1085–1088. [Google Scholar] [CrossRef]
- McKellar, D.W.; Mantri, M. Spatial mapping of the total transcriptome by in situ polyadenylation. Nat. Biotechnol. 2023, 41, 513–520. [Google Scholar] [CrossRef]
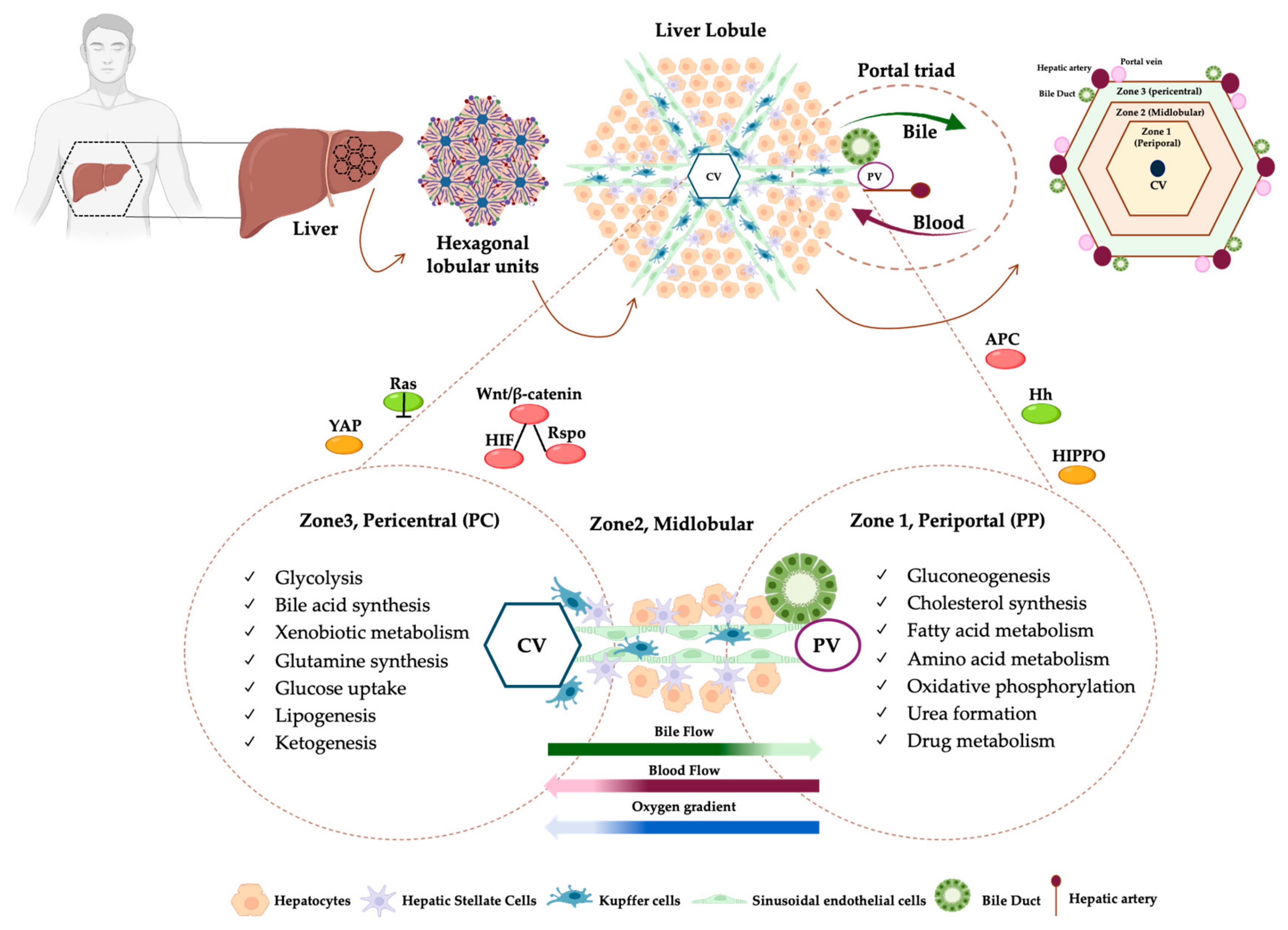
| Zone | |||
|---|---|---|---|
| Pericentral | Midlobular | Periportal | |
| Marker Genes | AHR, ALDH3A2, CSAD, CYP1A2, CYP2A5, CYP2C, CYP2E1, CYP3A, CYP3A4, CYP4A14, GCK, GLUL, GS, GSTM1, LDHD, LECT2, MUP17, OAT, SLC1A2, IGFBP1, NT5E, ADHA4, BCHE | HAMP, HAMP2, IGFBP2, CYP8B1, HINT1, COX7C, APOC1, FABP1, MT2A, MT1G, NDUFB1 | CPS1, SDS, CYP2F2, HAL, GLS2, ASS1, HSD17B13, HSD17B6, ALDH1B1, ARG1, PCK1, SULT5A1, MFSD2A, BTNL9, ANPEP, MTHFS, GATM, L-FABP, SOX9, E-CDH1, MUP1, GSTP1, HPX, ALDOB, FBP1, MUP21, IGF1, CDH1, CYP7A1, HSB3B7, HMGCS1 |
| Biological Process |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paolini, E.; Longo, M.; Meroni, M.; Dongiovanni, P. Hepatic Zonation in MASLD: Old Question, New Challenge in the Era of Spatial Omics. Int. J. Mol. Sci. 2025, 26, 10701. https://doi.org/10.3390/ijms262110701
Paolini E, Longo M, Meroni M, Dongiovanni P. Hepatic Zonation in MASLD: Old Question, New Challenge in the Era of Spatial Omics. International Journal of Molecular Sciences. 2025; 26(21):10701. https://doi.org/10.3390/ijms262110701
Chicago/Turabian StylePaolini, Erika, Miriam Longo, Marica Meroni, and Paola Dongiovanni. 2025. "Hepatic Zonation in MASLD: Old Question, New Challenge in the Era of Spatial Omics" International Journal of Molecular Sciences 26, no. 21: 10701. https://doi.org/10.3390/ijms262110701
APA StylePaolini, E., Longo, M., Meroni, M., & Dongiovanni, P. (2025). Hepatic Zonation in MASLD: Old Question, New Challenge in the Era of Spatial Omics. International Journal of Molecular Sciences, 26(21), 10701. https://doi.org/10.3390/ijms262110701







