Lipidomic Signature of Patients with Familial Hypercholesterolemia Carrying Pathogenic Variants Unveils a Cue of Increased Cardiovascular Risk
Abstract
1. Introduction
2. Results
2.1. Characteristics of the Study Population
2.2. Distinct Lipidomic Signatures in FH Groups and Healthy Controls
2.3. Distinct Metabolic NMR-Based Profiles in FH Patients and Healthy Individuals
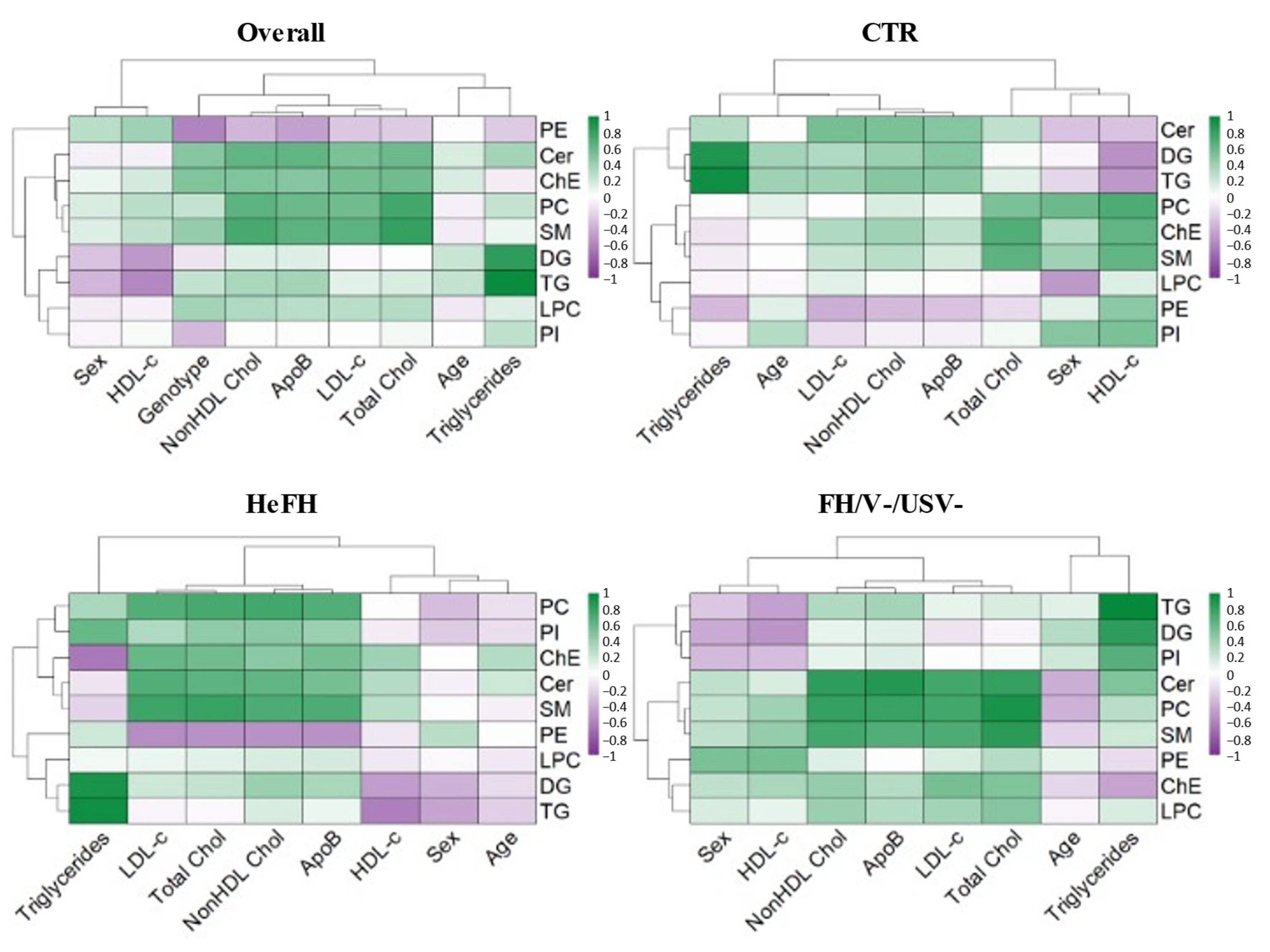
2.4. Lipidomic Signatures of FH Groups
2.5. Correlation of Lipidomic Profile with Clinical and Biochemical Data in FH Groups
3. Discussion
4. Materials and Methods
4.1. Study Population and Genetic Analysis
4.2. Lipid Extraction and Analysis
4.3. Lipid Identification
4.4. Metabolite Extraction and NMR Spectral Acquisition
4.5. NMR Data Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; Chapman, M.J.; De Backer, G.G.; Delgado, V.; Ference, B.A.; et al. 2019 ESC/EAS Guidelines for the Management of Dyslipidaemias: Lipid Modification to Reduce Cardiovascular Risk. Eur. Heart J. 2020, 41, 111–188. [Google Scholar] [CrossRef]
- Nordestgaard, B.G.; Chapman, M.J.; Humphries, S.E.; Ginsberg, H.N.; Masana, L.; Descamps, O.S.; Wiklund, O.; Hegele, R.A.; Raal, F.J.; Defesche, J.C.; et al. Familial Hypercholesterolaemia Is Underdiagnosed and Undertreated in the General Population: Guidance for Clinicians to Prevent Coronary Heart Disease: Consensus Statement of the European Atherosclerosis Society. Eur. Heart J. 2013, 34, 3478–3490a. [Google Scholar] [CrossRef]
- Watts, G.F.; Gidding, S.S.; Hegele, R.A.; Raal, F.J.; Sturm, A.C.; Jones, L.K.; Sarkies, M.N.; Al-Rasadi, K.; Blom, D.J.; Daccord, M.; et al. International Atherosclerosis Society Guidance for Implementing Best Practice in the Care of Familial Hypercholesterolaemia. Nat. Rev. Cardiol. 2023, 20, 845–869. [Google Scholar] [CrossRef]
- Abifadel, M.; Boileau, C. Genetic and Molecular Architecture of Familial Hypercholesterolemia. J. Intern. Med. 2023, 293, 144–165. [Google Scholar] [CrossRef] [PubMed]
- Di Taranto, M.D.; Fortunato, G. Genetic Heterogeneity of Familial Hypercholesterolemia: Repercussions for Molecular Diagnosis. Int. J. Mol. Sci. 2023, 24, 3224. [Google Scholar] [CrossRef]
- Defesche, J.C.; Gidding, S.S.; Harada-Shiba, M.; Hegele, R.A.; Santos, R.D.; Wierzbicki, A.S. Familial Hypercholesterolaemia. Nat. Rev. Dis. Primer 2017, 3, 17093. [Google Scholar] [CrossRef] [PubMed]
- Akioyamen, L.E.; Genest, J.; Shan, S.D.; Reel, R.L.; Albaum, J.M.; Chu, A.; Tu, J.V. Estimating the Prevalence of Heterozygous Familial Hypercholesterolaemia: A Systematic Review and Meta-Analysis. BMJ Open 2017, 7, e016461. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.C.; Alonso, R.; Diaz-Diaz, J.L.; Medeiros, A.M.; Jannes, C.E.; Merchan, A.; Vasques-Cardenas, N.A.; Cuevas, A.; Chacra, A.P.; Krieger, J.E.; et al. Phenotypical, Clinical, and Molecular Aspects of Adults and Children with Homozygous Familial Hypercholesterolemia in Iberoamerica. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 2508–2515. [Google Scholar] [CrossRef]
- Di Taranto, M.D.; Giacobbe, C.; Buonaiuto, A.; Calcaterra, I.; Palma, D.; Maione, G.; Iannuzzo, G.; Di Minno, M.N.D.; Rubba, P.; Fortunato, G. A Real-World Experience of Clinical, Biochemical and Genetic Assessment of Patients with Homozygous Familial Hypercholesterolemia. J. Clin. Med. 2020, 9, 219. [Google Scholar] [CrossRef]
- Tromp, T.R.; Hartgers, M.L.; Hovingh, G.K.; Vallejo-Vaz, A.J.; Ray, K.K.; Soran, H.; Freiberger, T.; Bertolini, S.; Harada-Shiba, M.; Blom, D.J.; et al. Worldwide Experience of Homozygous Familial Hypercholesterolaemia: Retrospective Cohort Study. Lancet 2022, 399, 719–728. [Google Scholar] [CrossRef]
- Di Taranto, M.D.; Giacobbe, C.; Fortunato, G. Familial Hypercholesterolemia: A Complex Genetic Disease with Variable Phenotypes. Eur. J. Med. Genet. 2020, 63, 103831. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.; Hartgers, M.L.; Reeskamp, L.F.; Zuurbier, L.; Defesche, J.; Kastelein, J.J.P.; Stroes, E.S.G.; Hovingh, G.K.; Huijgen, R. LDLR Variant Classification for Improved Cardiovascular Risk Prediction in Familial Hypercholesterolemia. Atherosclerosis 2024, 397, 117610. [Google Scholar] [CrossRef] [PubMed]
- Khera, A.V.; Won, H.-H.; Peloso, G.M.; Lawson, K.S.; Bartz, T.M.; Deng, X.; van Leeuwen, E.M.; Natarajan, P.; Emdin, C.A.; Bick, A.G.; et al. Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients With Severe Hypercholesterolemia. J. Am. Coll. Cardiol. 2016, 67, 2578–2589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dron, J.S.; Bellows, B.K.; Khera, A.V.; Liu, J.; Balte, P.P.; Oelsner, E.C.; Amr, S.S.; Lebo, M.S.; Nagy, A.; et al. Familial Hypercholesterolemia Variant and Cardiovascular Risk in Individuals With Elevated Cholesterol. JAMA Cardiol. 2024, 9, 263–271. [Google Scholar] [CrossRef]
- Anesi, A.; Di Minno, A.; Calcaterra, I.; Cavalca, V.; Tripaldella, M.; Porro, B.; Di Minno, M.N.D. An Untargeted Lipidomic Analysis Reveals Depletion of Several Phospholipid Classes in Patients with Familial Hypercholesterolemia on Treatment with Evolocumab. Biomedicines 2021, 9, 1941. [Google Scholar] [CrossRef]
- Rämö, J.T.; Ripatti, P.; Tabassum, R.; Söderlund, S.; Matikainen, N.; Gerl, M.J.; Klose, C.; Surma, M.A.; Stitziel, N.O.; Havulinna, A.S.; et al. Coronary Artery Disease Risk and Lipidomic Profiles Are Similar in Hyperlipidemias With Family History and Population-Ascertained Hyperlipidemias. J. Am. Heart Assoc. 2019, 8, e012415. [Google Scholar] [CrossRef]
- Gu, P.-S.; Su, K.-W.; Yeh, K.-W.; Huang, J.-L.; Lo, F.-S.; Chiu, C.-Y. Metabolomics Analysis Reveals Molecular Signatures of Metabolic Complexity in Children with Hypercholesterolemia. Nutrients 2023, 15, 1726. [Google Scholar] [CrossRef]
- OuYang, Y.; Zhou, L.; Jin, Y.; Hou, G.; Yang, P.; Chen, M.; Tian, Z. Reconstruction and Analysis of Correlation Networks Based on GC–MS Metabolomics Data for Hypercholesterolemia. Biochem. Biophys. Res. Commun. 2021, 553, 1–8. [Google Scholar] [CrossRef]
- Wang, X.-F.; Zhang, Y.-X.; Ma, H.-Y. Targeted Profiling of Amino Acid Metabolome in Serum by a Liquid Chromatography-Mass Spectrometry Method: Application to Identify Potential Markers for Diet-Induced Hyperlipidemia. Anal. Methods Adv. Methods Appl. 2020, 12, 2355–2362. [Google Scholar] [CrossRef]
- Cruzat, V.; Macedo Rogero, M.; Noel Keane, K.; Curi, R.; Newsholme, P. Glutamine: Metabolism and Immune Function, Supplementation and Clinical Translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Cheng, T. Q’s next: The Diverse Functions of Glutamine in Metabolism, Cell Biology and Cancer. Oncogene 2010, 29, 313–324. [Google Scholar] [CrossRef]
- Huang, H.; Vandekeere, S.; Kalucka, J.; Bierhansl, L.; Zecchin, A.; Brüning, U.; Visnagri, A.; Yuldasheva, N.; Goveia, J.; Cruys, B.; et al. Role of Glutamine and Interlinked Asparagine Metabolism in Vessel Formation. EMBO J. 2017, 36, 2334–2352. [Google Scholar] [CrossRef]
- Kim, B.; Li, J.; Jang, C.; Arany, Z. Glutamine Fuels Proliferation but Not Migration of Endothelial Cells. EMBO J. 2017, 36, 2321–2333. [Google Scholar] [CrossRef]
- Peyton, K.J.; Liu, X.-M.; Yu, Y.; Yates, B.; Behnammanesh, G.; Durante, W. Glutaminase-1 Stimulates the Proliferation, Migration, and Survival of Human Endothelial Cells. Biochem. Pharmacol. 2018, 156, 204–214. [Google Scholar] [CrossRef]
- Xi, P.; Jiang, Z.; Zheng, C.; Lin, Y.; Wu, G. Regulation of Protein Metabolism by Glutamine: Implications for Nutrition and Health. Front. Biosci. Landmark Ed. 2011, 16, 578–597. [Google Scholar] [CrossRef]
- Holmes, M.V.; Millwood, I.Y.; Kartsonaki, C.; Hill, M.R.; Bennett, D.A.; Boxall, R.; Guo, Y.; Xu, X.; Bian, Z.; Hu, R.; et al. Lipids, Lipoproteins, and Metabolites and Risk of Myocardial Infarction and Stroke. J. Am. Coll. Cardiol. 2018, 71, 620–632. [Google Scholar] [CrossRef]
- Zaric, B.L.; Radovanovic, J.N.; Gluvic, Z.; Stewart, A.J.; Essack, M.; Motwalli, O.; Gojobori, T.; Isenovic, E.R. Atherosclerosis Linked to Aberrant Amino Acid Metabolism and Immunosuppressive Amino Acid Catabolizing Enzymes. Front. Immunol. 2020, 11, 551758. [Google Scholar] [CrossRef] [PubMed]
- Goicoechea, L.; Conde de la Rosa, L.; Torres, S.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondrial Cholesterol: Metabolism and Impact on Redox Biology and Disease. Redox Biol. 2023, 61, 102643. [Google Scholar] [CrossRef]
- Shantha, G.P.S.; Wasserman, B.; Astor, B.C.; Coresh, J.; Brancati, F.; Sharrett, A.R.; Young, J.H. Association of Blood Lactate with Carotid Atherosclerosis: The Atherosclerosis Risk in Communities (ARIC) Carotid MRI Study. Atherosclerosis 2013, 228, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Rimbert, A.; Balder, W.; Zwinderman, A.H.; Kuivenhoven, J.A.; Dallinga-Thie, G.M.; Groen, A.K. Use of Plasma Metabolomics to Analyze Phenotype-Genotype Relationships in Young Hypercholesterolemic Females. J. Lipid Res. 2018, 59, 2174–2180. [Google Scholar] [CrossRef] [PubMed]
- Balder, J.-W.; Rimbert, A.; Zhang, X.; Viel, M.; Kanninga, R.; van Dijk, F.; Lansberg, P.; Sinke, R.; Kuivenhoven, J.A. Genetics, Lifestyle, and Low-Density Lipoprotein Cholesterol in Young and Apparently Healthy Women. Circulation 2018, 137, 820–831. [Google Scholar] [CrossRef] [PubMed]
- Di Minno, M.N.D.; Gentile, M.; Di Minno, A.; Iannuzzo, G.; Calcaterra, I.; Buonaiuto, A.; Di Taranto, M.D.; Giacobbe, C.; Fortunato, G.; Rubba, P.O.F. Changes in Carotid Stiffness in Patients with Familial Hypercholesterolemia Treated with Evolocumab®: A Prospective Cohort Study. Nutr. Metab. Cardiovasc. Dis. NMCD 2020, 30, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Rogozhina, A.A.; Minushkina, L.O.; Alessenko, A.V.; Gutner, U.A.; Shupik, M.A.; Kurochkin, I.N.; Maloshitskaya, O.A.; Sokolov, S.A.; Zateyshchikov, D.A. Lipidome Features in Patients with Different Probability of Family Hypercholesterolemia. Bull. Russ. State Med. Univ. 2019, 6, 84–91. [Google Scholar] [CrossRef]
- Stübiger, G.; Aldover-Macasaet, E.; Bicker, W.; Sobal, G.; Willfort-Ehringer, A.; Pock, K.; Bochkov, V.; Widhalm, K.; Belgacem, O. Targeted Profiling of Atherogenic Phospholipids in Human Plasma and Lipoproteins of Hyperlipidemic Patients Using MALDI-QIT-TOF-MS/MS. Atherosclerosis 2012, 224, 177–186. [Google Scholar] [CrossRef]
- Zhao, R.; Tang, Y.; Cao, W.; Zhao, L.; Wu, Z.; Chen, X.; Li, Y.; Jia, X.; Bai, H. Identification of Multiple Plasma Lipids as Diagnostic Biomarkers of Hypercholesterolemia and the Underlying Mechanisms Based on Pseudo-Targeted Lipidomics. Rapid Commun. Mass Spectrom. 2024, 38, e9723. [Google Scholar] [CrossRef]
- Jadhav, A.V.; Thompson, G.R. Reversible Abnormalities of Low Density Lipoprotein Composition in Familial Hypercholesterolaemia. Eur. J. Clin. Investig. 1979, 9, 63–67. [Google Scholar] [CrossRef]
- Jiang, X.; Paultre, F.; Pearson, T.A.; Reed, R.G.; Francis, C.K.; Lin, M.; Berglund, L.; Tall, A.R. Plasma Sphingomyelin Level as a Risk Factor for Coronary Artery Disease. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 2614–2618. [Google Scholar] [CrossRef]
- Schlitt, A.; Blankenberg, S.; Yan, D.; von Gizycki, H.; Buerke, M.; Werdan, K.; Bickel, C.; Lackner, K.J.; Meyer, J.; Rupprecht, H.J.; et al. Further Evaluation of Plasma Sphingomyelin Levels as a Risk Factor for Coronary Artery Disease. Nutr. Metab. 2006, 3, 5. [Google Scholar] [CrossRef]
- De Roos, B.; Caslake, M.J.; Milliner, K.; Benson, G.M.; Suckling, K.E.; Packard, C.J. Characterisation of the Lipoprotein Structure in the St. Thomas’ Mixed Hyperlipidaemic (SMHL) Rabbit. Atherosclerosis 2005, 181, 63–68. [Google Scholar] [CrossRef]
- Lee, H.; Young Jang, S.; Jung, Y.; Kwon, O.; Hwang, G.-S. Lipidomic Profiling Analysis of Human Plasma from Subjects with Hypercholesterolemia to Evaluate the Intake of Yellow Yeast Rice Fermented by Aspergillus Terreus DSMK01. Food Funct. 2022, 13, 7629–7637. [Google Scholar] [CrossRef]
- Zalloua, P.; Kadar, H.; Hariri, E.; Abi Farraj, L.; Brial, F.; Hedjazi, L.; Le Lay, A.; Colleu, A.; Dubus, J.; Touboul, D.; et al. Untargeted Mass Spectrometry Lipidomics Identifies Correlation between Serum Sphingomyelins and Plasma Cholesterol. Lipids Health Dis. 2019, 18, 38. [Google Scholar] [CrossRef] [PubMed]
- Mazière, J.C.; Mazière, C.; Mora, L.; Polonovski, J. Impairment of Exogenous Sphingomyelin Degradation in Cultured Fibroblasts from Familial Hypercholesterolemia. FEBS Lett. 1984, 173, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Pedro-Botet, J.; Climent, E.; Benaiges, D. Familial Hypercholesterolemia: Do HDL Play a Role? Biomedicines 2021, 9, 810. [Google Scholar] [CrossRef] [PubMed]
- Di Taranto, M.D.; de Falco, R.; Guardamagna, O.; Massini, G.; Giacobbe, C.; Auricchio, R.; Malamisura, B.; Proto, M.; Palma, D.; Greco, L.; et al. Lipid Profile and Genetic Status in a Familial Hypercholesterolemia Pediatric Population: Exploring the LDL/HDL Ratio. Clin. Chem. Lab. Med. 2019, 57, 1102–1110. [Google Scholar] [CrossRef]
- Tada, H.; Kojima, N.; Yamagami, K.; Nomura, A.; Nohara, A.; Usui, S.; Sakata, K.; Hayashi, K.; Fujino, N.; Takamura, M.; et al. Impact of Variants of Uncertain Significance of LDL Receptor on Phenotypes of Familial Hypercholesterolemia. J. Clin. Lipidol. 2022, 16, 863–869. [Google Scholar] [CrossRef]
- Rai, S.; Bhatnagar, S. Novel Lipidomic Biomarkers in Hyperlipidemia and Cardiovascular Diseases: An Integrative Biology Analysis. OMICS J. Integr. Biol. 2017, 21, 132–142. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhang, A.; Zong, W.; An, N.; Zhang, H.; Luan, Y.; Cao, H.; Sun, H.; Wang, X. Chemometrics Strategy Coupled with High Resolution Mass Spectrometry for Analyzing and Interpreting Comprehensive Metabolomic Characterization of Hyperlipemia. RSC Adv. 2016, 6, 112534–112543. [Google Scholar] [CrossRef]
- Christensen, J.J.; Ulven, S.M.; Retterstøl, K.; Narverud, I.; Bogsrud, M.P.; Henriksen, T.; Bollerslev, J.; Halvorsen, B.; Aukrust, P.; Holven, K.B. Comprehensive Lipid and Metabolite Profiling of Children with and without Familial Hypercholesterolemia: A Cross-Sectional Study. Atherosclerosis 2017, 266, 48–57. [Google Scholar] [CrossRef]
- Di Taranto, M.D.; Giacobbe, C.; Palma, D.; Iannuzzo, G.; Gentile, M.; Calcaterra, I.; Guardamagna, O.; Auricchio, R.; Di Minno, M.N.D.; Fortunato, G. Genetic Spectrum of Familial Hypercholesterolemia and Correlations with Clinical Expression: Implications for Diagnosis Improvement. Clin. Genet. 2021, 100, 529–541. [Google Scholar] [CrossRef]
- Cardiero, G.; Ferrandino, M.; Calcaterra, I.L.; Iannuzzo, G.; Di Minno, M.N.D.; Buganza, R.; Guardamagna, O.; Auricchio, R.; Di Taranto, M.D.; Fortunato, G. Impact of 12-SNP and 6-SNP Polygenic Scores on Predisposition to High LDL-Cholesterol Levels in Patients with Familial Hypercholesterolemia. Genes 2024, 15, 462. [Google Scholar] [CrossRef]
- Chora, J.R.; Medeiros, A.M.; Alves, A.C.; Bourbon, M. Analysis of Publicly Available LDLR, APOB, and PCSK9 Variants Associated with Familial Hypercholesterolemia: Application of ACMG Guidelines and Implications for Familial Hypercholesterolemia Diagnosis. Genet. Med. Off. J. Am. Coll. Med. Genet. 2018, 20, 591–598. [Google Scholar] [CrossRef]
- Chora, J.R.; Iacocca, M.A.; Tichý, L.; Wand, H.; Kurtz, C.L.; Zimmermann, H.; Leon, A.; Williams, M.; Humphries, S.E.; Hooper, A.J.; et al. The Clinical Genome Resource (ClinGen) Familial Hypercholesterolemia Variant Curation Expert Panel Consensus Guidelines for LDLR Variant Classification. Genet. Med. Off. J. Am. Coll. Med. Genet. 2022, 24, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid Extraction by Methyl-Tert-Butyl Ether for High-Throughput Lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Arcos, C.; Paris, D.; Mazzella, V.; Mutalipassi, M.; Costantini, M.; Buia, M.C.; von Elert, E.; Cutignano, A.; Zupo, V. Responses of the Macroalga Ulva Prolifera Müller to Ocean Acidification Revealed by Complementary NMR- and MS-Based Omics Approaches. Mar. Drugs 2022, 20, 743. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.D.; Meng, X.; Donovan, K.J.; Shaka, A.J. SOGGY: Solvent-Optimized Double Gradient Spectroscopy for Water Suppression. A Comparison with Some Existing Techniques. J. Magn. Reson. 2007, 184, 263–274. [Google Scholar] [CrossRef]
- Fan, T.W.-M. Metabolite Profiling by One- and Two-Dimensional NMR Analysis of Complex Mixtures. Prog. Nucl. Magn. Reson. Spectrosc. 1996, 28, 161–219. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Chung, R.W.S.; Wang, Z.; Bursill, C.A.; Wu, B.J.; Barter, P.J.; Rye, K.-A. Effect of Long-Term Dietary Sphingomyelin Supplementation on Atherosclerosis in Mice. PLoS ONE 2017, 12, e0189523. [Google Scholar] [CrossRef]
- Lee, T.-Y.; Lu, W.-J.; Changou, C.A.; Hsiung, Y.-C.; Trang, N.T.T.; Lee, C.-Y.; Chang, T.-H.; Jayakumar, T.; Hsieh, C.-Y.; Yang, C.-H.; et al. Platelet Autophagic Machinery Involved in Thrombosis through a Novel Linkage of AMPK-MTOR to Sphingolipid Metabolism. Autophagy 2021, 17, 4141–4158. [Google Scholar] [CrossRef]
- Ruuth, M.; Nguyen, S.D.; Vihervaara, T.; Hilvo, M.; Laajala, T.D.; Kondadi, P.K.; Gisterå, A.; Lähteenmäki, H.; Kittilä, T.; Huusko, J.; et al. Susceptibility of Low-Density Lipoprotein Particles to Aggregate Depends on Particle Lipidome, Is Modifiable, and Associates with Future Cardiovascular Deaths. Eur. Heart J. 2018, 39, 2562–2573. [Google Scholar] [CrossRef]
- Nilsson, Å.; Duan, R.-D. Absorption and Lipoprotein Transport of Sphingomyelin. J. Lipid Res. 2006, 47, 154–171. [Google Scholar] [CrossRef]
- Chong, S.Y.; Louise, R.A.; Huang, C.; Ting, H.J.; Seah, F.; Qi, X.; Pastorin, G.; Storm, G.; Wang, J.W. Inhibition of Ceramide de Novo Synthesis Mitigates Atherosclerosis. Eur. Heart J. 2024, 45, ehae666.3277. [Google Scholar] [CrossRef]
- Zhao, F.; Shao, M.; Li, M.; Li, T.; Zheng, Y.; Sun, W.; Ni, C.; Li, L. Sphingolipid Metabolites Involved in the Pathogenesis of Atherosclerosis: Perspectives on Sphingolipids in Atherosclerosis. Cell. Mol. Biol. Lett. 2025, 30, 18. [Google Scholar] [CrossRef]



| Characteristic | Controls (CTR) | Heterozygotes for a Pathogenic Variant (HeFH) | Without Pathogenic/ Uncertain Significance Variants (FH/V−/USV−) | HeFH vs. CTR | FH/V−/USV− vs. CTR | HeFH vs. FH/V−/USV− |
|---|---|---|---|---|---|---|
| (n = 22) | (n = 20) | (n = 19) | Adjusted p-Value | |||
| Demographics | ||||||
| Age (mean years ± SD) | 46.6 ± 12.0 | 46.1 ± 14.2 | 58.5 ± 13.1 | >0.9999 | 0.0268 | 0.0249 |
| Sex (% female) | 72.7% | 65.0% | 63.2% | 0.7828 | ||
| Biochemical Parameters | ||||||
| Total cholesterol (mg/dL) | 160.4 ± 15.8 | 203.2 ± 50.2 | 183.2 ± 63.5 | 0.0213 | >0.9999 | 0.2473 |
| LDL-C (mg/dL) | 94.5 ± 14.3 | 131.2 ± 52.4 | 104.8 ± 61.3 | 0.0500 | 0.9852 | 0.0800 |
| HDL-C (mg/dL) | 56.3 ± 12.0 | 49.0 ± 10.4 | 50.5 ± 15.6 | 0.1802 | 0.4096 | >0.9999 |
| Non-HDL-C (mg/dL) | 104.1 ± 16.5 | 154.1 ± 50.5 | 132.7 ± 59.6 | 0.0018 | 0.5278 | 0.1392 |
| Triglycerides (mg/dL) | 73.6 ± 24.4 | 106.3 ± 60.0 | 128.5 ± 90.0 | 0.1260 | 0.0064 | 0.8910 |
| ApoB (mg/dL) | 69.7 ± 10.7 | 103.8 ± 37.5 | 90.6 ± 38.1 | 0.0003 | 0.1482 | 0.1831 |
| Lipid Lowering Therapy *,# | ||||||
| Low/moderate-intensity | \ | 6 | 10 | 0.1005 | ||
| High-intensity | \ | 13 | 7 | |||
| PCSK9 inhibitors | \ | 0 | 2 | |||
| Metabolite | CTR | HeFH | FH/V−/USV− | Group Comparison padj * | ||
|---|---|---|---|---|---|---|
| Median (IQR) | Median (IQR) | Median (IQR) | HeFH vs. CTR | FH/V−/USV− vs. CTR | HeFH vs. FH/V−/USV− | |
| proline | 4.5 × 10−4 | 6.7 × 10−4 | 7.8 × 10−4 | 0.0010 | <0.0001 | 0.587 |
| (3.7 × 10−4–5.1 × 10−4) | (6.0 × 10−4–7.5 × 10−4) | (6.3 × 10−4–8.7 × 10−4) | ||||
| glutamate | 4.3 × 10−4 | 7.8 × 10−4 | 8.4 × 10−4 | <0.0001 | <0.0001 | 1 |
| (3.4 × 10−4–4.7 × 10−4) | (7.0 × 10−4–8.7 × 10−4) | (7.7 × 10−4–9.0 × 10−4) | ||||
| threonine | 9.8 × 10−4 | 0.002 | 0.003 | <0.0001 | <0.0001 | 1 |
| (8.9 × 10−4–11.7 × 10−4) | (0.002–0.003) | (0.002–0.003) | ||||
| 3-hydroxybutyrate | 10.3 × 10−4 | 0.001 | 0.002 | 0.0124 | 0.0001 | 0.724 |
| (9.3 × 10−4–14.9 × 10−4) | (0.001–0.002) | (0.001–0.002) | ||||
| 2-hydroxybutyrate | 2.1 × 10−4 | 2.7 × 10−4 | 2.7 × 10−4 | 0.0331 | 0.0015 | 1 |
| (1.6 × 10−4–2.6 × 10−4) | (2.1 × 10−4–3.7 × 10−4) | (2.4 × 10−4–3.4 × 10−4) | ||||
| isoleucine | 4.6 × 10−4 | 5.6 × 10−4 | 6.2 × 10−4 | 0.0013 | <0.0001 | 1 |
| (4.3 × 10−4–5.2 × 10−4) | (4.9 × 10−4–7.0 × 10−4) | (5.5 × 10−4–6.7 × 10−4) | ||||
| lysine | 4.9 × 10−4 | 4.7 × 10−4 | 7.3 × 10−4 | <0.0001 | <0.0001 | 1 |
| (4.2 × 10−4–5.5 × 10−4) | (6.0 × 10−4–8.1 × 10−4) | (6.4 × 10−4–8.0 × 10−4) | ||||
| lactate | 0.019 | 0.025 | 0.039 | 0.323 | <0.0001 | 0.118 |
| (0.017–0.024) | (0.021–0.037) | (0.027–0.052) | ||||
| glutamine | 6.1 × 10−4 | 3.4 × 10−4 | 3.8 × 10−4 | <0.0001 | 0.0001 | 0.877 |
| (5.4 × 10−4–6.4 × 10−4) | (3.0 × 10−4–4.2 × 10−4) | (2.5 × 10−4–5.2 × 10−4) | ||||
| acetate | 0.0012 | 8.2 × 10−4 | 0.0010 | <0.0001 | 0.0245 | 0.291 |
| (0.0010–0.0012) | (7.0 × 10−4–9.9 × 10−4) | (0.0007–0.0012) | ||||
| tyrosine | 9.2 × 10−4 | 6.7 × 10−4 | 6.8 × 10−4 | <0.0001 | <0.0001 | 1 |
| (8.8 × 10−4–9.9 × 10−4) | (5.9 × 10−4–7.5 × 10−4) | (5.5 × 10−4–7.2 × 10−4) | ||||
| glucose | 0.0021 | 0.0017 | 0.0021 | 0.005 | 1 | 0.09 |
| (0.0020–0.0023) | (0.0015–0.0019) | (0.0016–0.0025) | ||||
| HeFH | FH/V−/USV− | Adjusted p-Value * | FC | ||||
|---|---|---|---|---|---|---|---|
| Lipid | Class | Median (µg/mL) | IQR | Median (µg/mL) | IQR | ||
| Cer (d18:1/23:0) | Cer | 0.73 | 0.61–0.92 | 1.02 | 0.77–1.62 | 0.0283 | −1.41 |
| LPC (20:4) | LPC | 5.51 | 4.82–6.56 | 4.23 | 2.96–5.26 | 0.0243 | 1.30 |
| PC (18:0/20:4) | PC | 102.60 | 90.64–119.47 | 80.53 | 53.50–106.50 | 0.0168 | 1.27 |
| PE (18:0p/18:2) | PE | 0.38 | 0.01–1.06 | 1.17 | 0.48–1.76 | 0.0283 | −3.09 |
| SM (d32:0) | SM | 0.50 | 0.41–0.78 | 0.35 | 0.31–0.49 | 0.0256 | 1.44 |
| SM (d38:4) | SM | 0.76 | 0.64–0.93 | 0.47 | 0.44–0.65 | 0.0056 | 1.63 |
| SM (d43:2) | SM | 5.17 | 4.01–7.88 | 3.15 | 1.89–5.00 | 0.0116 | 1.64 |
| SM (d44:4) | SM | 6.47 | 3.83–7.52 | 3.98 | 2.67–5.75 | 0.0242 | 1.62 |
| Lipid | Presence of FH Pathogenic Variants (β, p-Value) | Lipid-Lowering Therapy (β, p-Value) | Sex (β, p-Value) | Age (β, p-Value) |
|---|---|---|---|---|
| Cer (d18:1/23:0) | −0.27, p = 0.0731 | −0.05, p = 0.7018 | 0, p = 0.9695 | 0, p = 0.8129 |
| LPC (20:4) | 1.11, p = 0.0733 | 1.45, p = 0.0134 | −0.64, p = 0.2509 | 0.02, p = 0.2380 |
| PC (18:0/20:4) | 21.51, p = 0.0193 | 20.18, p = 0.0180 | −6.29, p = 0.4382 | 0.23, p = 0.4473 |
| PE (18:0p/18:2) | −0.56, p = 0.0584 | −0.03, p = 0.9058 | 0.60, p = 0.0277 | 0, p = 0.7687 |
| SM (d32:0) | 0.22, p = 0.0084 | −0.10, p = 0.1654 | 0.01, p = 0.8882 | 0, p = 0.8075 |
| SM (d38:4) | 0.22, p = 0.0059 | 0.05, p = 0.5095 | −0.08, p = 0.2503 | 0, p = 0.7967 |
| SM (d43:2) | 2.99, p = 0.0003 | −1.23, p = 0.0788 | −0.20, p = 0.7711 | 0.03, p = 0.1599 |
| SM (d44:4) | 2.25, p = 0.0118 | −1.22, p = 0.1271 | −0.24, p = 0.7532 | 0, p = 0.8932 |
| SM (total) | 190.13, p = 0.0256 | −104.18, p = 0.1754 | 21.23, p = 0.7776 | 0.03, p = 0.9919 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Simone, G.; Di Taranto, M.D.; Paris, D.; Ferrandino, M.; Andolfi, M.; Iodice, A.; Cardiero, G.; De Luca, C.; Valletta, L.J.; Calcaterra, I.L.; et al. Lipidomic Signature of Patients with Familial Hypercholesterolemia Carrying Pathogenic Variants Unveils a Cue of Increased Cardiovascular Risk. Int. J. Mol. Sci. 2025, 26, 10688. https://doi.org/10.3390/ijms262110688
De Simone G, Di Taranto MD, Paris D, Ferrandino M, Andolfi M, Iodice A, Cardiero G, De Luca C, Valletta LJ, Calcaterra IL, et al. Lipidomic Signature of Patients with Familial Hypercholesterolemia Carrying Pathogenic Variants Unveils a Cue of Increased Cardiovascular Risk. International Journal of Molecular Sciences. 2025; 26(21):10688. https://doi.org/10.3390/ijms262110688
Chicago/Turabian StyleDe Simone, Giulia, Maria Donata Di Taranto, Debora Paris, Martina Ferrandino, Marco Andolfi, Annalaura Iodice, Giovanna Cardiero, Carmine De Luca, Luigi Junior Valletta, Ilenia Lorenza Calcaterra, and et al. 2025. "Lipidomic Signature of Patients with Familial Hypercholesterolemia Carrying Pathogenic Variants Unveils a Cue of Increased Cardiovascular Risk" International Journal of Molecular Sciences 26, no. 21: 10688. https://doi.org/10.3390/ijms262110688
APA StyleDe Simone, G., Di Taranto, M. D., Paris, D., Ferrandino, M., Andolfi, M., Iodice, A., Cardiero, G., De Luca, C., Valletta, L. J., Calcaterra, I. L., Iannuzzo, G., Di Minno, M. N. D., Fortunato, G., & Cutignano, A. (2025). Lipidomic Signature of Patients with Familial Hypercholesterolemia Carrying Pathogenic Variants Unveils a Cue of Increased Cardiovascular Risk. International Journal of Molecular Sciences, 26(21), 10688. https://doi.org/10.3390/ijms262110688









