Epigenetic Impact of Sleep Timing in Children: Novel DNA Methylation Signatures via SWAG Analysis
Abstract
1. Introduction
2. Results
2.1. Study Participants
2.2. Identification of Significantly Associated Target IDs
2.3. Top Hits of SWAG Analysis
2.4. Pathway Analysis
2.5. Cross-Comparison Analysis
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Anthropometric Measurements
4.3. Isolation of Salivary DNA
4.4. Sodium Bisulfite Conversion and Infinium Arrays
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABCG2 | ATP-binding cassette sub-family G member 2 |
| ABHD4 | Abhydrolase domain containing 4 |
| ACTG1 | Actin gamma 1 |
| ADHD | Attention-deficit/hyperactivity disorder |
| AIC | Akaike Information Criterion |
| AK3 | Adenylate kinase 3 |
| ANKRD11 | Ankyrin repeat domain-containing protein 11 |
| ATXN1 | Ataxin 1 |
| BMI | Body mass index |
| BMAL1 | Brain and muscle ARNT-like 1 (circadian clock gene, also ARNTL) |
| BRAT1 | BRCA1-associated ATM activator 1 |
| BY | Benjamini–Yekutieli (False Discovery Rate control method) |
| CAT | Catalase |
| CDC | Centers for Disease Control and Prevention |
| CDH4 | Cadherin 4 |
| CI | Confidence Interval |
| CLOCK | Circadian locomotor output cycles kaput (core circadian gene) |
| COG5 | Component of oligomeric Golgi complex 5 |
| CREM | cAMP responsive element modulator |
| CpG | Cytosine-phosphate-Guanine dinucleotide |
| CRP | C-reactive protein |
| CRY1 | Cryptochrome circadian regulator 1 |
| DNA | Deoxyribonucleic acid |
| DOK7 | Docking protein 7 |
| EWAS | Epigenome-wide association study |
| FDR | False Discovery Rate |
| FGFR1 | Fibroblast growth factor receptor 1 |
| FTO | Fat mass and obesity-associated gene |
| GABA | Gamma-aminobutyric acid |
| GABRB3 | Gamma-aminobutyric acid receptor subunit beta-3 |
| GO | Gene Ontology |
| HDAC4 | Histone deacetylase 4 |
| IDAT | Intensity Data File (Illumina) |
| IL-6 | Interleukin-6 |
| IRB | Institutional Review Board |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LMS | Lambda-Mu-Sigma method (for growth references) |
| MOBKL1A | MOB kinase activator-like 1A |
| NHANES | National Health and Nutrition Examination Survey |
| NR3C2 | Nuclear receptor subfamily 3 group C member 2 |
| NSCH | National Survey of Children’s Health |
| OCLN | Occludin |
| OR | Odds Ratio |
| PCS | Predictability, Computability, Stability framework |
| PER2 | Period circadian regulator 2 |
| PRAMEF4 | Preferentially expressed antigen in melanoma family member 4 |
| REM | Rapid eye movement |
| RNA | Ribonucleic acid |
| SCN9A | Sodium voltage-gated channel alpha subunit 9 |
| SDK1 | Sidekick cell adhesion molecule 1 |
| SDE2 | Silencing-defective 2 homolog |
| SE | Standard Error |
| SETD2 | SET domain containing 2 (lysine methyltransferase) |
| SWAG | Sparse Wrapper Algorithm |
| TBL1XR1 | Transducin beta-like 1 X-linked receptor 1 |
| TGF-β | Transforming Growth Factor Beta |
| UTR | Untranslated region |
| WHO | World Health Organization |
| WHtR | Waist-to-height ratio |
| β | Beta coefficient (regression estimate) |
Appendix A
| No. | Gene | Full Name/Protein | Key Function | Length (aa) | Methylation Direction (Late Bedtime) |
|---|---|---|---|---|---|
| 1 | ABCG2 | ATP-binding cassette sub-family G member 2 | Broad substrate transporter; exports porphyrins, heme, sphingosine-1-P, urate, and xenobiotics; contributes to detoxification and homeostasis | 655 | Hyper |
| 2 | ABHD4 | Abhydrolase domain-containing protein 4 | Lysophospholipase; hydrolyzes N-acyl phosphatidylethanolamines to generate precursors for endocannabinoids (e.g., anandamide) | 342 | Hypo |
| 3 | MOBKL1A | MOB kinase activator 1A | Activator in Hippo signaling; regulates organ size, cell proliferation, and apoptosis via LATS1/2–YAP1 pathway | 221 | Hypo |
| 4 | AK3 | Adenylate kinase 3, mitochondrial | Maintains nucleotide homeostasis; catalyzes GTP:AMP and ITP:AMP phosphotransferase reactions | 227 | Hypo |
| 5 | SDE2 | Stress response regulator SDE2 | DNA replication and cell cycle control; binds PCNA to modulate translesion DNA synthesis and DNA damage responses | 451 | Hypo |
| 6 | PRAMEF4 | PRAME family member 4 | Member of PRAME gene family; putative role in transcriptional regulation and oncogenesis | 478 | Hyper |
| 7 | CREM | cAMP response element modulator | Transcription factor binding CRE sites; functions as activator or repressor; essential for spermatogenesis | 300 | Hypo |
| 8 | CDH4 | Cadherin-4 (R-cadherin) | Calcium-dependent adhesion protein; mediates homophilic cell–cell adhesion; important in retinal development | 916 | Hypo |
| 9 | BRAT1 | BRCA1-associated ATM activator 1 | Regulates DNA damage response and mitochondrial function; stabilizes mTOR pathway proteins | 821 | Hyper |
| 10 | SDK1 | Sidekick cell adhesion molecule 1 | Large adhesion molecule; mediates lamina-specific synaptic connections in retina via homophilic interactions | 2213 | Hypo |
| No. | Gene | Full Name/Protein | Key Function | Length (aa) |
Methylation
Direction (Late Bedtime) |
|---|---|---|---|---|---|
| 1 | CDH4 | Cadherin-4 (R-cadherin) | Calcium-dependent adhesion protein; mediates homophilic cell–cell adhesion; important in retinal development | 916 | Hypo |
| 2 | NR3C2 | Mineralocorticoid receptor (MR) | Nuclear receptor for aldosterone and cortisol; regulates ion/water balance, blood pressure, and electrolyte homeostasis | 984 | Hyper |
| 3 | ACTG1 | Actin, cytoplasmic 2 | Ubiquitous cytoskeletal protein; essential for cell motility, structure, and intracellular transport | 375 | Hypo |
| 4 | COG5 | Conserved oligomeric Golgi subunit 5 | Component of the COG complex; required for normal Golgi trafficking and glycoprotein processing | 860 | Hyper |
| 5 | CAT | Catalase | Antioxidant enzyme; degrades hydrogen peroxide, protecting cells from oxidative stress; supports immune cell growth | 527 | Hypo |
| 6 | HDAC4 | Histone deacetylase 4 | Epigenetic regulator; deacetylates histones, repressing transcription; roles in development, muscle maturation, and cancer pathways | 1084 | Hyper |
| 7 | FTO | Fat mass and obesity-associated protein (RNA demethylase) | Demethylates N6-methyladenosine (m6A) in RNA; regulates gene expression, energy balance, adipogenesis, and obesity risk | 505 | Hyper |
| 8 | DOK7 | Docking protein 7 | Activates MUSK receptor; essential for neuromuscular junction formation and acetylcholine receptor clustering | 504 | Hypo |
| 9 | OCLN | Occludin | Integral tight junction protein; regulates paracellular permeability and barrier integrity | 522 | Hypo |
| 10 | ATXN1 | Ataxin-1 | Chromatin-binding protein; represses Notch signaling, regulates RNA metabolism; implicated in brain development and neurodegeneration | 815 | Hyper |
References
- American Academy of Sleep Medicine. Recharge with Sleep: Pediatric Sleep Recommendations Promoting Optimal Health. 2016. Available online: https://aasm.org/recharge-with-sleep-pediatric-sleep-recommendations-promoting-optimal-health/ (accessed on 1 June 2025).
- National Institutes of Health. Children’s Sleep Linked to Brain Development. 2015. Available online: https://www.nih.gov/news-events/nih-research-matters/children-s-sleep-linked-brain-development (accessed on 1 June 2025).
- Jalbrzikowski, M.; Hayes, R.A.; Scully, K.E.; Franzen, P.L.; Hasler, B.P.; Siegle, G.J.; Buysse, D.J.; Dahl, R.E.; Forbes, E.E.; Ladouceur, C.D.; et al. Associations between brain structure and sleep patterns across adolescent development. Sleep 2021, 44, zsab120. [Google Scholar] [CrossRef]
- Perrault, A.A.; Bayer, L.; Peuvrier, M.; Afyouni, A.; Ghisletta, P.; Brockmann, C.; Spiridon, M.; Vesely, S.H.; Haller, D.M.; Pichon, S.; et al. Reducing the use of screen electronic devices in the evening is associated with improved sleep and daytime vigilance in adolescents. Sleep 2019, 42, zsz125. [Google Scholar] [CrossRef] [PubMed]
- Bonsu, E.O.; Afetor, M.; Munkaila, L.; Okwei, R.; Nachibi, S.U.; Adjei, B.N.; Frimpong, E.; Arimiyaw, A.W.; Adu, C.; Peprah, P. Association of food insecurity and sleep difficulty among 189,619 school-going adolescents: A study from the global in-school students survey. Front. Public Health 2023, 11, 1212254. [Google Scholar] [CrossRef]
- Radošević-Vidaček, B.; Košćec, A.; Bakotić, M. Parents working non-standard schedules and schools operating in two shifts: Effects on sleep and daytime functioning of adolescents. In Social and Family Issues in Shift Work and Non Standard Working Hours; Iskra-Golec, I., Barnes-Farrell, J., Bohle, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 95–111. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Patterns and predictors of sleep among U.S. school-aged children and adolescents. Prev. Chronic. Dis. 2023, 20, 220408. Available online: https://www.cdc.gov/pcd/issues/2023/22_0408.htm (accessed on 1 June 2025).
- Venkatapoorna, C.M.K.; Ayine, P.; Selvaraju, V.; Parra, E.P.; Koenigs, T.; Babu, J.R.; Geetha, T. The relationship between obesity and sleep timing behavior, television exposure, and dinnertime among elementary school-age children. J. Clin. Sleep. Med. 2020, 16, 129–136. [Google Scholar] [CrossRef]
- Olds, T.S.; Maher, C.A.; Matricciani, L. Sleep duration or bedtime? Exploring the relationship between sleep habits and weight status and activity patterns. Sleep 2011, 34, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Gaine, M.E.; Chatterjee, S.; Abel, T. Sleep deprivation and the epigenome. Front. Neural Circuits 2018, 12, 14. [Google Scholar] [CrossRef]
- Simon, K.C.; Cadle, C.; Shuster, A.E.; Malerba, P. Sleep Across the Lifespan: A Neurobehavioral Perspective. Curr. Sleep Med. Rep. 2025, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Klibaner-Schiff, E.; Simonin, E.M.; Akdis, C.A.; Cheong, A.; Johnson, M.M.; Karagas, M.R.; Kirsh, S.; Kline, O.; Mazumdar, M.; Oken, E.; et al. Environmental exposures influence multigenerational epigenetic transmission. Clin. Epigenetics 2024, 16, 145. [Google Scholar] [CrossRef]
- Lahtinen, A.; Puttonen, S.; Vanttola, P.; Viitasalo, K.; Sulkava, S.; Pervjakova, N.; Joensuu, A.; Salo, P.; Toivola, A.; Härmä, M.; et al. A distinctive DNA methylation pattern in insufficient sleep. Sci. Rep. 2019, 9, 1193. [Google Scholar] [CrossRef] [PubMed]
- Massart, R.; Freyburger, M.; Suderman, M.; Paquet, J.; El Helou, J.; Belanger-Nelson, E.; Rachalski, A.; Koumar, O.C.; Carrier, J.; Szyf, M.; et al. The genome-wide landscape of DNA methylation and hydroxymethylation in response to sleep deprivation impacts on synaptic plasticity genes. Transl. Psychiatry 2014, 4, e347. [Google Scholar] [CrossRef] [PubMed]
- Maag, J.L.V.; Kaczorowski, D.C.; Panja, D.; Peters, T.J.; Bramham, C.R.; Wibrand, K.; Dinger, M.E. Widespread promoter methylation of synaptic plasticity genes in long-term potentiation in the adult brain in vivo. BMC Genom. 2017, 18, 250. [Google Scholar] [CrossRef]
- Dhar, G.A.; Saha, S.; Mitra, P.; Chaudhuri, R.N. DNA methylation and regulation of gene expression: Guardian of our health. Nucleus 2021, 64, 259–270. [Google Scholar] [CrossRef]
- Hari Gopal, S.; Alenghat, T.; Pammi, M. Early life epigenetics and childhood outcomes: A scoping review. Pediatr. Res. 2025, 97, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Sammallahti, S.; Koopman-Verhoeff, M.E.; Binter, A.C.; Mulder, R.H.; Cabré-Riera, A.; Kvist, T.; Malmberg, A.L.K.; Pesce, G.; Plancoulaine, S.; Heiss, J.A.; et al. Longitudinal associations of DNA methylation and sleep in children: A meta-analysis. Clin. Epigenetics 2022, 14, 83. [Google Scholar] [CrossRef]
- Richter, E.; Patel, P.; Babu, J.R.; Wang, X.; Geetha, T. The importance of sleep in overcoming childhood obesity and reshaping epigenetics. Biomedicines 2024, 12, 1334. [Google Scholar] [CrossRef]
- Rijo-Ferreira, F.; Takahashi, J.S. Genomics of circadian rhythms in health and disease. Genome Med. 2019, 11, 82. [Google Scholar] [CrossRef]
- Van Drunen, R.; Eckel-Mahan, K. Circadian rhythms as modulators of brain health during development and throughout aging. Front. Neural Circuits 2023, 16, 1059229. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.S.; Shin, W.J.; Lee, J.E.; Do, J.T. CpG and Non-CpG methylation in epigenetic gene regulation and brain function. Genes 2017, 8, 148. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Selvaraju, V.; Jeganathan, R.; Wang, X.; Geetha, T. Novel differentially methylated regions identified by genome-wide DNA methylation analyses contribute to racial disparities in childhood obesity. Genes 2023, 14, 1098. [Google Scholar] [CrossRef]
- Mo, W.; Zhang, J.T. Human ABCG2: Structure, function, and its role in multidrug resistance. Int. J. Biochem. Mol. Biol. 2012, 3, 1–27. [Google Scholar] [PubMed]
- Taryma-Leśniak, O.; Bińkowski, J.; Przybylowicz, P.K.; Sokolowska, K.E.; Borowski, K.; Wojdacz, T.K. Methylation patterns at the adjacent CpG sites within enhancers are a part of cell identity. Epigenetics Chromatin 2024, 17, 30. [Google Scholar] [CrossRef]
- Schulz, J.A.; Hartz, A.M.S.; Bauer, B. ABCB1 and ABCG2 regulation at the blood-brain barrier: Potential new targets to improve brain drug delivery. Pharmacol. Rev. 2023, 75, 815–853. [Google Scholar] [CrossRef] [PubMed]
- Furtado, A.; Duarte, A.C.; Costa, A.R.; Gonçalves, I.; Santos, C.R.A.; Gallardo, E.; Quintela, T. Circadian ABCG2 Expression Influences the Brain Uptake of Donepezil across the Blood-Cerebrospinal Fluid Barrier. Int. J. Mol. Sci. 2024, 25, 5014. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Guo, Z.; Xu, J.; Zhou, R.; Shi, B.; Chen, L.; Wu, C.; Wang, H.; Wang, X.; Wang, F.; et al. Regulation of sleep amount by CRTC1 via transcription of Crh in mice. J. Neurosci. 2025, 45, e0786242024. [Google Scholar] [CrossRef]
- Lee, H.C.; Simon, G.M.; Cravatt, B.F. ABHD4 regulates multiple classes of N-acyl phospholipids in the mammalian central nervous system. Biochemistry 2015, 54, 2539–2549. [Google Scholar] [CrossRef]
- Fujisawa, K. Regulation of Adenine Nucleotide Metabolism by Adenylate Kinase Isozymes: Physiological Roles and Diseases. Int. J. Mol. Sci. 2023, 24, 5561. [Google Scholar] [CrossRef]
- Fu, M.; Hu, Y.; Lan TGuan, K.L.; Luo, T.; Luo, M. The Hippo signalling pathway and its implications in human health and diseases. Sig. Transduct. Target. Ther. 2022, 7, 376, Correction in Sig. Transduct. Target. Ther. 2024, 9, 5. https://doi.org/10.1038/s41392-023-01682-3. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Reyes-Vigil, F.; Campo, M.; Brusés, J.L. Classical cadherins evolutionary constraints in primates is associated with their expression in the central nervous system. PLoS ONE 2024, 19, e0313428. [Google Scholar] [CrossRef] [PubMed]
- Lo, N.; Rageul, J.; Kim, H. Roles of SDE2 and TIMELESS at active and stalled DNA replication forks. Mol. Cell. Oncol. 2020, 8, 1855053. [Google Scholar] [CrossRef] [PubMed]
- Engel, C.; Valence, S.; Delplancq, G.; Maroofian, R.; Accogli, A.; Agolini, E.; Alkuraya, F.S.; Baglioni, V.; Bagnasco, I.; Becmeur-Lefebvre, M.; et al. BRAT1–related disorders: Phenotypic spectrum and phenotype-genotype correlations from 97 patients. Eur. J. Hum. Genet. 2023, 31, 1023–1031. [Google Scholar] [CrossRef]
- Li, Y.; Mo, Y.; Chen, C.; He, J.; Guo, Z. Research advances of polycomb group proteins in regulating mammalian development. Front. Cell Dev. Biol. 2024, 12, 1383200. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Li, Y.; Liao, X.; Tian, D.; Xu, Y.; Zhou, C.; Liu, J.; Li, S.; Zhou, J.; Nie, Y.; et al. FTO: A critical role in obesity and obesity-related diseases. Br. J. Nutr. 2023, 130, 1657–1664. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Hao, Y.; Li, X.; Wang, X.; Ji, B.; Wu, Y. HDAC4 in ischemic stroke: Mechanisms and therapeutic potential. Clin. Epigenetics 2018, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Qing, L.; Gao, C.; Ji, A.; Lü, X.; Zhou, L.; Nie, S. Association of mineralocorticoid receptor gene (NR3C2) hypermethylation in adult males with aggressive behavior. Behav. Brain Res. 2021, 398, 112980. [Google Scholar] [CrossRef] [PubMed]
- Plieger, T.; Felten, A.; Splittgerber, H.; Duke, É.; Reuter, M. The role of genetic variation in the glucocorticoid receptor (NR3C1) and mineralocorticoid receptor (NR3C2) in the association between cortisol response and cognition under acute stress. Psychoneuroendocrinology 2018, 87, 173–180. [Google Scholar] [CrossRef]
- Klok, M.D.; Giltay, E.J.; Van der Does, A.J.W.; Geleijnse, J.M.; Antypa, N.; Penninx, B.W.J.H.; de Geus, E.J.C.; Willemsen, G.; Boomsma, D.I.; van Leeuwen, N.; et al. A common and functional mineralocorticoid receptor haplotype enhances optimism and protects against depression in females. Transl. Psychiatry 2011, 1, e62. [Google Scholar] [CrossRef]
- Sundby, L.J.; Southern, W.M.; Hawbaker, K.M.; Trujillo, J.M.; Perrin, B.J.; Ervasti, J.M. Nucleotide- and Protein-Dependent Functions of Actg1. Mol. Biol. Cell 2022, 33, ar77. [Google Scholar] [CrossRef]
- Tabbarah, S.; Tavares, E.; Charish, J.; Vincent, A.; Paterson, A.; Di Scipio, M.; Yin, Y.; Mendoza-Londono, R.; Maynes, J.; Heon, E.; et al. COG5 variants lead to complex early onset retinal degeneration, upregulation of PERK and DNA damage. Sci. Rep. 2020, 10, 21269. [Google Scholar] [CrossRef] [PubMed]
- Konki, M.; Pasumarthy, K.; Malonzo, M.; Sainio, A.; Valensisi, C.; Söderström, M.; Emani, M.R.; Stubb, A.; Närvä, E.; Ghimire, B.; et al. Epigenetic Silencing of the Key Antioxidant Enzyme Catalase in Karyotypically Abnormal Human Pluripotent Stem Cells. Sci. Rep. 2016, 6, 22190. [Google Scholar] [CrossRef]
- Hallock, P.T.; Xu, C.F.; Park, T.J.; Neubert, T.A.; Curran, T.; Burden, S.J. Dok-7 regulates neuromuscular synapse formation by recruiting Crk and Crk-L. Genes. Dev. 2010, 24, 2451–2461. [Google Scholar] [CrossRef]
- Sugiyama, S.; Sasaki, T.; Tanaka, H.; Yan, H.; Ikegami, T.; Kanki, H.; Nishiyama, K.; Beck, G.; Gon, Y.; Okazaki, S.; et al. The tight junction protein occludin modulates blood-brain barrier integrity and neurological function after ischemic stroke in mice. Sci. Rep. 2023, 13, 2892. [Google Scholar] [CrossRef]
- Ma, Q.; Oksenberg, J.; Didonna, A. Epigenetic control of ataxin-1 in multiple sclerosis. Ann. Clin. Transl. Neurol. 2022, 9, 1186–1194. [Google Scholar] [CrossRef]
- Chawla, O.; Kundu, K.; Darbari, J.; Gupta, R. “Early to Bed and Early to Rise Makes You Healthy”: Results from A Cross-Sectional Study Among Adolescents. Sleep Vigil. 2025, 9, 27–37. [Google Scholar] [CrossRef]
- Larsen, M.; He, F.; Kawasawa, Y.I.; Berg, A.; Vgontzas, A.N.; Liao, D.; Bixler, E.O.; Fernandez-Mendoza, J. Objective and subjective measures of sleep initiation are differentially associated with DNA methylation in adolescents. Clin. Epigenetics 2023, 15, 136. [Google Scholar] [CrossRef] [PubMed]
- Bigini, E.G.; Chasens, E.R.; Conley, Y.P.; Imes, C.C. DNA methylation changes and improved sleep quality in adults with obstructive sleep apnea and diabetes. BMJ Open Diabetes Res. Care 2019, 7, e000707. [Google Scholar] [CrossRef] [PubMed]
- de Onis, M.; Onyango, A.W.; Borghi, E.; Siyam, A.; Nishida, C.; Siekmann, J. Development of a WHO growth reference for school-aged children and adolescents. Bull. World Health Organ. 2007, 85, 660–667. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; Metzger, D.L.; Daymont, C.; Hadjiyannakis, S.; Rodd, C.J. LMS tables for waist-circumference and waist-height ratio Z-scores in children aged 5-19 y in NHANES III: Association with cardio-metabolic risks. Pediatr. Res. 2015, 78, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Molinari, R.; Bakalli, G.; Guerrier, S.; Miglioli, C.; Orso, S.; Karemera, M.; Scaillet, O. SWAG: A Wrapper Method for Sparse Learning. arXiv 2020, arXiv:2006.12837. [Google Scholar] [CrossRef]
- Rudin, C.; Zhong, C.; Semenova, L.; Seltzer, M.; Parr, R.; Liu, J.; Katta, S.; Donnelly, J.; Chen, H.; Boner, Z. Amazing things come from having many good models. In Proceedings of the International Conference on Machine Learning (ICML), Vienna, Austria, 21–27 July 2024. [Google Scholar]
- Guerrier, S.; Mili, N.; Molinari, R.; Orso, S.; Avella-Medina, M.; Ma, Y. A predictive-based regression algorithm for gene network selection. Front. Genet. 2016, 7, 97. [Google Scholar] [CrossRef][Green Version]
- Mili, N.; Molinari, R.; Ma, Y.; Guerrier, S. P8 Differentiating inflammatory bowel diseases by using genomic data: Dimension of the problem and network organization. Human Genom. 2016, 10 (Suppl. 1), 12. [Google Scholar] [CrossRef]
- Kissel, N.; Mentch, L. Forward stability and model path selection. Stat. Comput. 2024, 34, 82. [Google Scholar] [CrossRef]
- Yu, B.; Kumbier, K. Veridical data science. Proc. Natl. Acad. Sci. USA 2020, 117, 3920–3929. [Google Scholar] [CrossRef] [PubMed]
- Branca, M.; Orso, S.; Molinari, R.C.; Xu, H.; Guerrier, S.; Zhang, Y.; Mili, N. Is nonmetastatic cutaneous melanoma predictable through genomic biomarkers? Melanoma Res. 2018, 28, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Parisi, N.; Janier-Dubry, A.; Ponzetto, E.; Pavlopoulos, C.; Bakalli, G.; Molinari, R.; Guerrier, S.; Mili, N. Non-applicability of validated predictive models for intensive care admission and death of COVID-19 patients in a secondary care hospital in Belgium. medRxiv 2020. [Google Scholar] [CrossRef]
- Miglioli, C.; Bakalli, G.; Orso, S.; Karemera, M.; Molinari, R.; Guerrier, S.; Mili, N. Evidence of antagonistic predictive effects of miRNAs in breast cancer cohorts through data-driven networks. Sci. Rep. 2022, 12, 5166. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, Y.Y.; Nukala, N.C.P.; Molinari, R.; Deshpande, G. A multi-model framework to explore ADHD diagnosis from neuroimaging data. J. Data Sci. 2024, 22, 199–207. [Google Scholar] [CrossRef]
- Fisher, A.; Rudin, C.; Dominici, F. All models are wrong, but many are useful: Learning a variable’s importance by studying an entire class of prediction models simultaneously. J. Mach. Learn. Res. 2019, 20, 1–81. [Google Scholar]
- Moons, K.G.M.; de Groot, J.A.H.; Bouwmeester, W.; Vergouwe, Y.; Mallett, S.; Altman, D.G.; Collins, G.S.; Reitsma, J.B. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: The CHARMS checklist. PLoS Med. 2014, 11, e1001744. [Google Scholar] [CrossRef]
- Pavlou, M.; Ambler, G.; Seaman, S.R.; De Iorio, M.; Omar, R.Z. Review and evaluation of penalised regression methods for risk prediction in low-dimensional data with few events. Stat. Med. 2016, 35, 1159–1177. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]



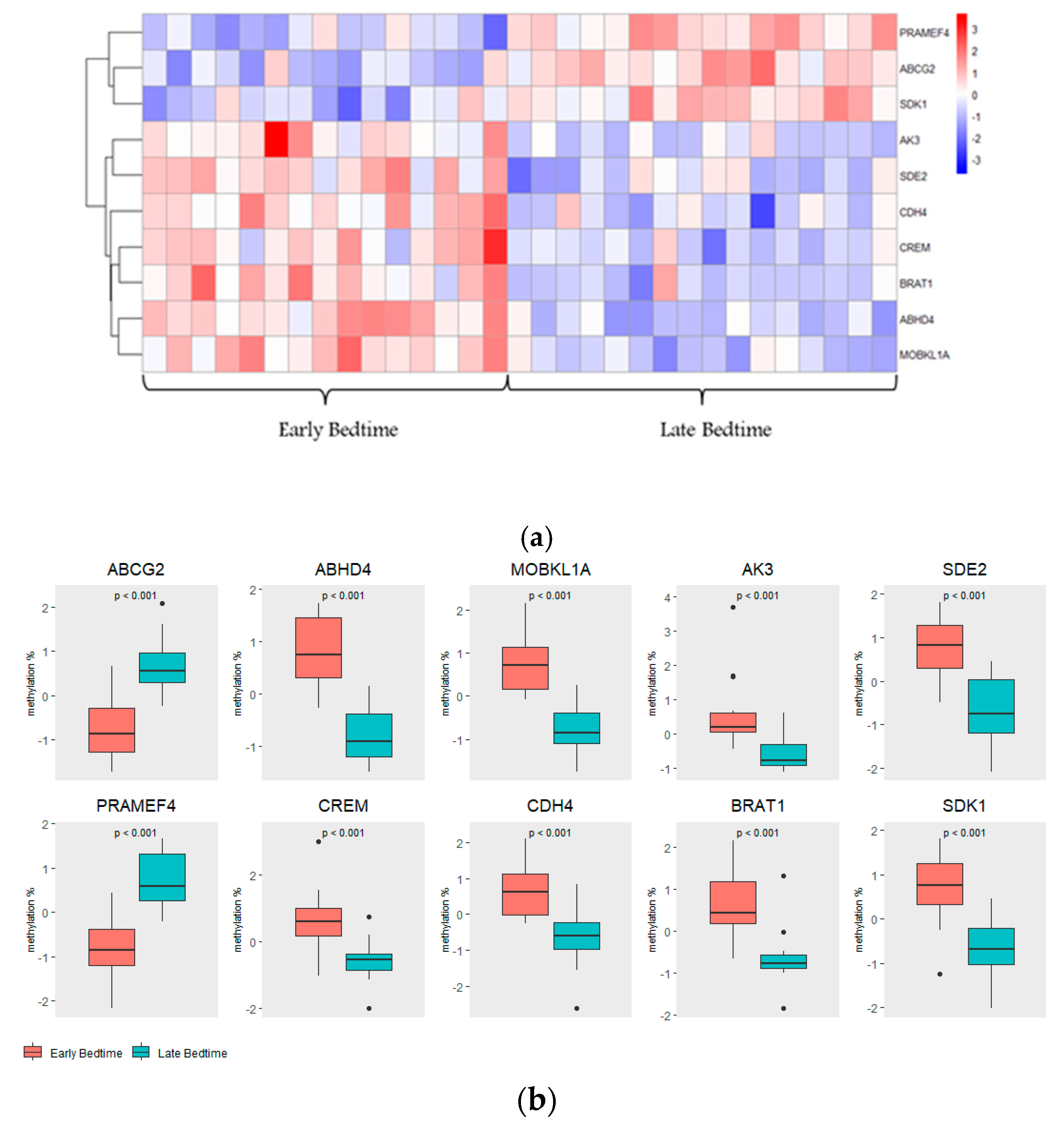
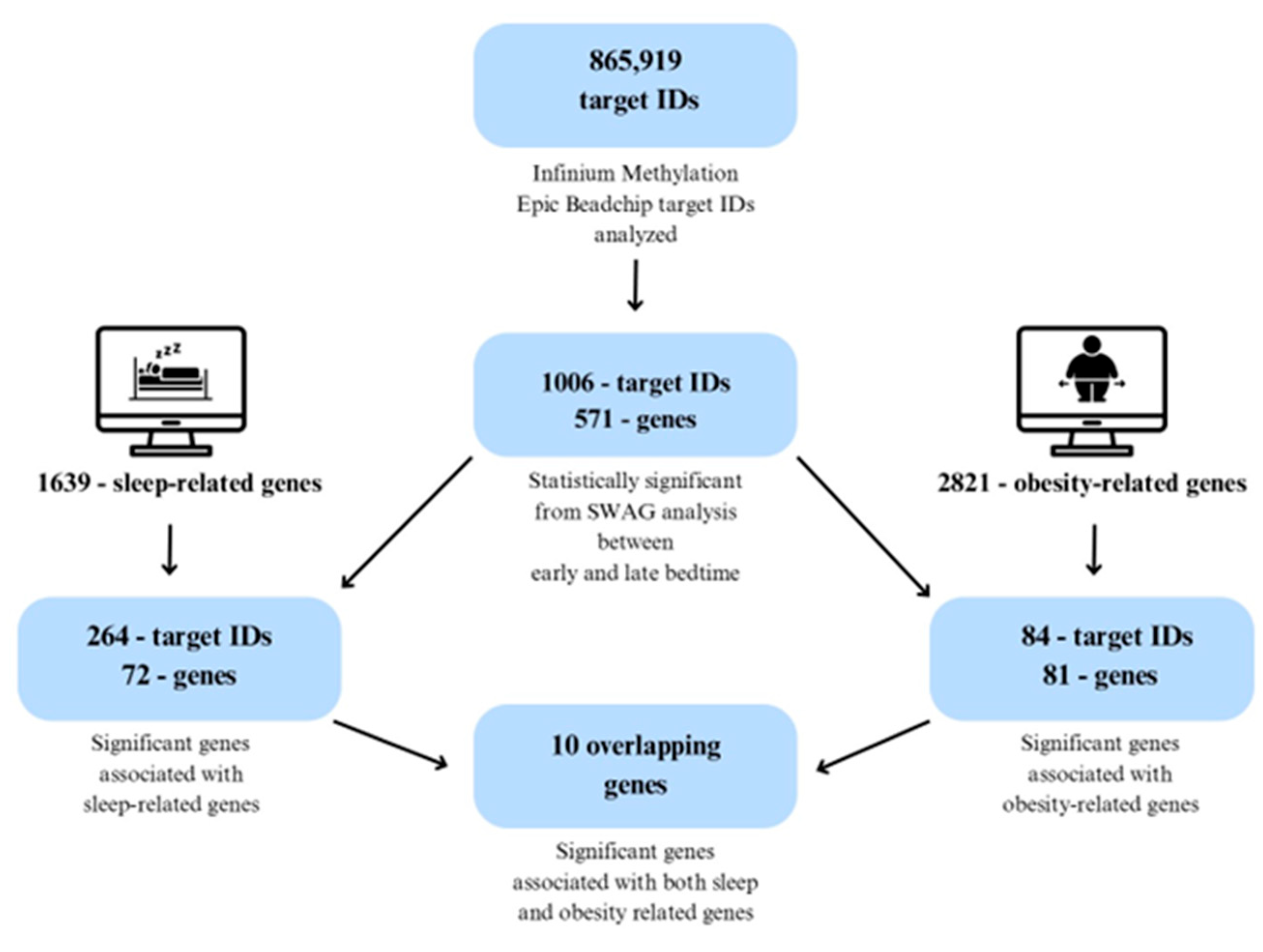
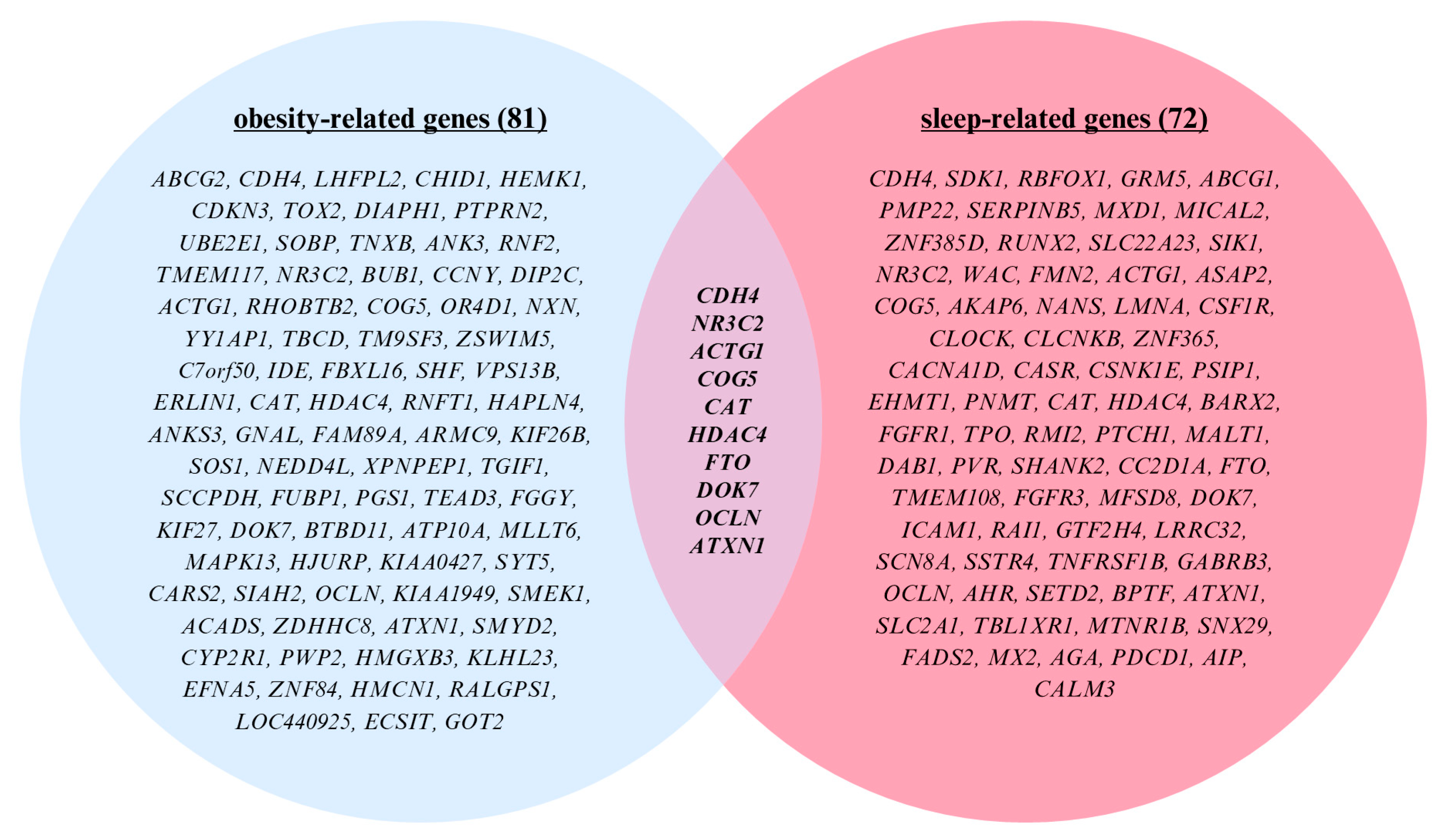
| Parameter | Total | Early Bedtime (Before 8:30 pm) | Late Bedtime (After 8:31 pm) | p-Value |
|---|---|---|---|---|
| Total Participants | 31 | 15 | 16 | - |
| Sex (male/female) | 17/14 | 9/6 | 8/8 | - |
| Age (years) | 8.55 ± 0.24 | 8.22 ± 0.39 | 8.86 ± 0.29 | 0.201 |
| Weight (kg) | 36.05 ± 2.27 | 33.58 ± 3.58 | 38.36 ± 2.84 | 0.305 |
| Height (cm) | 134.35 ± 2.23 | 132.24 ± 3.36 | 136.33 ± 2.97 | 0.369 |
| BMI (kg/m2) | 19.42 ± 0.64 | 18.51 ± 0.98 | 20.28 ± 0.81 | 0.174 |
| BMI z-score | 1.27 ± 022 | 0.91 ± 0.36 | 1.62 ± 0.23 | 0.111 |
| WC z-score | 0.91 ± 0.11 | 0.84 ± 0.18 | 0.97 ± 0.14 | 0.556 |
| WHtR z-score | 0.69 ± 0.12 | 0.63 ± 0.18 | 0.75 ± 0.17 | 0.637 |
| No. | Target ID | GENE | CHR | LOCATION |
|---|---|---|---|---|
| 1 | cg09760986 | ABCG2 | 4 | S_Shore |
| 2 | cg22792063 | ABHD4 | 14 | Island |
| 3 | cg00807892 | MOBKL1A | 4 | Island |
| 4 | cg25282780 | AK3 | 9 | Island |
| 5 | cg09321097 | SDE2 | 1 | Island |
| 6 | cg26811976 | PRAMEF4 | 1 | ~ |
| 7 | cg07891983 | CREM | 10 | ~ |
| 8 | cg04402799 | CDH4 | 20 | ~ |
| 9 | cg03232960 | BRAT1 | 7 | Island |
| 10 | cg00136968 | SDK1 | 7 | ~ |
| Index | Name | p-Value | Genes |
|---|---|---|---|
| 1 | Peptidyl-Lysine Dimethylation (GO:0018027) | 7.4 × 10−5 | SETD2, SETD7, SMYD2, EHMT1 |
| 2 | Amyloid Precursor Protein Catabolic Process (GO:0042987) | 0.00038 | ADAM19, APH1B, ADAM10, ABCG1 |
| 3 | Regulation Of Sodium Ion Transport (GO:0002028) | 0.00042 | NKAIN1, NEDD4L, SIK1, ATP1A1, ANK3, FGF12 |
| 4 | Regulation Of Transforming Growth Factor Beta Activation (GO:1901388) | 0.00074 | TNXB, LRRC32, LTBP1 |
| 5 | Positive Regulation of DNA Repair (GO:0045739) | 0.00099 | PRKCG, SMARCE1, TMEM161A, EYA2, DPF1, RUVBL1, RPS3, FMN2, SMARCA4 |
| 6 | Positive Regulation of Cardiac Muscle Hypertrophy (GO:0010613) | 0.00116 | TRPC3, HAND2, AKAP6, PRKCA |
| 7 | Peptidyl-Lysine Monomethylation (GO:0018026) | 0.00117 | SETD7, SMYD2, EHMT1 |
| 8 | High-Density Lipoprotein Particle Assembly (GO:0034380) | 0.00171 | ZDHHC8, PRKACA, PRKACB |
| 9 | Positive Regulation of Lipase Activity (GO:0060193) | 0.00171 | PDPK1, FGFR3, FGFR1 |
| 10 | Positive Regulation of Protein Sumoylation (GO:0033235) | 0.00171 | HDAC4, PIAS3, RWDD3 |
| Index | Name | p-Value | Genes |
|---|---|---|---|
| 1 | Circadian entrainment | 0.00002046 | PRKCG, KCNJ5, CREB1, MTNR1B, ADCY3, PRKCA, CACNA1D, CALM3, PRKACA, PRKACB, CACNA1H, GNG13 |
| 2 | Glutamatergic synapse | 0.0001022 | PRKCG, GRM5, HOMER2, ADCY3, HOMER3, PRKCA, CACNA1D, PRKACA, SLC1A6, PRKACB, SHANK2, GNG13 |
| 3 | Dopaminergic synapse | 0.0001058 | PRKCG, KCNJ5, PRKCA, CACNA1D, PPP2R5C, GNG13, MAPK1, 3 GNAL, CREB1, CALM3, PRKACA, PRKACB, CLOCK |
| 4 | Aldosterone synthesis and secretion | 0.0001107 | PRKCG, KCNJ5, CREB1, ADCY3, PRKCA, CACNA1D, CALM3, ATP1A1, PRKACA, PRKACB, CACNA1H |
| 5 | GABAergic synapse | 0.0002234 | GABRB3, PRKCG, SLC12A5, GABRA5, ADCY3, PRKCA, CACNA1D, PRKACA, PRKACB, GNG13 |
| 6 | Retrograde endocannabinoid signaling | 0.0003312 | PRKCG, GABRB3, KCNJ5, NDUFA12, GABRA5, ADCY3, PRKCA, CACNA1D, GNG13, MAPK13, GRM5, PRKACA, PRKACB |
| 7 | Aldosterone-regulated sodium reabsorption | 0.0005771 | PRKCG, PDPK1, NEDD4L, PRKCA, ATP1A1, NR3C2 |
| 8 | Growth hormone synthesis, secretion and action | 0.0006086 | MAP2K3, PRKCG, CREB1, IGFBP3, ADCY3, PRKCA, CACNA1D, PRKACA, SOS1, PRKACB, MAPK13 |
| 9 | Insulin secretion | 0.0007717 | PRKCG, CREB, SLC2A1, ADCY3, PRKCA, CACNA1D, ATP1A1, PRKACA, PRKACB |
| 10 | Hedgehog signaling pathway | 0.001028 | EVC, KIF3A, PTCH1, CSNK1E, PRKACA, PRKACB, GLI3 |
| Index | Name | p-Value | Genes |
|---|---|---|---|
| 1 | Neurodevelopmental Disorders | 0.00003113 | GABRB3, RBFOX1, SETD2, ANKRD11, EHMT1, VPS13B, ANK3, SSTR4, RNF2, SMARCA4, RHOBTB2, PARD3B, RAI1, APH1B, TBL1XR1, SCN8A, WAC, SHANK2 |
| 2 | Cognitive delay | 0.00003713 | GABRB3, SETD2, NUP107, NDUFA12, SLC2A1, EHMT1, FMN2, PEPD, SOBP, ACTG1, MFSD8, PMPCA, ACADS, MFSD2A, CEP135, HYMAI, ARMC9, VPS13B, PCCA, TBL1XR1, SCN8A, SIK1, CARS2, FTO, HDAC4, ANKRD11, BRAT1, NEDD4L, CACNA1D, RAI1, TPO, DPH1, LMNA, SLC13A5, RREB1, SEC23B, BUB1, SMARCE1, SLC12A5, SLC35A3, PTCH1, TBCD, SMARCA4, OCLN, OGDH, NF1, TAF6, FGFR3, CC2D1A, FGFR1 |
| 3 | Mental and motor retardation | 0.00003954 | GABRB3, SETD2, NUP107, NDUFA12, SLC2A1, EHMT1, FMN2, PEPD, SOBP, ACTG1, MFSD8, PMPCA, ACADS, MFSD2A, CEP135, HYMAI, ARMC9, VPS13B, PCCA, TBL1XR1, SCN8A, SIK1, CARS2, FTO, HDAC4, ANKRD11, BRAT1, NEDD4L, CACNA1D, RAI1, TPO, DPH1, LMNA, SLC13A5, RREB1, SEC23B, BUB1, BPTF, SMARCE1, SLC12A5, SLC35A3, PTCH1, TBCD, SMARCA4, DIAPH1, OCLN, OGDH, NF1, TAF6, FGFR3, CC2D1A, FGFR1 |
| 4 | Small midface; Decreased projection of midface; Hypotrophic midface; Midface retrusion | 0.00004885 | HDAC4, SF3B4, NXN, PTCH1, AGL, EHMT1, RUNX2, RAI1, TBL1XR1, LMNA, NF1, WAC, SOS, FGFR3, FGFR1 |
| 5 | Small head | 0.0001059 | SF3B4, FTO, HDAC4, RBM28, NUP107, ANKRD11, BRAT1, SLC2A1, EHMT1, ACTG1, RAP1A, AGA, KDSR, SLC13A5, NANS, BUB1, MFSD2A, SMARCE1, ENTPD1, CEP135, SLC35A3, VPS13B, SMARCA4, TUFM, DIAPH1, OCLN, TBL1XR1, SCN8A, NF1, TAF6, B9D2, FGFR3, FGFR1 |
| 6 | Decreased circulating renin level | 0.0001204 | KCNJ5, CYP11B1, CACNA1D, NR3C2 |
| 7 | Triangular head shape; Wedge shaped head | 0.0002024 | DPH1, PTCH1, GLI3, ACTG1, FGFR1 |
| 8 | Global developmental delay | 0.0002664 | GABRB3, SETD2, NUP107, NDUFA12, SLC2A1, EHMT1, FMN2, PEPD, SOBP, ACTG1, MFSD8, PMPCA, ACADS, MFSD2A, CEP135, HYMAI, ARMC9, VPS13B, PCCA, TBL1XR1, SCN8A, CAT, SIK1, CARS2, FTO, HDAC4, ANKRD11, BRAT1, NEDD4L, CACNA1D, RAI1, TPO, DPH1, LMNA, SLC13A5, RREB1, SEC23B, BUB1, SMARCE1, SLC12A5, SLC35A3, PTCH1, TBCD, SMARCA4, OCLN, OGDH, NF1, PMP22, TAF6, FGFR3, CC2D1A, FGFR1 |
| 9 | Triglycerides measurement | 0.0002834 | FTO, ABCC3, STARD13, DNAH17, PINX1, VPS13B, FMN2, AKR1C4, PEPD, INHBC, HAPLN4, TMEM241, AFF1, NR3C2, SUGCT, FADS2, RAP1A, TMEM117, DOK7, PSMD1, CLOCK, ABCG1 |
| 10 | Broad face | 0.0003828 | RAI1, TBL1XR1, PTCH1, AGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richter, E.; Patel, P.; Ozdemir, Y.Y.; Nnyaba, U.V.; Molinari, R.; Babu, J.R.; Geetha, T. Epigenetic Impact of Sleep Timing in Children: Novel DNA Methylation Signatures via SWAG Analysis. Int. J. Mol. Sci. 2025, 26, 10615. https://doi.org/10.3390/ijms262110615
Richter E, Patel P, Ozdemir YY, Nnyaba UV, Molinari R, Babu JR, Geetha T. Epigenetic Impact of Sleep Timing in Children: Novel DNA Methylation Signatures via SWAG Analysis. International Journal of Molecular Sciences. 2025; 26(21):10615. https://doi.org/10.3390/ijms262110615
Chicago/Turabian StyleRichter, Erika, Priyadarshni Patel, Yagmur Y. Ozdemir, Ukamaka V. Nnyaba, Roberto Molinari, Jeganathan R. Babu, and Thangiah Geetha. 2025. "Epigenetic Impact of Sleep Timing in Children: Novel DNA Methylation Signatures via SWAG Analysis" International Journal of Molecular Sciences 26, no. 21: 10615. https://doi.org/10.3390/ijms262110615
APA StyleRichter, E., Patel, P., Ozdemir, Y. Y., Nnyaba, U. V., Molinari, R., Babu, J. R., & Geetha, T. (2025). Epigenetic Impact of Sleep Timing in Children: Novel DNA Methylation Signatures via SWAG Analysis. International Journal of Molecular Sciences, 26(21), 10615. https://doi.org/10.3390/ijms262110615






