Rheumatoid Arthritis: Biomarkers and the Latest Breakthroughs
Abstract
1. Introduction
2. Current Biomarkers in RA
2.1. Protein Biomarkers
2.1.1. Autoantibodies
2.1.2. Inflammatory Biomarkers
2.2. Genetic Biomarkers
2.3. Epigenetic Biomarkers
2.4. Imaging Biomarkers
2.5. Limitations of Classical Biomarkers
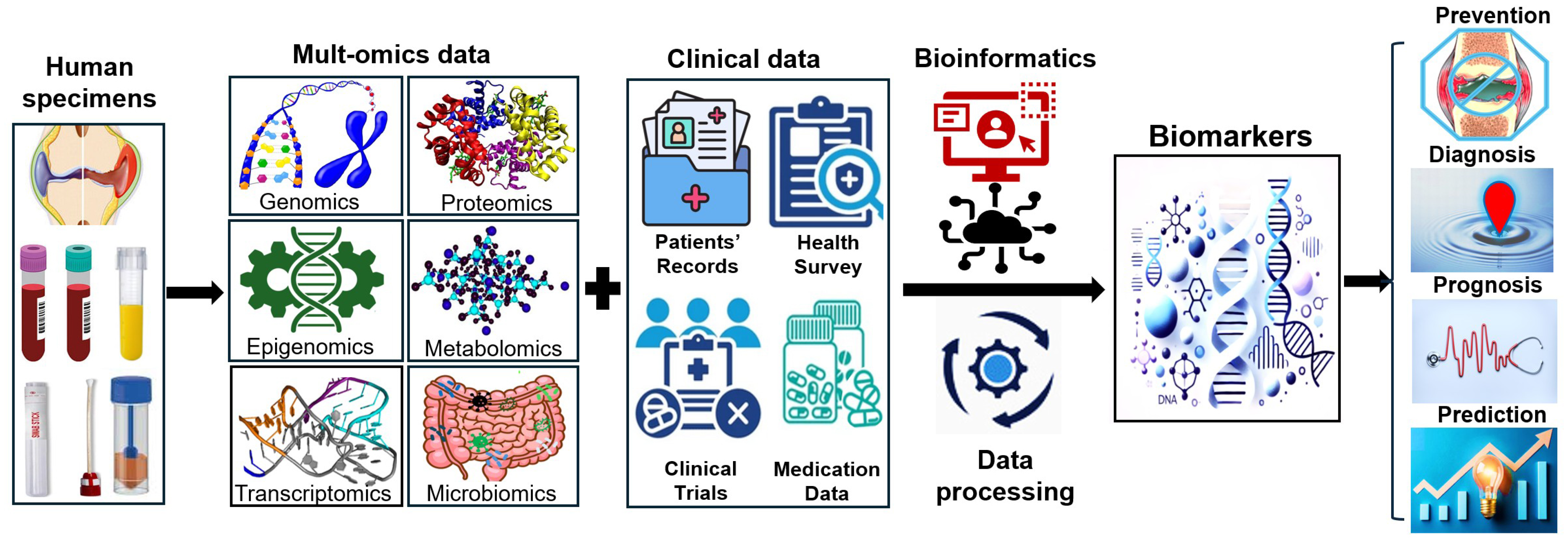
3. Breakthroughs in the Discovery of RA Biomarkers
3.1. Multi-Omics Approaches
3.1.1. Genomics
3.1.2. Epigenomics
3.1.3. Transcriptomics
3.1.4. Proteomics
3.1.5. Metabolomics
3.1.6. Microbiomics
3.1.7. Bioinformatics
3.1.8. Summary
3.1.9. Molecular Signatures and Synovial Biopsy-Based Biomarkers
4. Challenges and Future Directions of Biomarkers in RA
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RA | Rheumatoid arthritis |
| RF | Rheumatoid factor |
| ACPA | Anti-citrullinated protein antibodies |
| cs/bDMARDs | Conventional synthetic/biologic disease-modifying antirheumatic drugs |
| anti-CarP | Anti-carbamylated protein |
| anti-PAD4 | Anti-peptidyl arginine deiminase 4 |
| CRP | C-reactive protein |
| ESR | Erythrocyte sedimentation rate |
| SAA | Serum amyloid A |
| MMP | Matrix metalloproteinase |
| IL | Interleukin |
| TNF | Tumor necrosis factor |
| SNP | Single-nucleotide polymorphism |
| HDAC | Inhibition of histone deacetylase |
| ML | Machine learning |
| NGS | Next-generation sequencing |
| GWAS | Genome-wide association studies |
| SCFA | Short-chain fatty acid |
| VDBP | Vitamin D binding protein |
| IPA | Indole-3-propionate |
References
- van Vollenhoven, R.F. Sex differences in rheumatoid arthritis: More than meets the eye. BMC Med. 2009, 7, 12. [Google Scholar] [CrossRef]
- Almutairi, K.; Nossent, J.; Preen, D.; Keen, H.; Inderjeeth, C. The global prevalence of rheumatoid arthritis: A meta-analysis based on a systematic review. Rheumatol. Int. 2021, 41, 863–877. [Google Scholar] [CrossRef] [PubMed]
- Pongratz, G.; Frieser, R.; Brinks, R.; Schneider, M.; Hartung, W.; Fleck, M.; Ehrenstein, B. Association between autoantibody level and disease activity in rheumatoid arthritis is dependent on baseline inflammation. Clin. Exp. Rheumatol. 2020, 38, 691–698. [Google Scholar]
- Khader, Y.; Beran, A.; Ghazaleh, S.; Lee-Smith, W.; Altorok, N. Predictors of remission in rheumatoid arthritis patients treated with biologics: A systematic review and meta-analysis. Clin. Rheumatol. 2022, 41, 3615–3627. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, H.; Liu, L. Side effects of methotrexate therapy for rheumatoid arthritis: A systematic review. Eur. J. Med. Chem. 2018, 158, 502–516. [Google Scholar] [CrossRef]
- Burmester, G.R.; Pope, J.E. Novel treatment strategies in rheumatoid arthritis. Lancet 2017, 389, 2338–2348. [Google Scholar] [CrossRef]
- Takeuchi, T. Biomarkers as a treatment guide in rheumatoid arthritis. Clin. Immunol. 2018, 186, 59–62. [Google Scholar] [CrossRef]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O., 3rd; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D.; et al. 2010 rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann. Rheum. Dis. 2010, 69, 1580–1588. [Google Scholar] [CrossRef]
- Shi, J.; Knevel, R.; Suwannalai, P.; van der Linden, M.P.; Janssen, G.M.; van Veelen, P.A.; Levarht, N.E.; van der Helm-van Mil, A.H.; Cerami, A.; Huizinga, T.W. Autoantibodies recognizing carbamylated proteins are present in sera of patients with rheumatoid arthritis and predict joint damage. Proc. Natl. Acad. Sci. USA 2011, 108, 17372–17377. [Google Scholar] [CrossRef]
- Darrah, E.; Giles, J.T.; Davis, R.L.; Naik, P.; Wang, H.; Konig, M.F.; Cappelli, L.C.; Bingham, C.O., 3rd; Danoff, S.K.; Andrade, F. Autoantibodies to Peptidylarginine Deiminase 2 Are Associated With Less Severe Disease in Rheumatoid Arthritis. Front. Immunol. 2018, 9, 2696. [Google Scholar] [CrossRef]
- Julià, A.; López-Lasanta, M.; Blanco, F.; Gómez, A.; Haro, I.; Mas, A.J.; Erra, A.; Vivar, M.L.G.; Monfort, J.; Sánchez-Fernández, S.; et al. Interactions between rheumatoid arthritis antibodies are associated with the response to anti-tumor necrosis factor therapy. BMC Musculoskelet. Disord. 2021, 22, 372. [Google Scholar] [CrossRef] [PubMed]
- Petro, A.D.; Dougherty, J.; England, B.R.; Sayles, H.; Duryee, M.J.; Hunter, C.D.; Kremer, J.M.; Pappas, D.A.; Robinson, W.H.; Curtis, J.R.; et al. Associations between an expanded autoantibody profile and treatment responses to biologic therapies in patients with rheumatoid arthritis. Int. Immunopharmacol. 2021, 91, 107260. [Google Scholar] [CrossRef]
- de Moel, E.C.; Derksen, V.; Stoeken, G.; Trouw, L.A.; Bang, H.; Goekoop, R.J.; Speyer, I.; Huizinga, T.W.J.; Allaart, C.F.; Toes, R.E.M.; et al. Baseline autoantibody profile in rheumatoid arthritis is associated with early treatment response but not long-term outcomes. Arthritis Res. Ther. 2018, 20, 33. [Google Scholar] [CrossRef] [PubMed]
- Abdelhafiz, D.; Baker, T.; Glascow, D.A.; Abdelhafiz, A. Biomarkers for the diagnosis and treatment of rheumatoid arthritis—A systematic review. Postgrad. Med. 2023, 135, 214–223. [Google Scholar] [CrossRef]
- Sorić Hosman, I.; Kos, I.; Lamot, L. Serum Amyloid A in Inflammatory Rheumatic Diseases: A Compendious Review of a Renowned Biomarker. Front. Immunol. 2020, 11, 631299. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Bay-Jensen, A.C.; Karsdal, M.A.; Siebuhr, A.S.; Zheng, Q.; Maksymowych, W.P.; Christiansen, T.G.; Henriksen, K. The active form of MMP-3 is a marker of synovial inflammation and cartilage turnover in inflammatory joint diseases. BMC Musculoskelet. Disord. 2014, 15, 93. [Google Scholar] [CrossRef]
- Inciarte-Mundo, J.; Frade-Sosa, B.; Sanmartí, R. From bench to bedside: Calprotectin (S100A8/S100A9) as a biomarker in rheumatoid arthritis. Front. Immunol. 2022, 13, 1001025. [Google Scholar] [CrossRef]
- Li, P.; Wang, X.; Guo, X.; Wen, Y.; Liu, L.; Liang, X.; Du, Y.; Wu, C.; Wang, S.; Zhang, F. Integrative analysis of genome-wide association study and expression quantitative trait loci datasets identified various immune cell-related pathways for rheumatoid arthritis. Ann. Hum. Genet. 2020, 84, 72–79. [Google Scholar] [CrossRef]
- Dedmon, L.E. The genetics of rheumatoid arthritis. Rheumatology 2020, 59, 2661–2670. [Google Scholar] [CrossRef]
- Frisell, T.; Holmqvist, M.; Källberg, H.; Klareskog, L.; Alfredsson, L.; Askling, J. Familial risks and heritability of rheumatoid arthritis: Role of rheumatoid factor/anti-citrullinated protein antibody status, number and type of affected relatives, sex, and age. Arthritis Rheum. 2013, 65, 2773–2782. [Google Scholar] [CrossRef]
- Gregersen, P.K.; Silver, J.; Winchester, R.J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987, 30, 1205–1213. [Google Scholar] [CrossRef]
- Padyukov, L. Genetics of rheumatoid arthritis. Semin. Immunopathol. 2022, 44, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Bossini-Castillo, L.; de Kovel, C.; Kallberg, H.; van’t Slot, R.; Italiaander, A.; Coenen, M.; Tak, P.P.; Posthumus, M.D.; Wijmenga, C.; Huizinga, T.; et al. A genome-wide association study of rheumatoid arthritis without antibodies against citrullinated peptides. Ann. Rheum. Dis. 2015, 74, e15. [Google Scholar] [CrossRef] [PubMed]
- Eyre, S.; Bowes, J.; Diogo, D.; Lee, A.; Barton, A.; Martin, P.; Zhernakova, A.; Stahl, E.; Viatte, S.; McAllister, K.; et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 2012, 44, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Saevarsdottir, S.; Stefansdottir, L.; Sulem, P.; Thorleifsson, G.; Ferkingstad, E.; Rutsdottir, G.; Glintborg, B.; Westerlind, H.; Grondal, G.; Loft, I.C.; et al. Multiomics analysis of rheumatoid arthritis yields sequence variants that have large effects on risk of the seropositive subset. Ann. Rheum. Dis. 2022, 81, 1085–1095. [Google Scholar] [CrossRef]
- Martinez-Molina, C.; Gich, I.; Diaz-Torné, C.; Park, H.S.; Feliu, A.; Vidal, S.; Corominas, H. Patient-related factors influencing the effectiveness and safety of Janus Kinase inhibitors in rheumatoid arthritis: A real-world study. Sci. Rep. 2024, 14, 172. [Google Scholar] [CrossRef]
- Pombo-Suarez, M.; Sanchez-Piedra, C.; Gómez-Reino, J.; Lauper, K.; Mongin, D.; Iannone, F.; Pavelka, K.; Nordström, D.C.; Inanc, N.; Codreanu, C.; et al. After JAK inhibitor failure: To cycle or to switch, that is the question—Data from the JAK-pot collaboration of registries. Ann. Rheum. Dis. 2023, 82, 175–181. [Google Scholar] [CrossRef]
- Ciechomska, M.; Roszkowski, L.; Maslinski, W. DNA Methylation as a Future Therapeutic and Diagnostic Target in Rheumatoid Arthritis. Cells 2019, 8, 953. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, B.; Zhao, G.; Lee, Y.; Buzdin, A.; Mu, X.; Zhao, J.; Chen, H.; Li, X. Spatial transcriptomics: Technologies, applications and experimental considerations. Genomics 2023, 115, 110671. [Google Scholar] [CrossRef]
- Glant, T.T.; Mikecz, K.; Rauch, T.A. Epigenetics in the pathogenesis of rheumatoid arthritis. BMC Med. 2014, 12, 35. [Google Scholar] [CrossRef]
- Shi, Y.; Chang, C.; Xu, L.; Jiang, P.; Wei, K.; Zhao, J.; Xu, L.; Jin, Y.; Zhang, R.; Wang, H.; et al. Circulating DNA methylation level of CXCR5 correlates with inflammation in patients with rheumatoid arthritis. Immun. Inflamm. Dis. 2023, 11, e902. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Xu, L.; Chang, C.; Jiang, P.; Wei, K.; Shi, Y.; Xu, L.; Zheng, Y.; Shan, Y.; Bian, Y.; et al. Circulating methylation level of HTR2A is associated with inflammation and disease activity in rheumatoid arthritis. Front. Immunol. 2022, 13, 1054451. [Google Scholar] [CrossRef] [PubMed]
- Burchill, M.A.; Yang, J.; Vogtenhuber, C.; Blazar, B.R.; Farrar, M.A. IL-2 receptor [beta]-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 2007, 178, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef]
- Araki, Y.; Tsuzuki Wada, T.; Aizaki, Y.; Sato, K.; Yokota, K.; Fujimoto, K.; Kim, Y.T.; Oda, H.; Kurokawa, R.; Mimura, T. Histone Methylation and STAT-3 Differentially Regulate Interleukin-6-Induced Matrix Metalloproteinase Gene Activation in Rheumatoid Arthritis Synovial Fibroblasts. Arthritis Rheumatol. 2016, 68, 1111–1123. [Google Scholar] [CrossRef]
- Pai, P.; Vijeev, A.; Phadke, S.; Shetty, M.G.; Sundara, B.K. Epi-revolution in rheumatology: The potential of histone deacetylase inhibitors for targeted rheumatoid arthritis intervention. Inflammopharmacology 2024, 32, 2109–2123. [Google Scholar] [CrossRef]
- Araki, Y.; Mimura, T. The Histone Modification Code in the Pathogenesis of Autoimmune Diseases. Mediat. Inflamm. 2017, 2017, 2608605. [Google Scholar] [CrossRef]
- Šenolt, L.; Grassi, W.; Szodoray, P. Laboratory biomarkers or imaging in the diagnostics of rheumatoid arthritis? BMC Med. 2014, 12, 49. [Google Scholar] [CrossRef]
- Walter, W.R.; Samim, M. Imaging Updates in Rheumatoid Arthritis. Semin. Musculoskelet. Radiol. 2025, 29, 156–166. [Google Scholar] [CrossRef]
- Baker, J.F.; Conaghan, P.G.; Gandjbakhch, F. Update on magnetic resonance imaging and ultrasound in rheumatoid arthritis. Clin. Exp. Rheumatol. 2018, 36 (Suppl. 114), 16–23. [Google Scholar]
- Silvagni, E.; Zandonella Callegher, S.; Mauric, E.; Chiricolo, S.; Schreiber, N.; Tullio, A.; Zabotti, A.; Scirè, C.A.; Dejaco, C.; Sakellariou, G. Musculoskeletal ultrasound for treating rheumatoid arthritis to target-a systematic literature review. Rheumatology 2022, 61, 4590–4602. [Google Scholar] [CrossRef] [PubMed]
- D’Ignazio, E.; Corradini, D.; Cazenave, T.; Bixio, R.; Baldi, C.; Ubhi, H.K.; Smith, K.; Wakefield, R.J.; Emery, P.; Di Matteo, A. Ultrasound Beyond Joints: A Review of Extra-Articular Applications in Rheumatology. Curr. Rheumatol. Rep. 2025, 27, 20. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.J.; Conaghan, P.G. Imaging outcomes and their role in determining outcomes in osteoarthritis and rheumatoid arthritis. Curr. Opin. Rheumatol. 2006, 18, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Emery, P. Magnetic resonance imaging: Opportunities for rheumatoid arthritis disease assessment and monitoring long-term treatment outcomes. Arthritis Res. 2002, 4 (Suppl. 2), S6–S10. [Google Scholar] [CrossRef]
- Østergaard, M.; Boesen, M. Imaging in rheumatoid arthritis: The role of magnetic resonance imaging and computed tomography. Radiol. Med. 2019, 124, 1128–1141. [Google Scholar] [CrossRef]
- Singh, S.B.; Bhandari, S.; Bhandari, S.; Bhandari, S.; Singh, R.; Raynor, W.Y.; Hess, S.; Werner, T.J.; Alavi, A.; Revheim, M.E. Role of PET/CT in diagnosing and monitoring disease activity in rheumatoid arthritis: A review. Ann. Nucl. Med. 2024, 38, 165–175. [Google Scholar] [CrossRef]
- Diekhoff, T.; Ulas, S.T.; Poddubnyy, D.; Schneider, U.; Hermann, S.; Biesen, R.; Burmester, G.R.; Hamm, B.; Hermann, K.G. Ultra-low-dose CT detects synovitis in patients with suspected rheumatoid arthritis. Ann. Rheum. Dis. 2019, 78, 31–35. [Google Scholar] [CrossRef]
- Minopoulou, I.; Kleyer, A.; Yalcin-Mutlu, M.; Fagni, F.; Kemenes, S.; Schmidkonz, C.; Atzinger, A.; Pachowsky, M.; Engel, K.; Folle, L.; et al. Imaging in inflammatory arthritis: Progress towards precision medicine. Nat. Rev. Rheumatol. 2023, 19, 650–665. [Google Scholar] [CrossRef]
- Lord, M.S.; Farrugia, B.L.; Rnjak-Kovacina, J.; Whitelock, J.M. Current serological possibilities for the diagnosis of arthritis with special focus on proteins and proteoglycans from the extracellular matrix. Expert Rev. Mol. Diagn. 2015, 15, 77–95. [Google Scholar] [CrossRef]
- Curtis, J.R.; Weinblatt, M.E.; Shadick, N.A.; Brahe, C.H.; Østergaard, M.; Hetland, M.L.; Saevarsdottir, S.; Horton, M.; Mabey, B.; Flake, D.D., 2nd; et al. Validation of the adjusted multi-biomarker disease activity score as a prognostic test for radiographic progression in rheumatoid arthritis: A combined analysis of multiple studies. Arthritis Res. Ther. 2021, 23, 1. [Google Scholar] [CrossRef]
- Curtis, J.R.; Strand, V.; Golombek, S.; Zhang, L.; Wong, A.; Zielinski, M.C.; Akmaev, V.R.; Saleh, A.; Asgarian, S.; Withers, J.B. Patient outcomes improve when a molecular signature test guides treatment decision-making in rheumatoid arthritis. Expert Rev. Mol. Diagn. 2022, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Humby, F.; Lewis, M.; Ramamoorthi, N.; Hackney, J.A.; Barnes, M.R.; Bombardieri, M.; Setiadi, A.F.; Kelly, S.; Bene, F.; DiCicco, M.; et al. Synovial cellular and molecular signatures stratify clinical response to csDMARD therapy and predict radiographic progression in early rheumatoid arthritis patients. Ann. Rheum. Dis. 2019, 78, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Orr, C.; Vieira-Sousa, E.; Boyle, D.L.; Buch, M.H.; Buckley, C.D.; Canete, J.D.; Catrina, A.I.; Choy, E.H.S.; Emery, P.; Fearon, U.; et al. Synovial tissue research: A state-of-the-art review. Nat. Rev. Rheumatol. 2017, 13, 463–475, Erratum in Nat. Rev. Rheumatol. 2017, 13, 630; Erratum in Nat. Rev. Rheumatol. 2017, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Small, A.; Wechalekar, M.D. Synovial biopsies in inflammatory arthritis: Precision medicine in rheumatoid arthritis. Expert Rev. Mol. Diagn. 2020, 20, 315–325. [Google Scholar] [CrossRef]
- Triaille, C.; Lauwerys, B.R. Synovial Tissue: Turning the Page to Precision Medicine in Arthritis. Front. Med. 2019, 6, 46. [Google Scholar] [CrossRef]
- Gong, X.; Su, L.; Huang, J.; Liu, J.; Wang, Q.; Luo, X.; Yang, G.; Chi, H. An overview of multi-omics technologies in rheumatoid arthritis: Applications in biomarker and pathway discovery. Front. Immunol. 2024, 15, 1381272. [Google Scholar] [CrossRef]
- Zhao, R.; Zhang, Y.W.; Guo, J.C.; Qiao, J.; Song, S.; Zhang, T.T.; Zhang, H.Y.; Zhang, S.X. Genetic evidence reveals a causal relationship between rheumatoid arthritis and interstitial lung disease. Front. Genet. 2024, 15, 1395315. [Google Scholar] [CrossRef]
- Lo, H.J.; Tsai, C.H.; Huang, T.W. Apoptosis-associated genetic mechanisms in the transition from rheumatoid arthritis to osteoporosis: A bioinformatics and functional analysis approach. APL Bioeng. 2024, 8, 046107. [Google Scholar] [CrossRef]
- Hayashi, S.; Matsubara, T.; Fukuda, K.; Maeda, T.; Funahashi, K.; Hashimoto, M.; Kamenaga, T.; Takashima, Y.; Kuroda, R. A genome-wide association study identifying the SNPs predictive of rapid joint destruction in patients with rheumatoid arthritis. Biomed. Rep. 2021, 14, 31. [Google Scholar] [CrossRef]
- Zhao, R.; Zhang, Y.W.; Yao, J.Y.; Qiao, J.; Song, S.; Zhang, S.X.; Wang, C.H.; Li, X.F. Genetic association between interleukin-17 and susceptibility to rheumatoid arthritis. BMC Med. Genom. 2023, 16, 277. [Google Scholar] [CrossRef]
- Nasef, S.I.; Ellawindy, A.; Askar, A.M.; Hashem, A.A.; Omar, H.H. Assessment of Angiopoietin-2 Single Nucleotide Polymorphism in Patients with Rheumatoid Arthritis. Inflammation 2023, 46, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Leng, R.X.; Di, D.S.; Ni, J.; Wu, X.X.; Zhang, L.L.; Wang, X.F.; Liu, R.S.; Huang, Q.; Fan, Y.G.; Pan, H.F.; et al. Identification of new susceptibility loci associated with rheumatoid arthritis. Ann. Rheum. Dis. 2020, 79, 1565–1571. [Google Scholar] [CrossRef] [PubMed]
- Ishigaki, K.; Sakaue, S.; Terao, C.; Luo, Y.; Sonehara, K.; Yamaguchi, K.; Amariuta, T.; Too, C.L.; Laufer, V.A.; Scott, I.C.; et al. Multi-ancestry genome-wide association analyses identify novel genetic mechanisms in rheumatoid arthritis. Nat. Genet. 2022, 54, 1640–1651. [Google Scholar] [CrossRef]
- Hammaker, D.; Firestein, G.S. Epigenetics of inflammatory arthritis. Curr. Opin. Rheumatol. 2018, 30, 188–196. [Google Scholar] [CrossRef]
- Yang, C.; Li, D.; Teng, D.; Zhou, Y.; Zhang, L.; Zhong, Z.; Yang, G.J. Epigenetic Regulation in the Pathogenesis of Rheumatoid Arthritis. Front. Immunol. 2022, 13, 859400. [Google Scholar] [CrossRef]
- Prideaux, E.B.; Boyle, D.L.; Choi, E.; Buckner, J.H.; Robinson, W.H.; Holers, V.M.; Deane, K.D.; Firestein, G.S.; Wang, W. Epigenetic trajectory predicts development of clinical rheumatoid arthritis in ACPA+ individuals: Targeting Immune Responses for Prevention of Rheumatoid Arthritis (TIP-RA). bioRxiv 2025. [Google Scholar] [CrossRef]
- Hageman, I.; Mol, F.; Atiqi, S.; Joustra, V.; Sengul, H.; Henneman, P.; Visman, I.; Hakvoort, T.; Nurmohamed, M.; Wolbink, G.; et al. Novel DNA methylome biomarkers associated with adalimumab response in rheumatoid arthritis patients. Front. Immunol. 2023, 14, 1303231. [Google Scholar] [CrossRef]
- de la Calle-Fabregat, C.; Niemantsverdriet, E.; Cañete, J.D.; Li, T.; van der Helm-van Mil, A.H.M.; Rodríguez-Ubreva, J.; Ballestar, E. Prediction of the Progression of Undifferentiated Arthritis to Rheumatoid Arthritis Using DNA Methylation Profiling. Arthritis Rheumatol. 2021, 73, 2229–2239. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Q.; Liu, H.; Jin, L.; Feng, X.; Dai, B.; Chen, M.; Xin, F.; Wei, T.; Bai, B.; et al. The prognostic value of whole-genome DNA methylation in response to Leflunomide in patients with Rheumatoid Arthritis. Front. Immunol. 2023, 14, 1173187. [Google Scholar] [CrossRef]
- Feng, X.; Hao, X.; Shi, R.; Xia, Z.; Huang, L.; Yu, Q.; Zhou, F. Detection and Comparative Analysis of Methylomic Biomarkers of Rheumatoid Arthritis. Front. Genet. 2020, 11, 238. [Google Scholar] [CrossRef]
- Zhao, J.; He, B.; Shan, Y.; Wei, K.; Jiang, P.; Shi, Y.; Chang, C.; Zheng, Y.; Zhao, F.; Li, Y.; et al. Identifying AIM2 Circulating Methylation Levels as a Novel Diagnostic Biomarker for Rheumatoid Arthritis Using Targeted DNA Methylation Sequencing. Endocr. Metab. Immune Disord. Drug Targets 2025. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Wei, K.; Xu, L.; Chang, C.; Zhang, R.; Zhao, J.; Jin, Y.; Xu, L.; Shi, Y.; Qian, Y.; et al. DNA methylation change of HIPK3 in Chinese rheumatoid arthritis and its effect on inflammation. Front. Immunol. 2022, 13, 1087279. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Shen, Y.; Chang, C.; Shi, Y.; Wei, K.; Zhao, J.; Shan, Y.; Zheng, Y.; Zhao, F.; Guo, S.; et al. HIPK3 hypomethylation as a potential epigenetic biomarker in rheumatic immune diseases with emphasis on rheumatoid arthritis. Clin. Rheumatol. 2025, 44, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Tessarz, P.; Kouzarides, T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703–708. [Google Scholar] [CrossRef]
- Ntougkos, E.; Chouvardas, P.; Roumelioti, F.; Ospelt, C.; Frank-Bertoncelj, M.; Filer, A.; Buckley, C.D.; Gay, S.; Nikolaou, C.; Kollias, G. Genomic Responses of Mouse Synovial Fibroblasts During Tumor Necrosis Factor-Driven Arthritogenesis Greatly Mimic Those in Human Rheumatoid Arthritis. Arthritis Rheumatol. 2017, 69, 1588–1600. [Google Scholar] [CrossRef]
- Meng, W.; Zhu, Z.; Jiang, X.; Too, C.L.; Uebe, S.; Jagodic, M.; Kockum, I.; Murad, S.; Ferrucci, L.; Alfredsson, L.; et al. DNA methylation mediates genotype and smoking interaction in the development of anti-citrullinated peptide antibody-positive rheumatoid arthritis. Arthritis Res. Ther. 2017, 19, 71. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Campbell, C.; McKenney, P.T.; Konstantinovsky, D.; Isaeva, O.I.; Schizas, M.; Verter, J.; Mai, C.; Jin, W.B.; Guo, C.J.; Violante, S.; et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 2020, 581, 475–479. [Google Scholar] [CrossRef]
- Julià, A.; Gómez, A.; López-Lasanta, M.; Blanco, F.; Erra, A.; Fernández-Nebro, A.; Mas, A.J.; Pérez-García, C.; Vivar, M.L.G.; Sánchez-Fernández, S.; et al. Longitudinal analysis of blood DNA methylation identifies mechanisms of response to tumor necrosis factor inhibitor therapy in rheumatoid arthritis. eBioMedicine 2022, 80, 104053. [Google Scholar] [CrossRef]
- Simard, J.F.; Costenbader, K.H.; Hernán, M.A.; Liang, M.H.; Mittleman, M.A.; Karlson, E.W. Early life factors and adult-onset rheumatoid arthritis. J. Rheumatol. 2010, 37, 32–37. [Google Scholar] [CrossRef]
- Barik, R.R.; Bhatt, L.K. Emerging epigenetic targets in rheumatoid arthritis. Rheumatol. Int. 2021, 41, 2047–2067. [Google Scholar] [CrossRef]
- Chang, C.; Xu, L.; Zhang, R.; Jin, Y.; Jiang, P.; Wei, K.; Xu, L.; Shi, Y.; Zhao, J.; Xiong, M.; et al. MicroRNA-Mediated Epigenetic Regulation of Rheumatoid Arthritis Susceptibility and Pathogenesis. Front. Immunol. 2022, 13, 838884. [Google Scholar] [CrossRef]
- Kang, L.; Dai, C.; Wang, L.; Pan, X. Potential biomarkers that discriminate rheumatoid arthritis and osteoarthritis based on the analysis and validation of datasets. BMC Musculoskelet. Disord. 2022, 23, 319. [Google Scholar] [CrossRef] [PubMed]
- Mun, S.; Lee, J.; Park, A.; Kim, H.J.; Lee, Y.J.; Son, H.; Shin, M.; Lim, M.K.; Kang, H.G. Proteomics Approach for the Discovery of Rheumatoid Arthritis Biomarkers Using Mass Spectrometry. Int. J. Mol. Sci. 2019, 20, 4368. [Google Scholar] [CrossRef] [PubMed]
- Turkkila, M.; Andersson, K.M.; Amu, S.; Brisslert, M.; Erlandsson, M.C.; Silfverswärd, S.; Bokarewa, M.I. Suppressed diversity of survivin splicing in active rheumatoid arthritis. Arthritis Res. Ther. 2015, 17, 175. [Google Scholar] [CrossRef]
- Pathania, A.S. Crosstalk between Noncoding RNAs and the Epigenetics Machinery in Pediatric Tumors and Their Microenvironment. Cancers 2023, 15, 2833. [Google Scholar] [CrossRef]
- Tofigh, R.; Hosseinpourfeizi, M.; Baradaran, B.; Teimourian, S.; Safaralizadeh, R. Rheumatoid arthritis and non-coding RNAs; how to trigger inflammation. Life Sci. 2023, 315, 121367. [Google Scholar] [CrossRef]
- Wang, J.; Yan, S.; Yang, J.; Lu, H.; Xu, D.; Wang, Z. Non-coding RNAs in Rheumatoid Arthritis: From Bench to Bedside. Front. Immunol. 2019, 10, 3129. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, M.; Xie, H.; Hong, F.; Yang, S. Role of miRNAs in Rheumatoid Arthritis Therapy. Cells 2023, 12, 1749. [Google Scholar] [CrossRef]
- Zhao, X.; Lin, W.; Zhou, W. Clinical significance of long non-coding RNA NORAD in rheumatoid arthritis. Adv. Rheumatol. 2024, 64, 9. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhong, Z.; Miao, Q.; Zhang, Y.; Ni, B.; Zhang, M.; Tang, J. circPTPN22 as a novel biomarker and ceRNA in peripheral blood mononuclear cells of rheumatoid arthritis. Mol. Med. Rep. 2021, 24, 617. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.-J.; Zhang, W.-Y.; Fan, X.-X. Circular RNAs: Potential biomarkers and therapeutic targets for autoimmune diseases. Heliyon 2024, 10, e23694. [Google Scholar] [CrossRef] [PubMed]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef]
- Oliver, J.; Nair, N.; Orozco, G.; Smith, S.; Hyrich, K.L.; Morgan, A.; Isaacs, J.; Wilson, A.G.; Barton, A.; Plant, D. Transcriptome-wide study of TNF-inhibitor therapy in rheumatoid arthritis reveals early signature of successful treatment. Arthritis Res. Ther. 2021, 23, 80, Erratum in Arthritis Res. Ther. 2021, 23, 139. [Google Scholar] [CrossRef]
- Al-Amrani, S.; Al-Jabri, Z.; Al-Zaabi, A.; Alshekaili, J.; Al-Khabori, M. Proteomics: Concepts and applications in human medicine. World J. Biol. Chem. 2021, 12, 57–69. [Google Scholar] [CrossRef]
- Song, X.; Lin, Q. Genomics, transcriptomics and proteomics to elucidate the pathogenesis of rheumatoid arthritis. Rheumatol. Int. 2017, 37, 1257–1265. [Google Scholar] [CrossRef]
- Burska, A.; Boissinot, M.; Ponchel, F. Cytokines as biomarkers in rheumatoid arthritis. Mediat. Inflamm. 2014, 2014, 545493. [Google Scholar] [CrossRef]
- Myngbay, A.; Manarbek, L.; Ludbrook, S.; Kunz, J. The Role of Collagen Triple Helix Repeat-Containing 1 Protein (CTHRC1) in Rheumatoid Arthritis. Int. J. Mol. Sci. 2021, 22, 2426. [Google Scholar] [CrossRef]
- Nassef, E.M.; Elabd, H.A.; Elzomor, H.M.; El Nagger, B.; Ibrahim, A.S.; Ibrahim, A.H.; Kotb, H.G.; Hassan, D.A.; Abd ElAziz, R.E.M.; Mohamed, E.E.S. Serum Collagen Triple Helix Repeat Containing-1 Levels are Related to Radiological Affection and Disease Activity in Rheumatoid Arthritis. Open Access Rheumatol. 2022, 14, 291–299. [Google Scholar] [CrossRef]
- Tan, L.; Ouyang, T.; Li, X.; Chen, Y.; Ke, S.; Chen, J.; Liu, Y.; Zeng, F.; Chen, Y.; Long, J.; et al. Serum sirtuin-1 a potential marker in the diagnosis of rheumatoid arthritis. Autoimmunity 2023, 56, 2181234. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Dai, Z.; Xu, J.; Zhao, L.; Xu, Y.; Li, M.; Yu, J.; Zhang, L.; Deng, H.; Liu, L.; et al. Proteome Profiling Identifies Serum Biomarkers in Rheumatoid Arthritis. Front. Immunol. 2022, 13, 865425. [Google Scholar] [CrossRef] [PubMed]
- Ermencheva, P.; Kotov, G.; Shumnalieva, R.; Velikova, T.; Monov, S. Exploring the Role of the Microbiome in Rheumatoid Arthritis—A Critical Review. Microorganisms 2024, 12, 1387. [Google Scholar] [CrossRef] [PubMed]
- Hur, B.; Gupta, V.K.; Huang, H.; Wright, K.A.; Warrington, K.J.; Taneja, V.; Davis, J.M., III; Sung, J. Plasma metabolomic profiling in patients with rheumatoid arthritis identifies biochemical features predictive of quantitative disease activity. Arthritis Res. Ther. 2021, 23, 164. [Google Scholar] [CrossRef]
- Tsetseri, M.-N.; Silman, A.J.; Keene, D.J.; Dakin, S.G. The role of the microbiome in rheumatoid arthritis: A review. Rheumatol. Adv. Pract. 2023, 7, rkad034. [Google Scholar] [CrossRef]
- Zhou, D.; Jiao, W.; Shi, W.; Wang, Q.; Chen, M. Mendelian randomization identifies causal associations between GWAS-associated bacteria and their metabolites and rheumatoid arthritis. Front. Microbiol. 2024, 15, 1431367. [Google Scholar] [CrossRef]
- Yun, H.; Wang, X.; Wei, C.; Liu, Q.; Li, X.; Li, N.; Zhang, G.; Cui, D.; Liu, R. Alterations of the intestinal microbiome and metabolome in women with rheumatoid arthritis. Clin. Exp. Med. 2023, 23, 4695–4706. [Google Scholar] [CrossRef]
- Yang, X.Y.; Zheng, K.D.; Lin, K.; Zheng, G.; Zou, H.; Wang, J.M.; Lin, Y.Y.; Chuka, C.M.; Ge, R.S.; Zhai, W.; et al. Energy Metabolism Disorder as a Contributing Factor of Rheumatoid Arthritis: A Comparative Proteomic and Metabolomic Study. PLoS ONE 2015, 10, e0132695. [Google Scholar] [CrossRef]
- Chu, C.Q. The bacterial link with rheumatoid arthritis. Int. J. Rheum. Dis. 2023, 26, 603–604. [Google Scholar] [CrossRef]
- Dong, Y.; Yao, J.; Deng, Q.; Li, X.; He, Y.; Ren, X.; Zheng, Y.; Song, R.; Zhong, X.; Ma, J. Relationship between gut microbiota and rheumatoid arthritis: A bibliometric analysis. Front. Immunol. 2023, 14, 1131933. [Google Scholar] [CrossRef]
- Luo, Y.; Tong, Y.; Wu, L.; Niu, H.; Li, Y.; Su, L.C.; Wu, Y.; Bozec, A.; Zaiss, M.M.; Qing, P. Alteration of gut microbiota in individuals at high-risk for rheumatoid arthritis associated with disturbed metabolome and the initiation of arthritis through the triggering of mucosal immunity imbalance. Arthritis Rheumatol. 2023, 75, 1736–1748. [Google Scholar] [CrossRef]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013, 2, e01202. [Google Scholar] [CrossRef]
- Maeda, Y.; Kurakawa, T.; Umemoto, E.; Motooka, D.; Ito, Y.; Gotoh, K.; Hirota, K.; Matsushita, M.; Furuta, Y.; Narazaki, M.; et al. Dysbiosis Contributes to Arthritis Development via Activation of Autoreactive T Cells in the Intestine. Arthritis Rheumatol. 2016, 68, 2646–2661. [Google Scholar] [CrossRef]
- Pianta, A.; Arvikar, S.; Strle, K.; Drouin, E.E.; Wang, Q.; Costello, C.E.; Steere, A.C. Evidence of the Immune Relevance of Prevotella copri, a Gut Microbe, in Patients With Rheumatoid Arthritis. Arthritis Rheumatol. 2017, 69, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Ottman, N.; Reunanen, J.; Meijerink, M.; Pietilä, T.E.; Kainulainen, V.; Klievink, J.; Huuskonen, L.; Aalvink, S.; Skurnik, M.; Boeren, S.; et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS ONE 2017, 12, e0173004. [Google Scholar] [CrossRef] [PubMed]
- Stoll, M.L.; Pierce, M.K.; Watkins, J.A.; Zhang, M.; Weiss, P.F.; Weiss, J.E.; Elson, C.O.; Cron, R.Q.; Kumar, R.; Morrow, C.D.; et al. Akkermansia muciniphila is permissive to arthritis in the K/BxN mouse model of arthritis. Genes Immun. 2019, 20, 158–166. [Google Scholar] [CrossRef]
- Romero-Figueroa, M.D.S.; Ramírez-Durán, N.; Montiel-Jarquín, A.J.; Horta-Baas, G. Gut-joint axis: Gut dysbiosis can contribute to the onset of rheumatoid arthritis via multiple pathways. Front. Cell. Infect. Microbiol. 2023, 13, 1092118. [Google Scholar] [CrossRef]
- Lin, L.; Zhang, K.; Xiong, Q.; Zhang, J.; Cai, B.; Huang, Z.; Yang, B.; Wei, B.; Chen, J.; Niu, Q. Gut microbiota in pre-clinical rheumatoid arthritis: From pathogenesis to preventing progression. J. Autoimmun. 2023, 141, 103001. [Google Scholar] [CrossRef]
- Yang, Y.; Hong, Q.; Zhang, X.; Liu, Z. Rheumatoid arthritis and the intestinal microbiome: Probiotics as a potential therapy. Front. Immunol. 2024, 15, 1331486. [Google Scholar] [CrossRef]
- Ge, Y.; Zhou, L.; Chen, Z.; Mao, Y.; Li, T.; Tong, P.; Shan, L. Identification of differentially expressed genes, signaling pathways and immune infiltration in rheumatoid arthritis by integrated bioinformatics analysis. Hereditas 2021, 158, 5. [Google Scholar] [CrossRef]
- Zhang, R.; Zhou, X.; Jin, Y.; Chang, C.; Wang, R.; Liu, J.; Fan, J.; He, D. Identification of differential key biomarkers in the synovial tissue between rheumatoid arthritis and osteoarthritis using bioinformatics analysis. Clin. Rheumatol. 2021, 40, 5103–5110. [Google Scholar] [CrossRef]
- Chen, H.; Xu, J.; Wei, S.; Jia, Z.; Sun, C.; Kang, J.; Guo, X.; Zhang, N.; Tao, J.; Dong, Y.; et al. RABC: Rheumatoid Arthritis Bioinformatics Center. Nucleic Acids Res. 2023, 51, D1381–D1387. [Google Scholar] [CrossRef]
- Sun, F.; Zhou, J.L.; Peng, P.J.; Qiu, C.; Cao, J.R.; Peng, H. Identification of Disease-Specific Hub Biomarkers and Immune Infiltration in Osteoarthritis and Rheumatoid Arthritis Synovial Tissues by Bioinformatics Analysis. Dis. Markers 2021, 2021, 9911184. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.F.; Fang, R.Y.; Hsieh, H.L.; Chi, P.L.; Lin, C.C.; Hsiao, L.D.; Wu, C.C.; Wang, J.S.; Yang, C.M. Involvement of MAPKs and NF-kappaB in tumor necrosis factor alpha-induced vascular cell adhesion molecule 1 expression in human rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2010, 62, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Aterido, A.; Cañete, J.D.; Tornero, J.; Blanco, F.; Fernández-Gutierrez, B.; Pérez, C.; Alperi-López, M.; Olivè, A.; Corominas, H.; Martínez-Taboada, V.; et al. A Combined Transcriptomic and Genomic Analysis Identifies a Gene Signature Associated With the Response to Anti-TNF Therapy in Rheumatoid Arthritis. Front. Immunol. 2019, 10, 1459. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Ko, E.; Mersha, T.B. A roadmap for multi-omics data integration using deep learning. Brief. Bioinform. 2022, 23, bbab454. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef]
- Tao, W.; Concepcion, A.N.; Vianen, M.; Marijnissen, A.C.A.; Lafeber, F.; Radstake, T.; Pandit, A. Multiomics and Machine Learning Accurately Predict Clinical Response to Adalimumab and Etanercept Therapy in Patients with Rheumatoid Arthritis. Arthritis Rheumatol. 2021, 73, 212–222. [Google Scholar] [CrossRef]
- Sussulini, A.; Xia, J.; Orešič, M. Editorial: Multi-omics: Trends and applications in clinical research. Front. Mol. Biosci. 2022, 9, 994239. [Google Scholar] [CrossRef]
- Westerlind, H.; Maciejewski, M.; Frisell, T.; Jelinsky, S.A.; Ziemek, D.; Askling, J. What Is the Persistence to Methotrexate in Rheumatoid Arthritis, and Does Machine Learning Outperform Hypothesis-Based Approaches to Its Prediction? ACR Open Rheumatol. 2021, 3, 457–463. [Google Scholar] [CrossRef]
- Chang, M.J.; Feng, Q.F.; Hao, J.W.; Zhang, Y.J.; Zhao, R.; Li, N.; Zhao, Y.H.; Han, Z.Y.; He, P.F.; Wang, C.H. Deciphering the molecular landscape of rheumatoid arthritis offers new insights into the stratified treatment for the condition. Front. Immunol. 2024, 15, 1391848. [Google Scholar] [CrossRef]
- Jung, S.M.; Park, K.S.; Kim, K.J. Deep phenotyping of synovial molecular signatures by integrative systems analysis in rheumatoid arthritis. Rheumatology 2021, 60, 3420–3431. [Google Scholar] [CrossRef]
- Hur, B.; Cunningham, K.; Davis, J.; Sung, J. Integrative multi-omic profiling in blood reveals distinct immune and metabolic signatures between ACPA-negative and ACPA-positive rheumatoid arthritis. Front. Immunol. 2025, 16. [Google Scholar] [CrossRef]
- Yoosuf, N.; Maciejewski, M.; Ziemek, D.; Jelinsky, S.A.; Folkersen, L.; Müller, M.; Sahlström, P.; Vivar, N.; Catrina, A.; Berg, L.; et al. Early prediction of clinical response to anti-TNF treatment using multi-omics and machine learning in rheumatoid arthritis. Rheumatology 2022, 61, 1680–1689. [Google Scholar] [CrossRef] [PubMed]
- Kerle, I.A.; Gross, T.; Kögler, A.; Arnold, J.S.; Werner, M.; Eckardt, J.N.; Möhrmann, E.E.; Arlt, M.; Hutter, B.; Hüllein, J.; et al. Translational and clinical comparison of whole genome and transcriptome to panel sequencing in precision oncology. NPJ Precis. Oncol. 2025, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.; Humby, F.; Filer, A.; Ng, N.; Di Cicco, M.; Hands, R.E.; Rocher, V.; Bombardieri, M.; D’Agostino, M.A.; McInnes, I.B.; et al. Ultrasound-guided synovial biopsy: A safe, well-tolerated and reliable technique for obtaining high-quality synovial tissue from both large and small joints in early arthritis patients. Ann. Rheum. Dis. 2015, 74, 611–617. [Google Scholar] [CrossRef]
- Lliso-Ribera, G.; Humby, F.; Lewis, M.; Nerviani, A.; Mauro, D.; Rivellese, F.; Kelly, S.; Hands, R.; Bene, F.; Ramamoorthi, N.; et al. Synovial tissue signatures enhance clinical classification and prognostic/treatment response algorithms in early inflammatory arthritis and predict requirement for subsequent biological therapy: Results from the pathobiology of early arthritis cohort (PEAC). Ann. Rheum. Dis. 2019, 78, 1642–1652. [Google Scholar] [CrossRef]
- Nerviani, A.; Di Cicco, M.; Mahto, A.; Lliso-Ribera, G.; Rivellese, F.; Thorborn, G.; Hands, R.; Bellan, M.; Mauro, D.; Boutet, M.A.; et al. A Pauci-Immune Synovial Pathotype Predicts Inadequate Response to TNFα-Blockade in Rheumatoid Arthritis Patients. Front. Immunol. 2020, 11, 845. [Google Scholar] [CrossRef]
- Humby, F.; Durez, P.; Buch, M.H.; Lewis, M.J.; Rizvi, H.; Rivellese, F.; Nerviani, A.; Giorli, G.; Mahto, A.; Montecucco, C.; et al. Rituximab versus tocilizumab in anti-TNF inadequate responder patients with rheumatoid arthritis (R4RA): 16-week outcomes of a stratified, biopsy-driven, multicentre, open-label, phase 4 randomised controlled trial. Lancet 2021, 397, 305–317. [Google Scholar] [CrossRef]
- Rivellese, F.; Surace, A.E.A.; Goldmann, K.; Sciacca, E.; Çubuk, C.; Giorli, G.; John, C.R.; Nerviani, A.; Fossati-Jimack, L.; Thorborn, G.; et al. Rituximab versus tocilizumab in rheumatoid arthritis: Synovial biopsy-based biomarker analysis of the phase 4 R4RA randomized trial. Nat. Med. 2022, 28, 1256–1268. [Google Scholar] [CrossRef]
- Zhang, F.; Wei, K.; Slowikowski, K.; Fonseka, C.Y.; Rao, D.A.; Kelly, S.; Goodman, S.M.; Tabechian, D.; Hughes, L.B.; Salomon-Escoto, K.; et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 2019, 20, 928–942. [Google Scholar] [CrossRef]
- Zhang, F.; Jonsson, A.H.; Nathan, A.; Millard, N.; Curtis, M.; Xiao, Q.; Gutierrez-Arcelus, M.; Apruzzese, W.; Watts, G.F.M.; Weisenfeld, D.; et al. Deconstruction of rheumatoid arthritis synovium defines inflammatory subtypes. Nature 2023, 623, 616–624. [Google Scholar] [CrossRef]
- Julià, A.; Ávila, G.; Celis, R.; Sanmartí, R.; Ramírez, J.; Marsal, S.; Cañete, J.D. Lower peripheral helper T cell levels in the synovium are associated with a better response to anti-TNF therapy in rheumatoid arthritis. Arthritis Res. Ther. 2020, 22, 196. [Google Scholar] [CrossRef]
- Bo, L.; Jin, X.; Hu, Y.; Yang, R. Role of Liquid Biopsies in Rheumatoid Arthritis. Methods Mol. Biol. 2023, 2695, 237–246. [Google Scholar]
- Zerrouk, N.; Augé, F.; Niarakis, A. Building a modular and multi-cellular virtual twin of the synovial joint in Rheumatoid Arthritis. NPJ Digit. Med. 2024, 7, 379. [Google Scholar] [CrossRef]
| Autoantibodies | Sensitivity | Specificity | Presence in Other Conditions | Clinical Application in RA | Typical Algorithms Used |
|---|---|---|---|---|---|
| Rheumatoid Factor | 60–80% | 70–80% | Healthy individuals, Sjögren’s syndrome, lupus, chronic infections | Diagnostic and prognostic marker, prediction of treatment responses to rituximab, sarilumab, tofacitinib | Ensemble Tree-Based Models, Multivariate Logistic Regression, Feature Importance Algorithms, ACR/EULAR Classification Criteria (2010) |
| Anti-Citrullinated Protein Antibodies | 60–70% | >95% | Rarely in other conditions | Diagnostic and prognostic marker, prediction of treatment responses to rituximab, sarilumab, tofacitinib | Classification Algorithms, Receiver Operating Characteristic (ROC) Analysis, Random Forest, Feedforward Neural Networks, ACR/EULAR Classification Criteria (2010) |
| Anti-Carbamylated Protein Antibodies | 35–50% | ~90% | Rarely in other conditions | Useful for the diagnosis of seronegative RA, associated with a more aggressive disease | Multivariate Logistic Regression, Meta-Analysis. |
| Anti-Peptidyl Arginine Deiminase 4 | 20–30% | >90% | Rarely in other conditions | Associated with a more severe and erosive disease; may predict the need for biologic treatments | ROC Analysis, Correlation Analysis, Simple Classification Algorithms |
| Anti-Sa Antibodies | 20–30% | >95% | Rarely in other conditions | Prediction of disease severity and joint erosions | Multivariate Regression Models, Correlation Analysis |
| Anti-Mutated Citrullinated Vimentin antibodies | 70–80% | 85–95% | Rarely in other conditions | Highly useful for the diagnosis of seronegative RA, associated with disease activity and severity | ROC Analysis, Classification Algorithms |
| Anti-Nuclear Antibodies | 20–30% | 30–50% | Lupus, Sjögren’s syndrome, Scleroderma | The presence may indicate an overlap with another autoimmune disease | Artificial Intelligence /Machine Learning models |
| Anti-Ro/SSA Antibodies | 3–15% | 30–50% | Sjögren’s syndrome, Lupus, Systemic sclerosis | The presence may indicate an overlap with Sjögren’s syndrome | Clustering Algorithms |
| Anti-La/SSB Antibodies | 3–10% | 30–50% | Sjögren’s syndrome, Lupus, Systemic sclerosis | The presence usually indicates an overlap with Sjögren’s syndrome | Clustering Algorithms |
| Inflammatory Biomarkers | Sensitivity | Specificity | Presence in Other Conditions | Clinical Application in RA | Typical Algorithm |
|---|---|---|---|---|---|
| CRP | 40–60% | ~40% | Elevated in a wide range of inflammatory, infectious, and tissue-damaging conditions | A key component of disease activity scores, monitoring disease activity and treatment response | DAS28-CRP, ACR/EULAR Classification Criteria (2010) |
| ESR | 40–60% | ~40% | Elevated in many inflammatory and infectious conditions, and certain cancers | A key component of disease activity scores, monitoring disease activity and treatment response | DAS28-ESR, ACR/EULAR Classification Criteria (2010) |
| SAA | 40–60% | 40–60% | Elevated in various inflammatory conditions | Monitoring disease activity, predicting treatment response | Multi-Biomarker Disease Activity (MBDA) Score |
| MMP-3 | 40–80% | 50–70% | Osteoarthritis, other joint diseases, and lupus | A marker of joint destruction and cartilage breakdown | MBDA Score |
| Calprotectin | 60–80% | ~90% | Inflammatory bowel disease, psoriatic arthritis, and infections | Correlated with disease activity, predicting a poor radiographic outcome | Receiver Operating Characteristic (ROC) Analysis, MBDA Scores |
| 14-3-3η | 60–70% | 80–90% | Osteoporosis, other autoimmune diseases | Early diagnostic and prognostic marker, predicting a more erosive disease | ROC Analysis |
| TNF-α | 40–70% | 40–60% | Various autoimmune /inflammatory diseases | A key therapeutic target, monitoring disease activity | MBDA Score |
| IL-6 | 40–70% | 40–60% | Various autoimmune/inflammatory diseases | A key therapeutic target, monitoring disease activity | MBDA Score |
| Genetic Biomarkers | Sensitivity | Specificity | Presence in Other Conditions | Clinical Application in RA | Typical Algorithm |
|---|---|---|---|---|---|
| HLA-DRB1 | 80–90% | 60–70% | A key shared risk factor for multiple autoimmune diseases | May be used for risk assessment and early diagnosis | Polygenic Risk Scores (PRS) Models |
| PTPN22 | 50–60% | 60–80% | A key shared risk factor for multiple autoimmune diseases | Potential use in predicting disease risk and tailoring treatment | Machine learning (ML) Models (e.g., Logistic Regression, Decision Trees, XGBoost) |
| STAT4 | 50–70% | 40–60% | Associated with a variety of autoimmune and inflammatory conditions | Limited clinical application, could be a potential target for new therapies | PRS / ML Models |
| TRAF1/C5 | 40–60% | 40–60% | Associated with a variety of autoimmune and inflammatory conditions | Not used in clinical practice for diagnosis or prognosis | PRS Models |
| PADI4 | ~70% | 40–60% | Rarely in other conditions | Potential use for predicting a more aggressive and erosive disease | ML Models |
| TNFAIP3 | 40–60% | 40–60% | Associated with numerous other autoimmune conditions | Not used in clinical practice for diagnosis or prognosis | ML Models |
| IL-2RA | 40–50% | 40–50% | Associated with numerous other autoimmune conditions | Not used in clinical practice for diagnosis or prognosis | ML Models |
| CD40 | 40–60% | 40–60% | Associated with numerous other autoimmune conditions | Not used in clinical practice for diagnosis or prognosis | PRS Models |
| CTLA4 | 40–70% | 40–60% | A well-known risk factor for numerous autoimmune diseases | Not used in clinical practice for diagnosis or prognosis | ML Models |
| Metabolite Biomarkers | Sensitivity | Specificity | Presence in Other Conditions | Clinical Application in RA | Typical Algorithm Used |
|---|---|---|---|---|---|
| Glyceric Acid | Limited data available | Limited data available | Glyceric Aciduria, some cardiovascular diseases | A potential marker for disease activity | Machine Learning (ML) Classifiers (e.g., Random Forest, Logistic Regression) |
| Lactic Acid | 30–40% | 30–50% | Elevated in a wide range of conditions | A general marker for increased tissue inflammation | Statistical Analysis (Correlation/Regression), ML |
| 3-Hydroxyisovaleric Acid | Limited data available | Limited data available | Leucine deficiency and other metabolic disorders | Not used clinically for diagnosis or monitoring. | ML Classifiers |
| Angiotensinogen | 40–60% | 40–60% | Elevated in hypertension and metabolic syndrome | A potential diagnostic marker for seronegative RA | Statistical Analysis, ML |
| Serum Amyloid A-4 Protein | Limited data available | Limited data available | Elevated in various inflammatory conditions | A potential prescreening marker when used in combination with other markers | Statistical Analysis, ML |
| Vitamin D-Binding Protein | Limited data available | Limited data available | Liver disease, kidney disease, and sepsis | A component of a multi-biomarker panel for the diagnosis of seronegative RA | Statistical Analysis, ML |
| Retinol-Binding Protein-4 | 40–60% | 40–60% | Metabolic syndrome and cardiovascular diseases | A potential component of a diagnostic panel for seronegative RA | Statistical Analysis, ML |
| Microbiota Biomarkers | Sensitivity | Specificity | Changes in RA | Changes in Other Conditions | Clinical Application for RA | Typical Algorithm Used |
|---|---|---|---|---|---|---|
| Prevotella copri | 70% | ~70% | ↑ | Inflammatory bowel disease, psoriatic arthritis, and other autoimmune diseases | A diagnostic marker for new-onset RA, predict response to MTX therapy | Statistical Analysis, Linear Discriminant Analysis, Effect Size, Machine Learning (ML) Classifiers (Random Forest) |
| Collinsella | 30–50% | 30–50% | ↑ | Psoriasis, ankylosing spondylitis, and other spondyloarthropathies | Associated with high ACPA levels, used to understand pathogenesis | Differential Abundance Analysis (DAA), Correlation Analysis |
| Lactobacillus | Varies | Varies | ↓ | Inflammatory bowel disease, metabolic disorders, allergies, and cardiovascular disease | Can be used for potential probiotic interventions | DAA, Aitchison Distance (Beta-diversity) |
| Bacteroides | 20–50% | 20–50% | ↓ | Obesity, diabetes, and Inflammatory bowel disease | A component of a predictive model, associated with a poorer response to MTX | Statistical Analysis, Regression Models |
| Faecalibacterium | 20–50% | 20–50% | ↓ | Inflammatory bowel diseases and chronic fatigue syndrome | A general marker of dysbiosis, a potential probiotic treatment target | Statistical Analysis, Functional Prediction Tools (e.g., PICRUSt or Tax4Fun) |
| Eggerthellales | 20–50% | 20–50% | ↑ | Some species are associated with gut infections and inflammation | A potential marker for disease severity and a potential probiotic treatment target | DAA, Correlation Analysis |
| Enterococcus | 20–50% | 20–50% | ↓ | A wide range of infections, including urinary tract infections | The general decrease is a marker of dysbiosis | DAA, Principal Coordinate Analysis/Non-metric Multidimensional Scaling |
| Bifidobacterium species | 20–50% | 20–50% | ↓ | Depleted in various inflammatory and metabolic diseases | Monitoring gut health and potential probiotic treatments | Statistical Analysis, Correlation Analysis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, M.; Wang, H.; Campos, F.; Jackson, C.J.; March, L. Rheumatoid Arthritis: Biomarkers and the Latest Breakthroughs. Int. J. Mol. Sci. 2025, 26, 10594. https://doi.org/10.3390/ijms262110594
Xue M, Wang H, Campos F, Jackson CJ, March L. Rheumatoid Arthritis: Biomarkers and the Latest Breakthroughs. International Journal of Molecular Sciences. 2025; 26(21):10594. https://doi.org/10.3390/ijms262110594
Chicago/Turabian StyleXue, Meilang, Hui Wang, Frida Campos, Christopher J. Jackson, and Lyn March. 2025. "Rheumatoid Arthritis: Biomarkers and the Latest Breakthroughs" International Journal of Molecular Sciences 26, no. 21: 10594. https://doi.org/10.3390/ijms262110594
APA StyleXue, M., Wang, H., Campos, F., Jackson, C. J., & March, L. (2025). Rheumatoid Arthritis: Biomarkers and the Latest Breakthroughs. International Journal of Molecular Sciences, 26(21), 10594. https://doi.org/10.3390/ijms262110594








