Multi-Omics Investigation into Why Viable Oogonial Stem Cells Can Still Be Isolated and Cultured from Post-Mortem Paralichthys olivaceus
Abstract
1. Introduction
2. Results
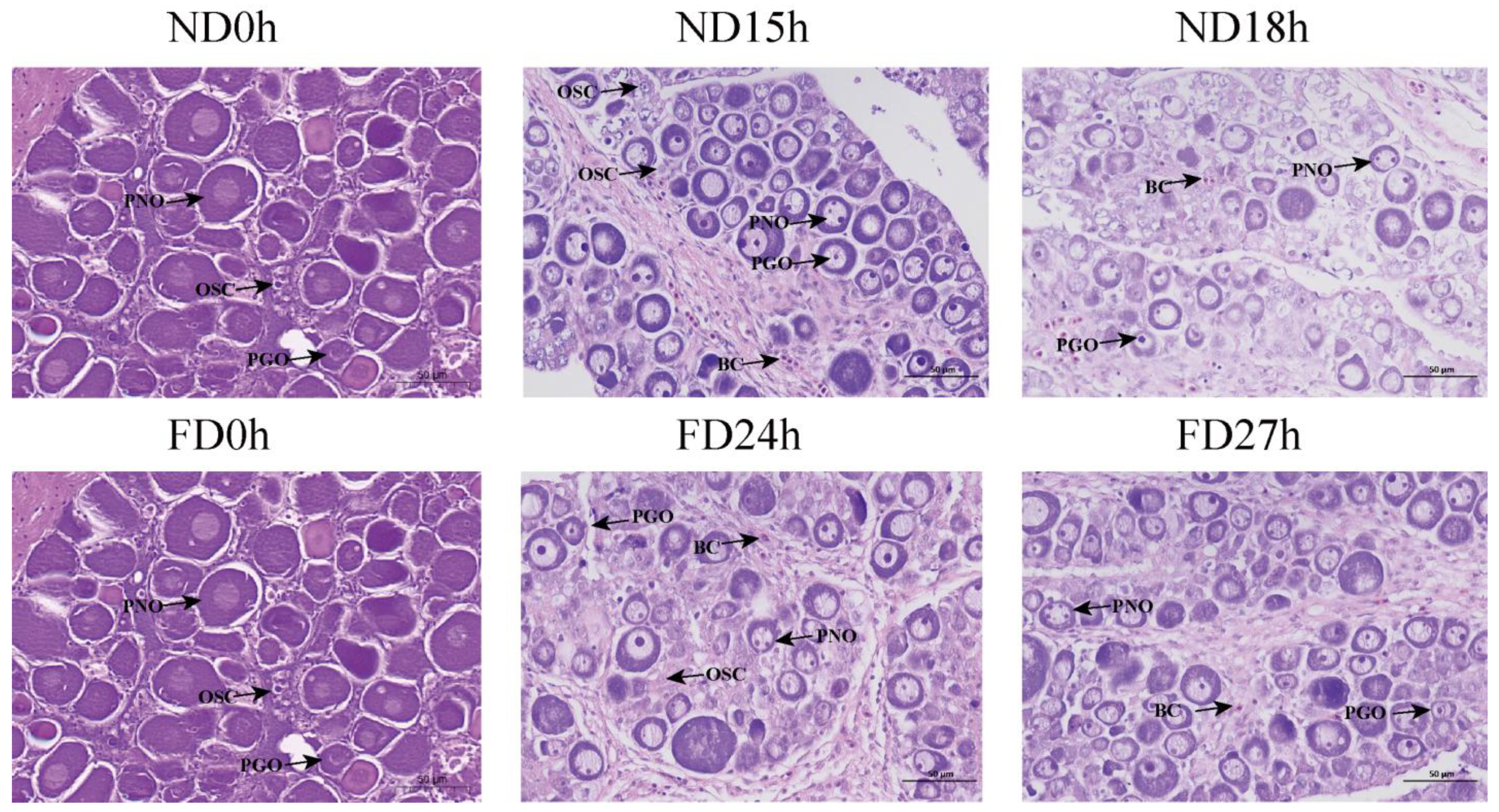
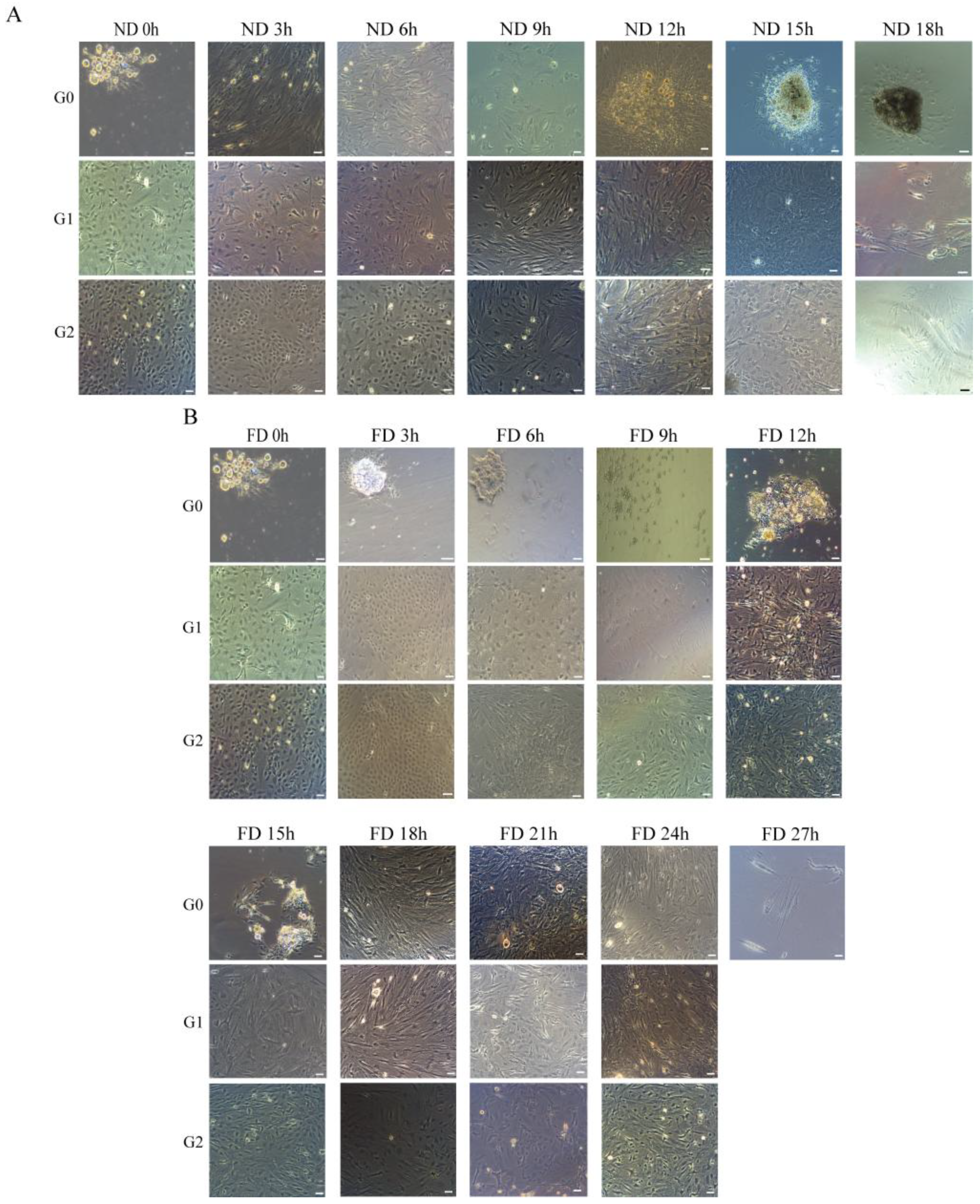
2.1. In Vitro Culture of OSCs at Different Time Intervals After Death
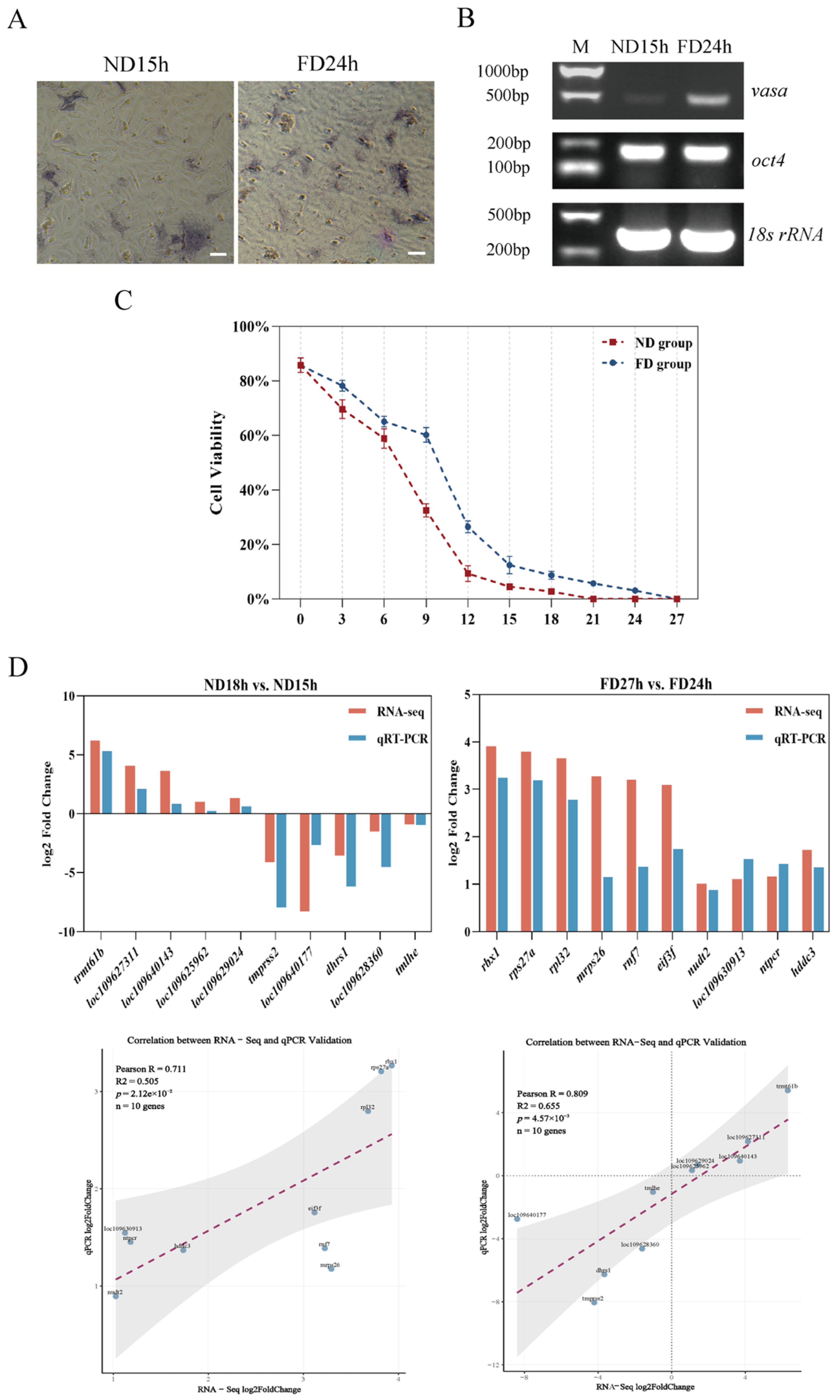
2.2. Detection of Stem Cells Using Alkaline Phosphatase and RT-PCR
2.3. Identification of DEGs at Different Times of ND Group and FD Group
2.4. Enrichment Analysis of DEGs and Non DEGs
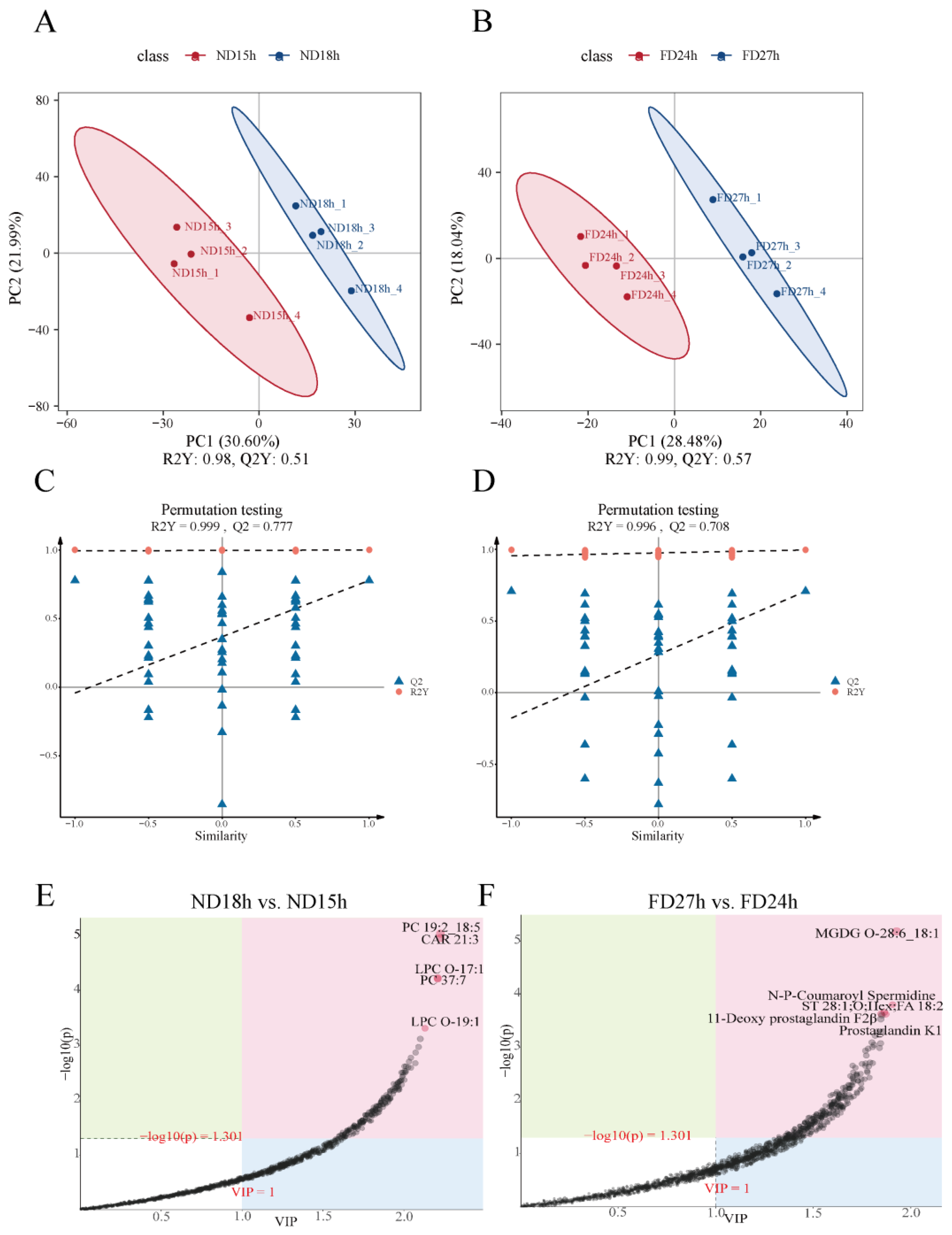
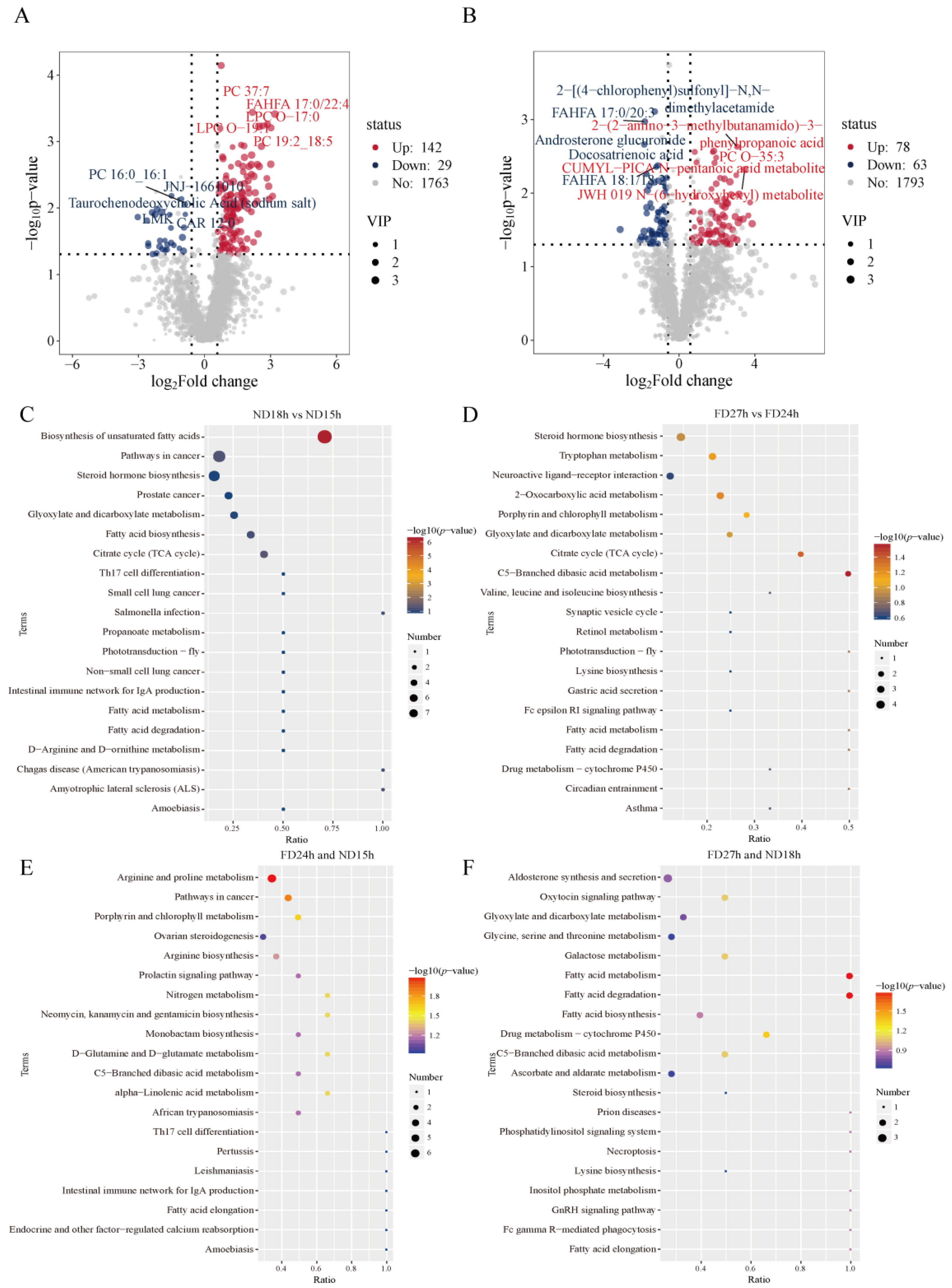
2.5. Identification of DEMs at Different Times of ND Group and FD Group
2.6. Enrichment Analysis of DEMs
2.7. Joint Analysis of Transcriptome and Metabolomics
3. Discussion
3.1. Post-Mortem Energy Metabolism Disruption and Its Impact on Cells
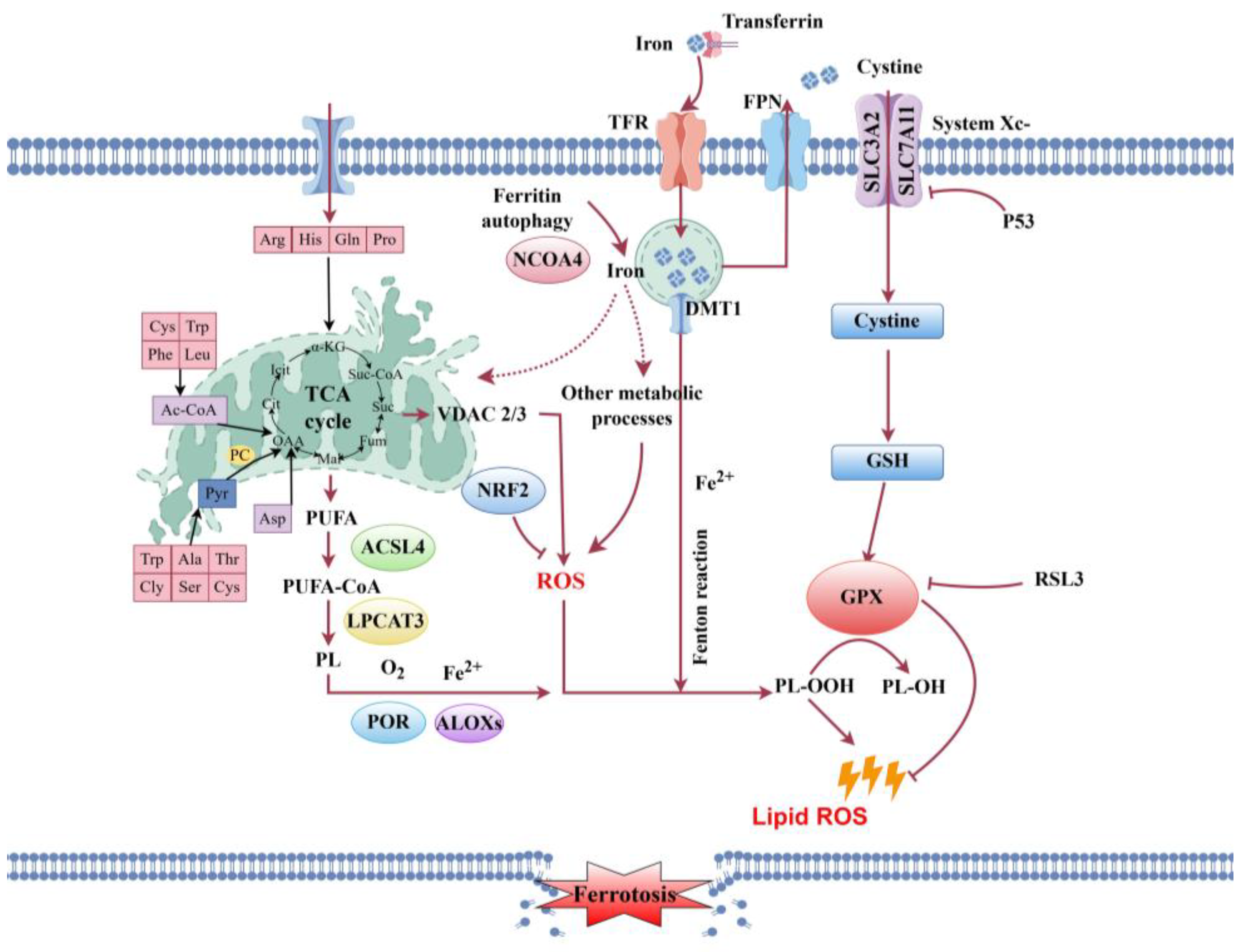
3.2. Post-Mortem Lipid Metabolism Disruption and Its Impact on Cellular Integrity
4. Materials and Methods
4.1. Experimental Animal and Sampling
4.2. Detection of Cell Survival Rates
4.3. Alkaline Phosphatase and RT-PCR Analysis
4.4. RNA Sample Collection, Library Construction, and Sequencing
4.5. Transcriptome Assembly, Functional Annotation, and Data Analysis
4.6. Metabolite Extraction and LC-MS/MS Analysis
4.7. Metabolomics Data Analysis
4.8. Combined Analysis of Transcriptomics and Metabolomics
4.9. qRT-PCR Validation of DEGs
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| ND Group | 0 h | 3 h | 6 h | 9 h | 12 h | 15 h | 18 h | |
|---|---|---|---|---|---|---|---|---|
| Generation | ||||||||
| G1 | 25 d | 43 d | 30 d | 55 d | 55 d | 90 d | / | |
| G2 | 7 d | 7 d | 15 d | 11 d | 9 d | 15 d | / | |
| G3 | 7 d | 7 d | 4 d | 7 d | 7 d | 7 d | / | |
| FD Group | 3 h | 6 h | 9 h | 12 h | 15 h | 18 h | 21 h | 24 h | 27 h | |
|---|---|---|---|---|---|---|---|---|---|---|
| Generation | ||||||||||
| G1 | 45 d | 32 d | 18 d | 17 d | 36 d | 30 d | 21 d | 36 d | / | |
| G2 | 15 d | 3 d | 4 d | 25 d | 5 d | 7 d | 15 d | 5 d | / | |
| G3 | 7 d | 5 d | 8 d | 3 d | 3 d | 7 d | 5 d | 5 d | / | |
| Sample | Total | Unmapped (%) | Unique Mapped (%) | Multiple Mapped (%) | Total Mapped (%) |
|---|---|---|---|---|---|
| ND15h-1 | 40,801,018 | 16,440,151 (40.29%) | 37,617,362 (92.2%) | 1,054,338 (2.58%) | 38,671,700 (94.78%) |
| ND15h-2 | 40,310,714 | 16,339,041 (40.53%) | 37,239,993 (92.38%) | 1,103,402 (2.74%) | 38,343,395 (95.12%) |
| ND15h-3 | 46,885,594 | 18,325,940 (39.09%) | 43,250,261 (92.25%) | 1,373,811 (2.93%) | 44,624,072 (95.18%) |
| ND18h-1 | 38,825,814 | 15,930,534 (41.03%) | 35,820,303 (92.26%) | 1,124,155 (2.9%) | 36,944,458 (95.15%) |
| ND18h-2 | 42,378,838 | 18,639,800 (43.98%) | 37,455,412 (88.38%) | 978,946 (2.31%) | 38,434,358 (90.69%) |
| ND18h-3 | 37,283,930 | 15,355,381 (41.18%) | 32,976,563 (88.45%) | 931,946 (2.5%) | 33,908,509 (90.95%) |
| FD24h-1 | 40,445,378 | 16,445,352 (40.66%) | 37,346,216 (92.34%) | 1,119,270 (2.77%) | 38,465,486 (95.1%) |
| FD24h-2 | 48,930,460 | 19,085,661 (39.01%) | 42,725,682 (87.32%) | 1,374,441 (2.81%) | 44,100,123 (90.13%) |
| FD24h-3 | 52,648,168 | 20,366,600 (38.68%) | 46,566,537 (88.45%) | 1,486,062 (2.82%) | 48,052,599 (91.27%) |
| FD27h-1 | 41,534,978 | 17,939,371 (43.19%) | 37,768,396 (90.93%) | 1,139,589 (2.74%) | 38,907,985 (93.68%) |
| FD27h-2 | 43,442,624 | 17,732,956 (40.82%) | 39,693,588 (91.37%) | 1,199,797 (2.76%) | 40,893,385 (94.13%) |
| FD27h-3 | 43,529,080 | 18,317,491 (42.08%) | 39,967,505 (91.82%) | 1,151,657 (2.65%) | 41,119,162 (94.46%) |
| Sample | Raw Bases (bp) | Clean Bases (bp) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|
| ND15h-1 | 6,400,731,900 | 40,801,018 | 39,732,031 (97.38%) | 38,042,869 (93.24%) | 20,849,320 (51.10%) |
| ND15h-2 | 6,585,703,200 | 40,310,714 | 39,302,946 (97.50%) | 37,686,487 (93.49%) | 20,526,216 (50.92%) |
| ND15h-3 | 7,423,662,900 | 46,885,594 | 45,727,520 (97.53%) | 43,819,276 (93.46%) | 24,061,687 (51.32%) |
| ND18h-1 | 6,238,026,600 | 38,825,814 | 37,820,225 (97.41%) | 36,232,250 (93.32%) | 19,719,631 (50.79%) |
| ND18h-2 | 6,912,105,300 | 42,378,838 | 41,336,319 (97.54%) | 39,564,883 (93.36%) | 20,841,913 (49.18%) |
| ND18h-3 | 6,019,846,500 | 37,283,930 | 36,404,029 (97.64%) | 34,890,302 (93.58%) | 18,656,879 (50.04%) |
| FD24h-1 | 6,327,326,700 | 40,445,378 | 39,414,021 (97.45%) | 37,784,072 (93.42%) | 20,675,677 (51.12%) |
| FD24h-2 | 7,364,892,300 | 48,930,460 | 47,795,273 (97.68%) | 45,872,306 (93.75%) | 24,797,957 (50.68%) |
| FD24h-3 | 7,921,212,600 | 52,648,168 | 51,463,584 (97.75%) | 49,399,776 (93.83%) | 26,724,210 (50.76%) |
| FD27h-1 | 6,653,903,100 | 41,534,978 | 40,313,850 (97.06%) | 38,511,232 (92.72%) | 20,796,563 (50.07%) |
| FD27h-2 | 6,899,318,700 | 43,442,624 | 42,208,853 (97.16%) | 40,362,542 (92.91%) | 21,968,935 (50.57%) |
| FD27h-3 | 6,795,099,300 | 43,529,080 | 42,392,971 (97.39%) | 40,595,220 (93.26%) | 21,847,245 (50.19%) |
| ID | Class | Description | p-Value | Q-Value | Ratio | Num | Up | Down |
|---|---|---|---|---|---|---|---|---|
| pov00330 | Metabolism; Amino acid metabolism | Arginine and proline metabolism | 0.002313977 | 0.141152599 | 0.09 | 3 | 2 | 1 |
| pov00010 | Glycolysis/Gluconeogenesis | Glycolysis/Gluconeogenesis | 0.006390935 | 0.194923503 | 0.09 | 3 | 2 | 1 |
| pov04218 | Cellular senescence | Cellular senescence | 0.019128015 | 0.304633194 | 0.13 | 4 | 2 | 2 |
| pov00260 | Glycine, serine and threonine metabolism | Glycine, serine and threonine metabolism | 0.022065588 | 0.304633194 | 0.06 | 2 | 1 | 1 |
| pov00620 | Metabolism; Carbohydrate metabolism | Pyruvate metabolism | 0.024969934 | 0.304633194 | 0.06 | 2 | 2 | 0 |
| pov04217 | Necroptosis | Necroptosis | 0.04034051 | 0.35638506 | 0.09 | 3 | 1 | 2 |
| pov00310 | Metabolism; Amino acid metabolism | Lysine degradation | 0.05046653 | 0.35638506 | 0.06 | 2 | 1 | 1 |
| pov04020 | Calcium signaling pathway | Calcium signaling pathway | 0.054192134 | 0.35638506 | 0.13 | 4 | 1 | 3 |
| pov00561 | Metabolism; Lipid metabolism | Glycerolipid metabolism | 0.055808883 | 0.35638506 | 0.06 | 2 | 2 | 0 |
| pov04115 | p53 signaling pathway | p53 signaling pathway | 0.067057946 | 0.35638506 | 0.06 | 2 | 2 | 0 |
| pov04146 | Peroxisome | Peroxisome | 0.074443165 | 0.35638506 | 0.06 | 2 | 0 | 2 |
| pov04110 | Cell cycle | Cell cycle | 0.074697428 | 0.35638506 | 0.09 | 3 | 3 | 0 |
| pov01230 | Biosynthesis of amino acids | Biosynthesis of amino acids | 0.075950914 | 0.35638506 | 0.06 | 2 | 1 | 1 |
| pov00430 | Biosynthesis of amino acids | Taurine and hypotaurine metabolism | 0.085790241 | 0.359435202 | 0.03 | 1 | 0 | 1 |
| pov01232 | Nucleotide metabolism | Nucleotide metabolism | 0.089950194 | 0.359435202 | 0.06 | 2 | 2 | 0 |
| pov00053 | Metabolism; Carbohydrate metabolism | Ascorbate and aldarate metabolism | 0.095401578 | 0.359435202 | 0.03 | 1 | 1 | 0 |
| pov00340 | Metabolism; Amino acid metabolism | Histidine metabolism | 0.100170466 | 0.359435202 | 0.03 | 1 | 1 | 0 |
| pov00601 | Metabolism; Amino acid metabolism | Glycosphingolipid biosynthesis—lacto and neolacto series | 0.109635285 | 0.371541799 | 0.03 | 1 | 0 | 1 |
| pov00982 | Metabolism; Xenobiotics biodegradation and metabolism | Drug metabolism—cytochrome P450 | 0.128276447 | 0.404567961 | 0.03 | 1 | 0 | 1 |
| pov00515 | Mannose type O-glycan biosynthesis | Mannose type O-glycan biosynthesis | 0.13287733 | 0.404567961 | 0.03 | 1 | 0 | 1 |
| ID | Class | Description | p-Value | Q-Value | Ratio | Num | Up | Down |
|---|---|---|---|---|---|---|---|---|
| pov03010 | Genetic Information Processing; Translation | Ribosome | 7.00 × 10−57 | 1.06 × 10−54 | 0.092576419 | 106 | 106 | 0 |
| pov00190 | Metabolism; Energy metabolism | Oxidative phosphorylation | 2.73 × 10−18 | 2.07 × 10−16 | 0.059388646 | 68 | 68 | 0 |
| pov04260 | Cardiac muscle contraction | Cardiac muscle contraction | 4.21 × 10−7 | 2.13 × 10−5 | 0.036681223 | 42 | 25 | 17 |
| pov03020 | Genetic Information Processing; Transcription | RNA polymerase | 0.000310433 | 0.01179644 | 0.013100437 | 15 | 13 | 2 |
| pov03018 | Genetic Information Processing; Folding, sorting and degradation | RNA degradation | 0.000539594 | 0.016403653 | 0.025327511 | 29 | 19 | 10 |
| pov03050 | Genetic Information Processing; Folding, sorting and degradation | Proteasome | 0.000994358 | 0.025190408 | 0.016593886 | 19 | 19 | 0 |
| pov03420 | Genetic Information Processing; Replication and repair | Nucleotide excision repair | 0.005929872 | 0.128762935 | 0.017467249 | 20 | 14 | 6 |
| pov03013 | Genetic Information Processing; Translation | Nucleocytoplasmic transport | 0.014518762 | 0.246774467 | 0.026200873 | 30 | 8 | 22 |
| pov03060 | Genetic Information Processing; Folding, sorting and degradation | Protein export | 0.014611646 | 0.246774467 | 0.007860262 | 9 | 8 | 1 |
| pov03040 | Genetic Information Processing; Transcription | Spliceosome | 0.02216678 | 0.334145793 | 0.031441048 | 36 | 28 | 8 |
| pov03030 | Genetic Information Processing; Replication and repair | DNA replication | 0.024181603 | 0.334145793 | 0.011353712 | 13 | 8 | 5 |
| pov04814 | Cellular Processes; Cell motility | Motor proteins | 0.043827537 | 0.555148808 | 0.043668122 | 50 | 15 | 35 |
| pov03460 | Genetic Information Processing; Replication and repair | Fanconi anemia pathway | 0.104087353 | 1 | 0.012227074 | 14 | 6 | 8 |
| pov03022 | Genetic Information Processing; Transcription | Basal transcription factors | 0.142019371 | 1 | 0.009606987 | 11 | 8 | 3 |
| pov00310 | Metabolism; Amino acid metabolism | Lysine degradation | 0.150289521 | 1 | 0.014847162 | 17 | 4 | 13 |
| pov00100 | Metabolism; Lipid metabolism | Steroid biosynthesis | 0.16535451 | 1 | 0.005240175 | 6 | 4 | 2 |
| pov00860 | Metabolism; Metabolism of cofactors and vitamins | Porphyrin metabolism | 0.172198533 | 1 | 0.0069869 | 8 | 7 | 1 |
| pov04110 | Cellular Processes; Cell growth and death | Cell cycle | 0.176882551 | 1 | 0.03580786 | 41 | 14 | 27 |
| pov04150 | Environmental Information Processing; Signal transduction | mTOR signaling pathway | 0.184488124 | 1 | 0.033187773 | 38 | 13 | 25 |
| pov04623 | Organismal Systems; Immune system | Cytosolic DNA-sensing pathway | 0.195926873 | 1 | 0.011353712 | 13 | 10 | 3 |
| ID | Class | Description | p-Value | Q-Value | Ratio | Num |
|---|---|---|---|---|---|---|
| pov04080 | Environmental Information Processing; Signaling molecules and interaction | Neuroactive ligand-receptor interaction | 2.2803047891782 × 10−74 | 3.39765413587552 × 10−72 | 0.178278689 | 261 |
| pov04060 | Environmental Information Processing; Signaling molecules and interaction | Cytokine-cytokine receptor interaction | 1.54860757163338 × 10−26 | 1.15371264086687 × 10−24 | 0.073770492 | 108 |
| pov04020 | Environmental Information Processing; Signal transduction | Calcium signaling pathway | 3.69346909045443 × 10−22 | 1.83442298159237 × 10−20 | 0.099043716 | 145 |
| pov04514 | Environmental Information Processing; Signaling molecules and interaction | Cell adhesion molecules | 2.20278383435677 × 10−12 | 8.20536978297897 × 10−11 | 0.056693989 | 83 |
| pov04744 | Organismal Systems; Sensory system | Phototransduction | 3.28572791644228 × 10−7 | 9.791469190998 × 10−6 | 0.017759563 | 26 |
| pov04672 | Organismal Systems; Immune system | Intestinal immune network for IgA production | 9.55222811284292 × 10−5 | 0.00220169939771074 | 0.012295082 | 18 |
| pov04270 | Organismal Systems; Circulatory system | Vascular smooth muscle contraction | 0.00010343554217433 | 0.00220169939771074 | 0.035519126 | 52 |
| pov04261 | Organismal Systems; Circulatory system | Adrenergic signaling in cardiomyocytes | 0.00013256443189031 | 0.00246901254395702 | 0.049863388 | 73 |
| pov00533 | Metabolism; Glycan biosynthesis and metabolism | Glycosaminoglycan biosynthesis—keratan sulfate | 0.000449363736190256 | 0.00743946629914979 | 0.008196721 | 12 |
| pov04350 | Environmental Information Processing; Signal transduction | TGF-beta signaling pathway | 0.000642067969688125 | 0.00956681274835306 | 0.030737705 | 45 |
| pov00230 | Metabolism; Nucleotide metabolism | Purine metabolism | 0.00104623266514309 | 0.0141716970096655 | 0.035519126 | 52 |
| pov04010 | Environmental Information Processing; Signal transduction | MAPK signaling pathway | 0.00146441859372183 | 0.0181831975387127 | 0.071721311 | 105 |
| pov00140 | Metabolism; Lipid metabolism | Steroid hormone biosynthesis | 0.00186736245711854 | 0.0214028466238971 | 0.010245902 | 15 |
| pov00604 | Metabolism; Glycan biosynthesis and metabolism | Glycosphingolipid biosynthesis—ganglio series | 0.00242041178360021 | 0.0257600968397451 | 0.006147541 | 9 |
| pov00590 | Metabolism; Lipid metabolism | Arachidonic acid metabolism | 0.00817442252971953 | 0.0778334418749539 | 0.012978142 | 19 |
| pov00532 | Metabolism; Glycan biosynthesis and metabolism | Glycosaminoglycan biosynthesis—chondroitin sulfate/dermatan sulfate | 0.00835795348992793 | 0.0778334418749539 | 0.007513661 | 11 |
| pov04916 | Organismal Systems; Endocrine system | Melanogenesis | 0.00930495422181556 | 0.0815551870029717 | 0.028688525 | 42 |
| pov04310 | Environmental Information Processing; Signal transduction | Wnt signaling pathway | 0.0151342015408406 | 0.12527755719918 | 0.040983607 | 60 |
| pov00910 | Metabolism; Energy metabolism | Nitrogen metabolism | 0.0260673964408138 | 0.203009420682821 | 0.006147541 | 9 |
| pov00380 | Metabolism; Amino acid metabolism | Tryptophan metabolism | 0.027249586668835 | 0.203009420682821 | 0.010928962 | 16 |
| ID | Class | Description | p-Value | Q-Value | Ratio | Num |
|---|---|---|---|---|---|---|
| pov04080 | Environmental Information Processing; Signaling molecules and interaction | Neuroactive ligand-receptor interaction | 6.15224639748607 × 10−59 | 9.16684713225424 × 10−57 | 0.171344165 | 232 |
| pov04020 | Environmental Information Processing; Signal transduction | Calcium signaling pathway | 5.83044313395946 × 10−20 | 4.3436801347998 × 10−18 | 0.098966027 | 134 |
| pov04060 | Environmental Information Processing; Signaling molecules and interaction | Cytokine-cytokine receptor interaction | 2.18024245322315 × 10−16 | 1.0828537517675 × 10−14 | 0.064992614 | 88 |
| pov04514 | Environmental Information Processing; Signaling molecules and interaction | Cell adhesion molecules | 2.28511681686016 × 10−11 | 8.51206014280408 × 10−10 | 0.056868538 | 77 |
| pov04744 | Organismal Systems; Sensory system | Phototransduction | 6.20894376944006 × 10−8 | 1.85026524329314 × 10−6 | 0.019202363 | 26 |
| pov04672 | Organismal Systems; Immune system | Intestinal immune network for IgA production | 6.58195396633777 × 10−6 | 0.000163451856830721 | 0.014032496 | 19 |
| pov04010 | Environmental Information Processing; Signal transduction | MAPK signaling pathway | 8.10034590883145 × 10−6 | 0.000172421648630841 | 0.080502216 | 109 |
| pov04270 | Organismal Systems; Circulatory system | Vascular smooth muscle contraction | 0.000236162899145753 | 0.00439853399658965 | 0.035450517 | 48 |
| pov00230 | Metabolism; Nucleotide metabolism | Purine metabolism | 0.00188063675192268 | 0.0311349862262755 | 0.035450517 | 48 |
| pov04261 | Organismal Systems; Circulatory system | Adrenergic signaling in cardiomyocytes | 0.0030049955859055 | 0.044774434229992 | 0.046528804 | 63 |
| pov00910 | Metabolism; Energy metabolism | Nitrogen metabolism | 0.00446926225877797 | 0.0605381887779925 | 0.007385524 | 10 |
| pov00604 | Metabolism; Glycan biosynthesis and metabolism | Glycosphingolipid biosynthesis—ganglio series | 0.0066887673854356 | 0.0795442582859311 | 0.005908419 | 8 |
| pov04916 | Organismal Systems; Endocrine system | Melanogenesis | 0.00694010307192687 | 0.0795442582859311 | 0.029542097 | 40 |
| pov04350 | Environmental Information Processing; Signal transduction | TGF-beta signaling pathway | 0.0102746435496404 | 0.109351563492602 | 0.028064993 | 38 |
| pov04810 | Cellular Processes; Cell motility | Regulation of actin cytoskeleton | 0.0125372806301676 | 0.124536987592998 | 0.052437223 | 71 |
| pov00532 | Metabolism; Glycan biosynthesis and metabolism | Glycosaminoglycan biosynthesis—chondroitin sulfate/dermatan sulfate | 0.0147359052616125 | 0.137228117748767 | 0.007385524 | 10 |
| pov00533 | Metabolism; Glycan biosynthesis and metabolism | Glycosaminoglycan biosynthesis—keratan sulfate | 0.0158685919402005 | 0.139083541122934 | 0.006646972 | 9 |
| pov00561 | Metabolism; Lipid metabolism | Glycerolipid metabolism | 0.0256715684407054 | 0.212503538759172 | 0.017725258 | 24 |
| pov04814 | Cellular Processes; Cell motility | Motor proteins | 0.0301713061938436 | 0.236606559099089 | 0.043574594 | 59 |
| pov04912 | Organismal Systems; Endocrine system | GnRH signaling pathway | 0.0320475961612918 | 0.238754591401624 | 0.025110783 | 34 |
| Group (n = 3) | Body Length (cm) | Overall Length (cm) | Body Weight (g) | Gonad Weight (g) |
|---|---|---|---|---|
| 0h | 16.67 ± 0.29 | 26.00 ± 0.00 | 226.96 ± 36.13 | 1.23 ± 0.46 |
| ND3h | 13.00 ± 1.00 | 21.90 ± 1.49 | 140.07 ± 38.12 | 0.25 ± 0.03 |
| ND6h | 14.60 ± 0.85 | 19.67 ± 0.58 | 121.17 ± 9.02 | 0.21 ± 0.04 |
| ND9h | 11.83 ± 1.04 | 19.33 ± 0.76 | 135.04 ± 27.57 | 0.22 ± 0.03 |
| ND12h | 15.23 ± 0.64 | 23.67 ± 1.15 | 158.22 ± 32.66 | 0.25 ± 0.06 |
| ND15h | 15.83 ± 0.58 | 20.83 ± 1.44 | 112.22 ± 10.53 | 0.27 ± 0.07 |
| ND18h | 16.00 ± 1.73 | 25.50 ± 2.29 | 223.36 ± 34.30 | 0.80 ± 0.12 |
| FD3h | 13.83 ± 1.26 | 22.50 ± 1.32 | 163.70 ± 34.14 | 0.24 ± 0.04 |
| FD6h | 12.67 ± 0.58 | 21.00 ± 1.73 | 118.42 ± 11.46 | 0.23 ± 0.02 |
| FD9h | 14.00 ± 1.00 | 21.67 ± 1.26 | 143.88 ± 49.04 | 0.25 ± 0.04 |
| FD12h | 10.83 ± 1.04 | 18.50 ± 1.00 | 130.89 ± 26.28 | 0.24 ± 0.02 |
| FD15h | 13.27 ± 0.40 | 20.93 ± 0.12 | 143.05 ± 5.14 | 0.25 ± 0.04 |
| FD18h | 12.67 ± 0.29 | 20.50 ± 1.32 | 151.41 ± 42.16 | 0.39 ± 0.15 |
| FD21h | 13.00 ± 1.00 | 23.33 ± 0.58 | 167.30 ± 38.56 | 0.55 ± 0.17 |
| FD24h | 14.83 ± 1.76 | 24.83 ± 2.02 | 211.88 ± 7.61 | 0.68 ± 0.17 |
| FD27h | 13.33 ± 1.04 | 22.83 ± 0.76 | 201.65 ± 8.40 | 0.69 ± 0.35 |
| NCBI Reference Sequence | Forward | Reverse | Gene Name | Usage |
|---|---|---|---|---|
| XM_069535616.1 | GCCTGACTCCAGCACA | CCATCGCCTACCCAA | trmt61b | qRT-PCR |
| XM_069530630.1 | GGCTGAGGATGAGGTG | GGCTGGTGGCTTGTAG | loc109627311 | |
| XM_020103989.1 | TCACGGGCCATACCAG | TGACCGAGGCAGAGCA | loc109640143 | |
| XM_020081586.1 | CTAACCCACGGCTTTGT | GCTGCGGATTTGAACC | loc109625962 | |
| XM_020086501.1 | TTGGCAAACTCGGGAC | GCAGCGTTGAGGAAAG | loc109629024 | |
| XM_020085544.2 | CACGACTCCCAGACCCA | GCAAGATGCCGACCA | tmprss2 | |
| XM_020104037.2 | CGGCGGGCTTCATTT | AGAACGGCGTGGAGGT | loc109640177 | |
| XM_069511009.1 | TGCCTACGCTGGAGT | CGAGAAGAAATAGTGCC | dhrs1 | |
| XM_020085444.1 | GGTTGATGGCATTTAGGT | CTGCTGTTTACAAGGCTCT | loc109628360 | |
| XM_020085379.1 | AAGCACCACATTCTTCCATA | AGCCACGCTTCCTTTG | tmlhe | |
| XM_020108792.2 | TGGGCTTGGGACATT | GGAAGGCGTGATTGC | rbx1 | |
| XM_020106609.2 | AGGACAAAGAGGGAATC | CAGCACCAAGTGAAGG | rps27a | |
| XM_020096236.2 | GGCGGTTCAAGGGTCA | TGGGCGATCTCAGCAC | rpl32 | |
| XM_020098086.2 | GGAACATAGTGGAGGCT | ATCTGGGTGGTGGAGTA | mrps26 | |
| XM_020094857.2 | CAGCAACACCTCCTCAG | ACAAGCGTCCATCACC | rnf7 | |
| XM_020112725.2 | CGAGCGGAGAAATGA | CCAGCCGATGATGAC | eif3f | |
| XM_020086721.1 | CAAGGCTGTGGTGAGGT | TCGTCTGGCGAGTTCTAT | nudt2 | |
| XM_020089424.1 | GAGTGCGACAAGAAGGAT | GGAGGAGCAGATTAGGG | loc109630913 | |
| XM_020108373.1 | TCCGCACCTCTTCCAC | TTTTCAGCCAGTCGTTC | ntpcr | |
| XM_020106718.1 | CAGGTCCCTCAGGTTGT | AGGCTGCTCTGCTTCA | hddc3 | |
| XM_069524822.1 | GGAAATCGTGCGTGACATTAAG | CCTCTGGACAACGGAACCTCT | β-actin | |
| JQ070418.1 | CTAACCACGAGGGAACTA | GCTGGATGAGGCTGAC | vasa | RT-PCR |
| KJ522774.2 | AGACTTTCTTCCCATTCCC | TGACGGTGTCAGATACTGTTG | oct4 | |
| XR_011242831.1 | CCTGAGAAACGGCTACCACAT | ATCCCGAGGTCCAACTACGAG | 18s rRNA |
References
- Liu, Y.; Blackburn, H.; Taylor, S.S.; Tiersch, T.R. Development of germplasm repositories to assist conservation of endangered fishes: Examples from small-bodied livebearing fishes. Theriogenology 2019, 135, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Goswami, M.; Mishra, A.; AS, N.; VL, T.; WS, L. Bio-banking: An Emerging Approach for Conservation of Fish Germplasm. Poult. Fish. Wildl. Sci. 2016, 4, 1000143. [Google Scholar] [CrossRef]
- Wang, T.; Li, Z.; Zheng, D.; Liu, W.; Huang, P.; Zeng, Z.; Xu, C.; Wang, B.; Wei, J. Establishment and characterization of a fibroblast cell line from postmortem skin of an adult Chinese muntjac (Muntiacus reevesi). Vitr. Cell. Dev. Biology Anim. 2020, 56, 97–102. [Google Scholar] [CrossRef]
- Shigunov, P.; Dallagiovanna, B. Stem Cell Ribonomics: RNA-Binding Proteins and Gene Networks in Stem Cell Differentiation. Front. Mol. Biosci. 2015, 2, 74. [Google Scholar] [CrossRef]
- Yoshizaki, G.; Ichikawa, M.; Hayashi, M.; Iwasaki, Y.; Miwa, M.; Shikina, S.; Okutsu, T. Sexual plasticity of ovarian germ cells in rainbow trout. Development 2010, 137, 1227–1230. [Google Scholar] [CrossRef]
- Lee, S.; Iwasaki, Y.; Yoshizaki, G. Long-term (5 years) cryopreserved spermatogonia have high capacity to generate functional gametes via interspecies transplantation in salmonids. Cryobiology 2016, 73, 286–290. [Google Scholar] [CrossRef]
- Ye, H.; Li, C.J.; Yue, H.M.; Du, H.; Yang, X.G.; Yoshino, T.; Hayashida, T.; Takeuchi, Y.; Wei, Q.W. Establishment of intraperitoneal germ cell transplantation for critically endangered Chinese sturgeon Acipenser sinensis. Theriogenology 2017, 94, 37–47. [Google Scholar] [CrossRef]
- Yoshikawa, H.; Ino, Y.; Shigenaga, K.; Katayama, T.; Kuroyanagi, M.; Yoshiura, Y. Production of tiger puffer Takifugu rubripes from cryopreserved testicular germ cells using surrogate broodstock technology. Aquaculture 2018, 493, 302–313. [Google Scholar] [CrossRef]
- Ren, Y.; Sun, Z.; Wang, Y.; Yu, Q.; Wang, G.; He, Z.; Liu, Y.; Jiang, X.; Kang, X.; Hou, J. Production of donor-derived offsprings by allogeneic transplantation of oogonia in the adult Japanese flounder (Paralichthys olivaceus). Aquaculture 2021, 543, 736977. [Google Scholar] [CrossRef]
- Yoshizaki, G.; Takashiba, K.; Shimamori, S.; Fujinuma, K.; Shikina, S.; Okutsu, T.; Kume, S.; Hayashi, M. Production of germ cell-deficient salmonids by dead end gene knockdown, and their use as recipients for germ cell transplantation. Mol. Reprod. Dev. 2016, 83, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Pšenička, M.; Saito, T.; Rodina, M.; Dzyuba, B. Cryopreservation of early stage Siberian sturgeon Acipenser baerii germ cells, comparison of whole tissue and dissociated cells. Cryobiology 2016, 72, 119–122. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, X.; Liu, Q.; Yang, J.; Xu, S.; Wu, Z.; Wang, Y.; You, F.; Song, Z.; Li, J. Successful Spermatogonial Stem Cells Transplantation within Pleuronectiformes: First Breakthrough at inter-family Level in Marine Fish. Int. J. Biol. Sci. 2021, 17, 4426–4441. [Google Scholar] [CrossRef]
- Marinovic, Z.; Li, Q.; Lujic, J.; Iwasaki, Y.; Csenki, Z.; Urbanyi, B.; Yoshizaki, G.; Horvath, A. Preservation of zebrafish genetic resources through testis cryopreservation and spermatogonia transplantation. Sci. Rep. 2019, 9, 13861. [Google Scholar] [CrossRef] [PubMed]
- de Queiroz Neta, L.B.; Lira, G.P.D.O.; Borges, A.A.; Santos, M.V.D.O.; Silva, M.B.; de Oliveira, L.R.M.; Silva, A.R.; de Oliveira, M.F.; Pereira, A.F. Influence of storage time and nutrient medium on recovery of fibroblast-like cells from refrigerated collared peccary (Pecari tajacu Linnaeus, 1758) skin. Vitr. Cell. Dev. Biol. Anim. 2018, 54, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, A.; Yamada, C.; Tani, M.; Hirano, S.; Tokumoto, Y.; Miyake, J. Caspase inhibitors increase the rate of recovery of neural stem/progenitor cells from post-mortem rat brains stored at room temperature. J. Biosci. Bioeng. 2009, 107, 652–657. [Google Scholar] [CrossRef]
- Saito, T.; Sato, T.; Suzuki, K. Isolation and culture of human adipose-derived mesenchymal stromal/stem cells harvested from postmortem adipose tissues. J. Forensic Leg. Med. 2020, 69, 101875. [Google Scholar] [CrossRef]
- Porretti, L.; Gatti, S.; Gramignoli, R.; Colombo, F.; Lopa, R.; Cattaneo, A.; Scalamogna, M.; Colombo, G.; Rossi, G.; Bonino, F.; et al. Animal model for liver cell banking from non-heart beating donors after prolonged ischaemia time. Dig. Liver Dis. 2006, 38, 905–911. [Google Scholar] [CrossRef]
- Erker, L.; Azuma, H.; Lee, A.Y.; Guo, C.; Orloff, S.; Eaton, L.; Benedetti, E.; Jensen, B.; Finegold, M.; Willenbring, H.; et al. Therapeutic Liver Reconstitution With Murine Cells Isolated Long After Death. Gastroenterology 2010, 139, 1019–1029. [Google Scholar] [CrossRef]
- Shikh Alsook, M.K.; Gabriel, A.; Piret, J.; Waroux, O.; Tonus, C.; Connan, D.; Baise, E.; Antoine, N. Tissues from equine cadaver ligaments up to 72 hours of post-mortem: A promising reservoir of stem cells. Stem Cell Res. Ther. 2015, 6, 253. [Google Scholar] [CrossRef]
- Tolbert, M.; Finley, S.J.; Visonà, S.D.; Soni, S.; Osculati, A.; Javan, G.T. The thanatotranscriptome: Gene expression of male reproductive organs after death. Gene 2018, 675, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, P.G.; Munoz-Aguirre, M.; Reverter, F.; Sa, G.C.; Sousa, A.; Amadoz, A.; Sodaei, R.; Hidalgo, M.R.; Pervouchine, D.; Carbonell-Caballero, J.; et al. The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nat. Commun. 2018, 9, 490. [Google Scholar] [CrossRef]
- Javan, G.T.; Kwon, I.; Finley, S.J.; Lee, Y. Progression of thanatophagy in cadaver brain and heart tissues. Biochem. Biophys. Rep. 2016, 5, 152–159. [Google Scholar] [CrossRef][Green Version]
- Zapico, S.C.; Menéndez, S.T.; Núñez, P. Cell death proteins as markers of early postmortem interval. Cell. Mol. Life Sci. 2014, 71, 2957–2962. [Google Scholar] [CrossRef]
- Javan, G.T.; Can, I.; Finley, S.J.; Soni, S. The apoptotic thanatotranscriptome associated with the liver of cadavers. Forensic Sci. Med. Pathol. 2015, 11, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, Z.; Jiang, J.; Gao, J.; Wang, J.; Zhou, X.; Zhang, Q. Cloning, expression promoter analysis of vasa gene in Japanese flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2014, 167, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, X.; Zhang, Q. Evolutionary Conservation of pou5f3 Genomic Organization and Its Dynamic Distribution during Embryogenesis and in Adult Gonads in Japanese Flounder Paralichthys olivaceus. Int. J. Mol. Sci. 2017, 18, 231. [Google Scholar] [CrossRef]
- Fujihara, M.; Shiraishi, J.I.; Onuma, M.; Ohta, Y.; Inoue-Murayama, M. Cryopreservation Competence of Chicken Oocytes as a Model of Endangered Wild Birds: Effects of Storage Time and Temperature on the Ovarian Follicle Survival. Animals 2022, 12, 1434. [Google Scholar] [CrossRef]
- Zhang, G.L.; Ma, J.Y.; Sun, Q.; Hu, M.W.; Yang, X.Y.; Gao, S.H.; Jiang, G.J. Effects of postmortem interval on mouse ovary oocyte survival and maturation. PLoS ONE 2014, 9, e98384. [Google Scholar] [CrossRef]
- Chaves, R.N.; Martins, F.S.; Saraiva, M.V.; Celestino, J.J.; Lopes, C.A.; Correia, J.C.; Verde, I.B.; Matos, M.H.; Bao, S.N.; Name, K.P.; et al. Chilling ovarian fragments during transportation improves viability and growth of goat preantral follicles cultured in vitro. Reprod. Fertil. Dev. 2008, 20, 640–647. [Google Scholar] [CrossRef] [PubMed]
- Churchill, T.A.; Cheetham, K.M.; Fuller, B.J. Glycolysis and Energy Metabolism in Rat Liver during Warm and Cold Ischemia: Evidence of an Activation of the Regulatory Enzyme Phosphofructokinase. Cryobiology 1994, 31, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, K.; Mutter, M.; Gustafsson, E.; Gustafsson, L.; Vaegler, M.; Schultheiss, M.; Muller, S.; Yoeruek, E.; Schrader, M.; Munch, T.A. Hypothermia Promotes Survival of Ischemic Retinal Ganglion Cells. Investig. Ophthalmol. Vis. Sci. 2016, 57, 658–663. [Google Scholar] [CrossRef]
- de Araujo, J.M.; Oliveira, R.A.; Capobianco, N.E.; Cunha, A.; Dode, M.; Martins, C.F. Effects of Refrigeration at 5 degrees C for Long Periods of Time on Bovine Ear Skin as a Strategy to Transport Biological Material and Isolate Fibroblasts to Use in the Nuclear Transfer. Biopreserv. Biobank. 2022, 20, 323–330. [Google Scholar] [CrossRef]
- Silvestre, M.A.; Sánchez, J.P.; Gómez, E.A. Vitrification of goat, sheep, and cattle skin samples from whole ear extirpated after death and maintained at different storage times and temperatures. Cryobiology 2004, 49, 221–229. [Google Scholar] [CrossRef]
- Walcott, B.; Singh, M. Recovery of proliferative cells up to 15- and 49-day postmortem from bovine skin stored at 25 °C and 4 °C, respectively. Cogent Biol. 2017, 3, 1333760. [Google Scholar] [CrossRef]
- Liu, W.; Han, L.; Yuan, F.; Liu, Q.; Cheng, H.; Jin, X.; Sun, Y. Mechanism of blocking the glutamate pathway to exacerbate oxidative stress, ammonia toxicity and metabolic disorders in crucian carp (Carassius auratus) under saline-alkaline exposure. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2025, 291, 110146. [Google Scholar] [CrossRef]
- Wang, W.; Mou, S.; Xiu, W.; Li, Y.; Liu, Z.; Feng, Y.; Ma, J.; Li, X. Fenpropathrin disrupted the gills of common carp (Cyprinus carpio L.) through oxidative stress, inflammatory responses, apoptosis, and transcriptional alterations. Ecotoxicol. Environ. Saf. 2024, 271, 116007. [Google Scholar] [CrossRef] [PubMed]
- Becerril, M.R.; Savin, T.Z. Bisphenol A Induces Reactive Oxygen Species Production and Apoptosis-Related Gene Expression in Pacific Red Snapper Lutjanus peru Leukocytes. Mar. Biotechnol. 2024, 26, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Dirk, R.; Stuart, C.G. Rethinking the Citric Acid Cycle: Connecting Pyruvate Carboxylase and Citrate Synthase to the Flow of Energy and Material. Int. J. Mol. Sci. 2021, 22, 604. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Wu, R.; Niu, S.; Miao, B.; Liang, Z.; Zhai, Y. Liver transcriptome analysis reveals changes in energy metabolism, oxidative stress, and apoptosis in pearl gentian grouper exposed to acute hypoxia. Aquaculture 2022, 561, 738635. [Google Scholar] [CrossRef]
- Li, P.; Mai, K.; Trushenski, J.; Wu, G. New developments in fish amino acid nutrition: Towards functional and environmentally oriented aquafeeds. Amino Acids 2009, 37, 43–53. [Google Scholar] [CrossRef]
- Paulusma, C.C.; Lamers, W.H.; Broer, S.; van de Graaf, S. Amino acid metabolism, transport and signalling in the liver revisited. Biochem. Pharmacol. 2022, 201, 115074. [Google Scholar] [CrossRef]
- Abdel-Rhman, A.; Morsy, W.; Selim, N.; Abdel-Hady, E.A. L-arginine supplementation attenuates ovarian oxidative stress in female rats subjected to chronic intermittent hypoxia. Physiol. Int. 2023, 110, 326–341. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, S.; Wang, J.; Nie, L.; Li, L.; Zhu, X.; Zhang, L. Multi-omics analysis provides insight into liver metabolism in yellow catfish (Pelteobagrus fulvidraco) under hypoxic stress. Aquaculture 2024, 583, 740531. [Google Scholar] [CrossRef]
- Wen, J.; Feng, Y.; Xue, L.; Yuan, S.; Chen, Q.; Luo, A.; Wang, S.; Zhang, J. High-fat diet-induced L-saccharopine accumulation inhibits estradiol synthesis and damages oocyte quality by disturbing mitochondrial homeostasis. Gut Microbes 2024, 16, 2412381. [Google Scholar] [CrossRef]
- Ma, J.L.; Qiang, J.; Tao, Y.F.; Bao, J.W.; Zhu, H.J.; Li, L.G.; Xu, P. Multi-omics analysis reveals the glycolipid metabolism response mechanism in the liver of genetically improved farmed Tilapia (GIFT, Oreochromis niloticus) under hypoxia stress. BMC Genom. 2021, 22, 105. [Google Scholar] [CrossRef]
- Qi, M.; Wu, Q.; Liu, T.; Hou, Y.; Miao, Y.; Hu, M.; Liu, Q. Hepatopancreas Transcriptome Profiling Analysis Reveals Physiological Responses to Acute Hypoxia and Reoxygenation in Juvenile Qingtian Paddy Field Carp Cyprinus carpio var qingtianensis. Front. Physiol. 2020, 11, 1110. [Google Scholar] [CrossRef] [PubMed]
- Stecyk, J.A.W.; Stensløkken, K.; Farrell, A.P.; Nilsson, G.E. Maintained Cardiac Pumping in Anoxic Crucian Carp. Science 2004, 306, 77. [Google Scholar] [CrossRef] [PubMed]
- Anja, R.; A, M.S.; Thomas, H.; Thorsten, B. Globins and hypoxia adaptation in the goldfish, Carassius auratus. FEBS J. 2008, 275, 3633–3643. [Google Scholar] [CrossRef]
- Li, Y.; Yao, C.; Xu, F.; Qu, Y.; Li, J.; Lin, Y.; Cao, Z.; Lin, P.; Xu, W.; Zhao, S.; et al. APC/CCDH1 synchronizes ribose-5-phosphate levels and DNA synthesis to cell cycle progression. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Huang, Z.; Xie, N.; Illes, P.; Di Virgilio, F.; Ulrich, H.; Semyanov, A.; Verkhratsky, A.; Sperlagh, B.; Yu, S.G.; Huang, C.; et al. From purines to purinergic signalling: Molecular functions and human diseases. Signal Transduct. Target. Ther. 2021, 6, 162. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zheng, J.; Ge, L.; Tang, H.; Hu, J.; Li, X.; Wang, X.; Zhang, Y.; Shi, Q. Integrated metabolomic and transcriptomic analyses reveal the roles of alanine, aspartate and glutamate metabolism and glutathione metabolism in response to salt stress in tomato. Sci. Hortic. 2024, 328, 112911. [Google Scholar] [CrossRef]
- Chen, Q.L.; Luo, Z.; Huang, C.; Pan, Y.X.; Wu, K. De novo characterization of the liver transcriptome of javelin goby Synechogobius hasta and analysis of its transcriptomic profile following waterborne copper exposure. Fish. Physiol. Biochem. 2016, 42, 979–994. [Google Scholar] [CrossRef] [PubMed]
- Tsukahara, T.; Matsuda, Y.; Haniu, H. Lysophospholipid-Related Diseases and PPARγ Signaling Pathway. Int. J. Mol. Sci. 2017, 18, 2730. [Google Scholar] [CrossRef] [PubMed]
- Doll, S.; Proneth, B.; Tyurina, Y.Y.; Panzilius, E.; Kobayashi, S.; Ingold, I.; Irmler, M.; Beckers, J.; Aichler, M.; Walch, A.; et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017, 13, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; Stockwell, B.R. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016, 26, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Winter, G.E.; Musavi, L.S.; Lee, E.D.; Snijder, B.; Rebsamen, M.; Superti-Furga, G.; Stockwell, B.R. Human Haploid Cell Genetics Reveals Roles for Lipid Metabolism Genes in Nonapoptotic Cell Death. ACS Chem. Biol. 2015, 10, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Shchepinov, M.S.; Pratt, D.A. Resolving the Role of Lipoxygenases in the Initiation and Execution of Ferroptosis. ACS Cent. Sci. 2018, 4, 387–396. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Ding, W.; Ji, X.; Ao, X.; Liu, Y.; Yu, W.; Wang, J. Molecular mechanisms of ferroptosis and its role in cancer therapy. J. Cell. Mol. Med. 2019, 23, 4900–4912. [Google Scholar] [CrossRef]
- Chen, W.; Jiang, L.; Hu, Y.; Tang, N.; Liang, N.; Li, X.F.; Chen, Y.W.; Qin, H.; Wu, L. Ferritin reduction is essential for cerebral ischemia-induced hippocampal neuronal death through p53/SLC7A11-mediated ferroptosis. Brain Res. 2021, 1752, 147216. [Google Scholar] [CrossRef]
- Jiang, L.; Kon, N.; Li, T.; Wang, S.J.; Su, T.; Hibshoosh, H.; Baer, R.; Gu, W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015, 520, 57–62. [Google Scholar] [CrossRef]
- Bridges, R.J.; Natale, N.R.; Patel, S.A. System xc− cystine/glutamate antiporter: An update on molecular pharmacology and roles within the CNS. Br. J. Pharmacol. 2012, 165, 20–34. [Google Scholar] [CrossRef]
- Hu, K.; Li, K.; Lv, J.; Feng, J.; Chen, J.; Wu, H.; Cheng, F.; Jiang, W.; Wang, J.; Pei, H.; et al. Suppression of the SLC7A11/glutathione axis causes synthetic lethality in KRAS-mutant lung adenocarcinoma. J. Clin. Investig. 2020, 130, 1752–1766. [Google Scholar] [CrossRef] [PubMed]
- Lei, G.; Zhuang, L.; Gan, B. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer 2022, 22, 381–396. [Google Scholar] [CrossRef]
- Gao, M.; Yi, J.; Zhu, J.; Minikes, A.M.; Monian, P.; Thompson, C.B.; Jiang, X. Role of Mitochondria in Ferroptosis. Mol. Cell 2019, 73, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lu, S.; Wu, L.L.; Yang, L.; Yang, L.; Wang, J. The diversified role of mitochondria in ferroptosis in cancer. Cell Death Dis. 2023, 14, 519. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ren, Y.; Zhang, Y.; Wang, G.; He, Z.; Liu, Y.; Cao, W.; Wang, Y.; Chen, S.; Fu, Y.; et al. A New Cell Line Derived from the Spleen of the Japanese Flounder (Paralichthys olivaceus) and Its Application in Viral Study. Biology 2022, 11, 1697. [Google Scholar] [CrossRef]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic. Acids. Res. 2009, 37, e123. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Wilson, I.D.; Gika, H.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Holmes, E.; Nicholson, J.K. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- The R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Volume 1. [Google Scholar]
- Python Software Foundation. Python 2.7.6 Documentation. 2014. Available online: https://www.python.org/doc/versions/ (accessed on 17 September 2025).
- CentOS Project. CentOS 6.6 Release Notes. 2014. Available online: https://wiki.centos.org/Manuals(2f)ReleaseNotes(2f)CentOS6(2e)6.html (accessed on 17 September 2025).
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, Y.; Yang, Y.; He, N.; Wang, G.; He, Z.; Liu, Y.; Cao, W.; Zhang, X.; Zhang, Y.; San, L.; et al. Multi-Omics Investigation into Why Viable Oogonial Stem Cells Can Still Be Isolated and Cultured from Post-Mortem Paralichthys olivaceus. Int. J. Mol. Sci. 2025, 26, 10679. https://doi.org/10.3390/ijms262110679
Ren Y, Yang Y, He N, Wang G, He Z, Liu Y, Cao W, Zhang X, Zhang Y, San L, et al. Multi-Omics Investigation into Why Viable Oogonial Stem Cells Can Still Be Isolated and Cultured from Post-Mortem Paralichthys olivaceus. International Journal of Molecular Sciences. 2025; 26(21):10679. https://doi.org/10.3390/ijms262110679
Chicago/Turabian StyleRen, Yuqin, Yucong Yang, Nuan He, Guixing Wang, Zhongwei He, Yufeng Liu, Wei Cao, Xiaoyan Zhang, Yitong Zhang, Lize San, and et al. 2025. "Multi-Omics Investigation into Why Viable Oogonial Stem Cells Can Still Be Isolated and Cultured from Post-Mortem Paralichthys olivaceus" International Journal of Molecular Sciences 26, no. 21: 10679. https://doi.org/10.3390/ijms262110679
APA StyleRen, Y., Yang, Y., He, N., Wang, G., He, Z., Liu, Y., Cao, W., Zhang, X., Zhang, Y., San, L., Han, Z., & Hou, J. (2025). Multi-Omics Investigation into Why Viable Oogonial Stem Cells Can Still Be Isolated and Cultured from Post-Mortem Paralichthys olivaceus. International Journal of Molecular Sciences, 26(21), 10679. https://doi.org/10.3390/ijms262110679






