The Complete Mitochondrial Genome of the Stingless Bee Meliplebeia beccarii (Hymenoptera: Apidae: Meliponini) and Insights into Unusual Gene Rearrangement
Abstract
1. Introduction
2. Results and Discussion
2.1. General Mitogenome Features and Nucleotide Composition
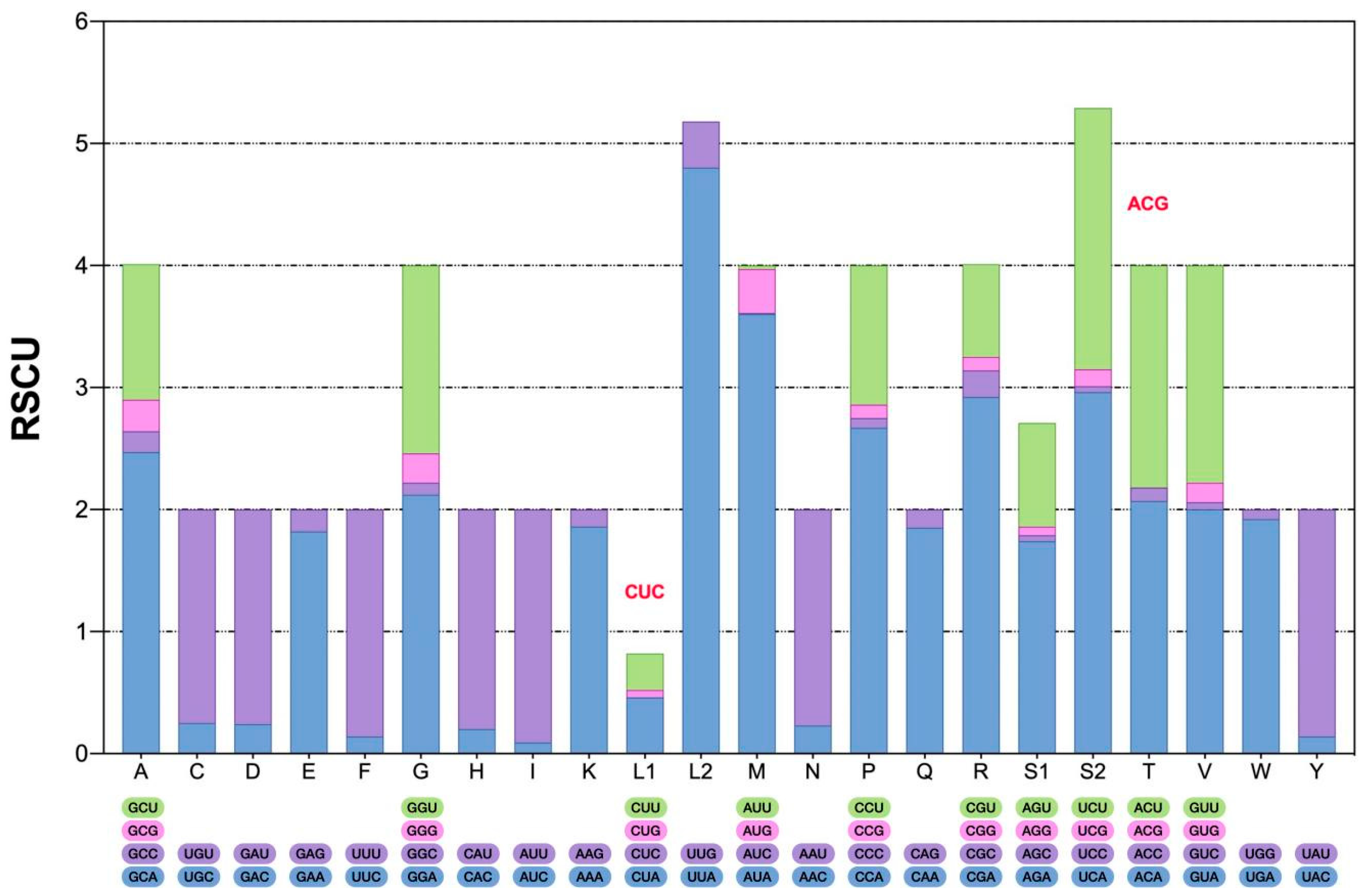
2.2. PCGs and Codon Usage Bias
2.3. Ribosomal and Transfer RNAs
2.4. Phylogenetic Analysis
2.5. Gene Order and Rearrangement
3. Materials and Methods
3.1. Stingless Bee Acquisition and DNA Extraction
3.2. Sequencing and Assembly
3.3. Phylogeny Analysis
3.4. Gene Rearrangement Evaluation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roubik, D.W. Stingless bee (Apidae: Apinae: Meliponini) ecology. Annu. Rev. Entomol. 2023, 68, 231–256. [Google Scholar] [CrossRef]
- Bueno, F.G.B.; Kendall, L.; Alves, D.A.; Tamara, M.L.; Heard, T.; Latty, T.; Gloag, R. Stingless bee floral visitation in the global tropics and subtropics. Glob. Ecol. Conserv. 2023, 43, e02454. [Google Scholar] [CrossRef]
- Witter, S.; Nunes-Silva, P.; Lisboa, B.B.; Tirelli, F.P.; Sattler, A.; Both Hilgert-Moreira, S.; Blochtein, B. Stingless bees as alternative pollinators of canola. J. Econ. Entomol. 2015, 108, 880–886. [Google Scholar] [CrossRef]
- Lourino, M.C.; Fonseca, V.L.I.; Roubik, D.W.; Dollin, A.; Heard, T.; Aguilar, I.B.; Venturieri, G.C.; Eardley, C.; Neto, P.N. Global meliponiculture: Challenges and opportunities. Apidologie 2006, 37, 275–292. [Google Scholar] [CrossRef]
- Rasmussen, C.; Cameron, S.A. A molecular phylogeny of the Old World stingless bees (Hymenoptera: Apidae: Meliponini) and the non-monophyly of the large genus Trigona. Syst. Entomol. 2007, 32, 26–39. [Google Scholar] [CrossRef]
- Iwasaki, J.M.; Hogendoorn, K. Mounting evidence that managed and introduced bees have negative impacts on wild bees: An updated review. Curr. Res. Insect Sci. 2022, 2, 100043. [Google Scholar] [CrossRef]
- Bayeta, A.G.; Hora, Z.A. Evaluation of different hive designs for domestication and conservation of native stingless bee (Apidae: Meliponula beccarii) in Ethiopia. Int. J. Trop. Insect Sc. 2021, 41, 1791–1798. [Google Scholar] [CrossRef]
- Mduda, C.A.; Hussein, J.M.; Muruke, M.H. Discrimination of honeys produced by Tanzanian stingless bees (Hymenoptera, Apidae, Meliponini) based on physicochemical properties and sugar profiles. J. Agric. Food Res. 2023, 14, 100803. [Google Scholar] [CrossRef]
- Kidane, A.A.; Tegegne, F.M.; Tack, A.J.M. Indigenous knowledge of ground-nesting stingless bees in southwestern Ethiopia. Int. J. Trop. Insect Sci. 2021, 41, 2617–2626. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Galtier, N.; Nabholz, B.; Glémin, S.; Hurst, G.D. Mitochondrial DNA as a marker of molecular diversity: A reappraisal. Mol. Ecol. 2009, 18, 4541–4550. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.R.; Wang, Z.W.; Corlett, R.T.; Yu, W.B. Comparative analyses of mitogenomes in the social bees with insights into evolution of long inverted repeats in the Meliponini. Zool. Res. 2024, 45, 160–175. [Google Scholar] [CrossRef] [PubMed]
- Tihelka, E.; Cai, C.; Pisani, D.; Donoghue, P.C. Mitochondrial genomes illuminate the evolutionary history of the Western honey bee (Apis mellifera). Sci. Rep. 2020, 10, 14515. [Google Scholar] [CrossRef]
- Wang, C.Y.; Zhao, M.; Xu, H.L.; Zhang, F.L.; Zhong, Y.H.; Feng, Y.; Wang, S.J. Complete mitochondrial genome of the stingless bee Lepidotrigona terminata (Hymenoptera: Meliponinae) and phylogenetic analysis. Mitochondrial DNA B 2020, 5, 752–753. [Google Scholar] [CrossRef]
- Wang, C.Y.; Zhao, M.; Wang, S.J.; Xu, H.L.; Yang, Y.M.; Liu, L.N.; Feng, Y. The complete mitochondrial genome of Lepidotrigona flavibasis (Hymenoptera: Meliponini) and high gene rearrangement in Lepidotrigona mitogenomes. J. Insect Sci. 2021, 21, 10. [Google Scholar] [CrossRef]
- Wang, C.Y.; Yang, P.L.; Zhao, M.; Xu, H.L.; Liu, L.N.; Feng, Y.; Wang, S.J. Unusual mitochondrial tRNA rearrangements in stingless bee Tetragonula pagdeni and phylogenetic analysis. Entomol. Sci. 2022, 25, e12526. [Google Scholar] [CrossRef]
- Dong, Z.; Wang, Y.; Li, C.; Li, L.; Men, X. Mitochondrial DNA as a molecular marker in insect ecology: Current status and future prospects. Ann. Entomol. Soc. Am. 2021, 114, 470–476. [Google Scholar] [CrossRef]
- Zhang, D.X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Ramirez, S.R.; Roubik, D.W.; Skov, C.; Pierce, N.E. Phylogeny, diversification patterns and historical biogeography of euglossine orchid bees (Hymenoptera: Apidae). Biol. J. Linn. Soc. 2010, 100, 552–572. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic. Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.M.; Salinas-Giegé, T.; Triant, D.A.; Taylor, D.R.; Drouard, L.; Sloan, D.B. Rapid shifts in mitochondrial tRNA import in a plant lineage with extensive mitochondrial tRNA gene loss. Mol. Biol. Evol. 2021, 38, 5735–5751. [Google Scholar] [CrossRef]
- Shao, R.; Dowton, M.; Murrell, A.; Barker, S.C. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol. Biol. Evol. 2003, 20, 1612–1619. [Google Scholar] [CrossRef]
- Boore, J.L.; Collins, T.M.; Stanton, D.; Daehler, L.L.; Brown, W.M. Deducing the pattern of arthropod phylogeny from mitochondrial DNA rearrangements. Nature 1995, 376, 163–165. [Google Scholar] [CrossRef]
- Silvestre, D.; Dowton, M.; Arias, M.C. The mitochondrial genome of the stingless bee Melipona bicolor (Hymenoptera, Apidae, Meliponini): Sequence, gene organization and a unique tRNA translocation event conserved across the tribe Meliponini. Genet. Mol. Biol. 2008, 31, 451–460. [Google Scholar] [CrossRef]
- Moritz, C.; Dowling, T.E.; Brown, W.M. Evolution of animal mitochondrial DNA: Relevance for population biology and systematics. Annu. Rev. Ecol. Syst. 1987, 18, 269–292. [Google Scholar] [CrossRef]
- San Mauro, D.; Gower, D.J.; Zardoya, R.; Wilkinson, M. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol. Biol. Evol. 2006, 23, 227–234. [Google Scholar] [CrossRef]
- Smith, M.J.; Arndt, A.; Gorski, S.; Fajber, E. The phylogeny of echinoderm classes based on mitochondrial gene arrangements. J. Mol. Evol. 1993, 36, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Dowton, M.; Austin, A.D. Evolutionary dynamics of a mitochondrial rearrangement ‘‘hotspot’’ in the Hymenoptera. Mol. Biol. Evol. 1999, 16, 298–309. [Google Scholar] [CrossRef]
- Miller, A.D.; Nguyen, T.T.T.; Burridge, C.P.; Austin, C.M. Complete mitochondrial DNA sequence of the Australian freshwater crayfish, Cherax destructor (Crustacea: Decapoda: Parastacidae): A novel gene order revealed. Gene 2004, 331, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Shao, R.; Barker, S.; Mitani, H.; Takahashi, M.; Fukunaga, M. Molecular mechanisms for the variation of mitochondrial gene content and gene arrangement among chigger mites of the genus Leptotrombidium (Acari: Acariformes). J. Mol. Evol. 2006, 63, 251–261. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, W686–W689. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
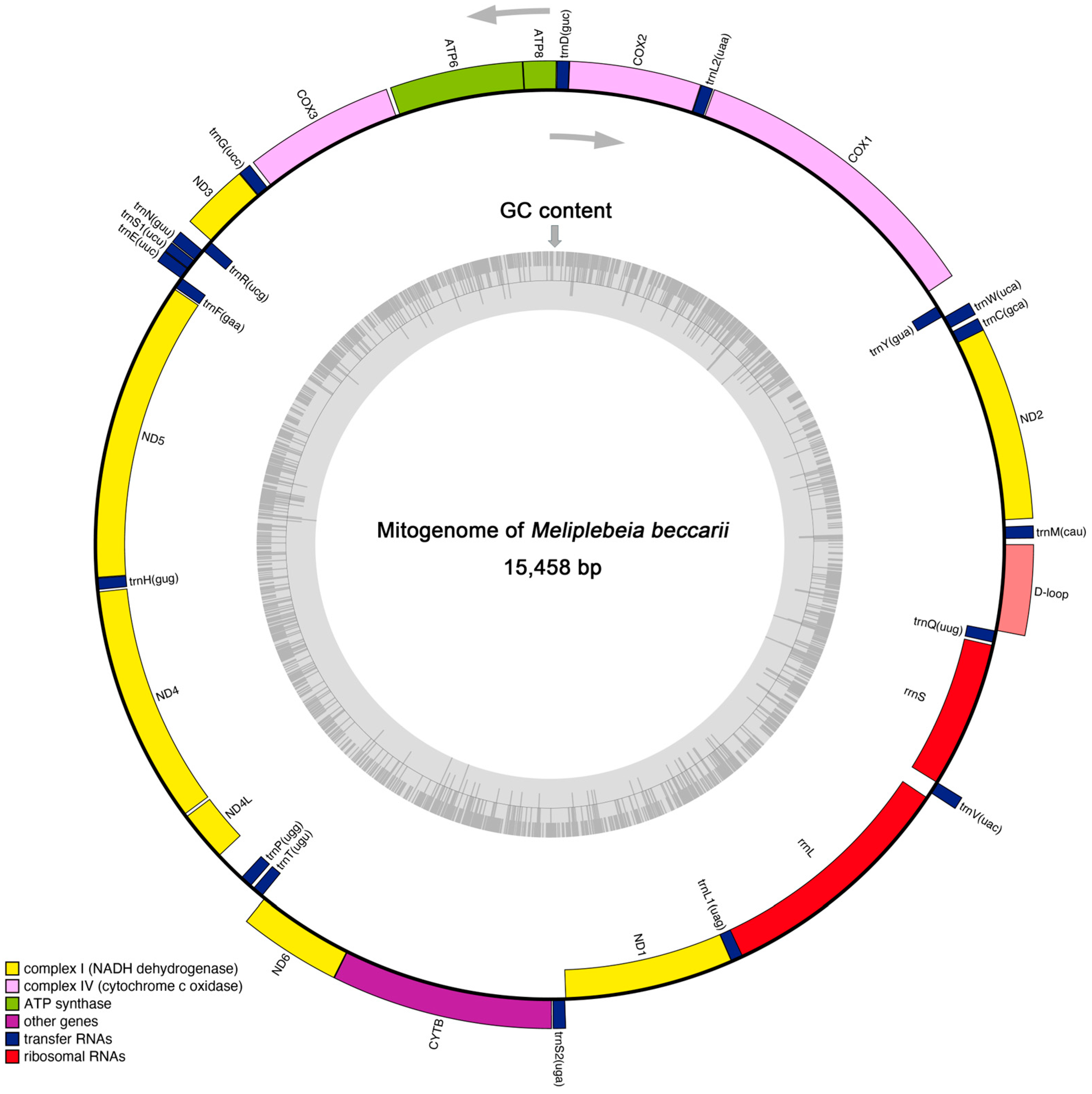



| Gene | Strand | Nucleotide Number | Intergenic Length | Start Codon | Stop Codon | Anticodon |
|---|---|---|---|---|---|---|
| trnM | J | 30–96 | 34 | CAU | ||
| ND2 | J | 131–1147 | −20 | ATT | TAA | |
| trnC | J | 1128–1197 | 20 | GCA | ||
| trnW | J | 1218–1287 | 4 | UCA | ||
| trnY | N | 1292–1358 | 88 | GUA | ||
| COX1 | J | 1447–3012 | 4 | ATC | TAA | |
| trnL2 | J | 3017–3081 | 0 | UAA | ||
| COX2 | J | 3082–3765 | −2 | |||
| trnD | J | 3764–3829 | 0 | GUC | ||
| ATP8 | J | 3830–4003 | −1 | ATT | TAA | |
| ATP6 | J | 4003–4692 | 18 | ATA | TAA | |
| COX3 | J | 4711–5490 | 19 | ATG | TAA | |
| trnG | J | 5510–5577 | 0 | UCC | ||
| ND3 | J | 5578–5931 | 4 | ATA | TAA | |
| trnR | N | 5936–6000 | −1 | UCG | ||
| trnN | J | 6000–6068 | 2 | GUU | ||
| trnS1 | J | 6071–6127 | 0 | UCU | ||
| trnE | J | 6128–6192 | −6 | UUC | ||
| trnF | N | 6187–6251 | 6 | GAA | ||
| ND5 | N | 6258–7910 | 65 | ATT | TAA | |
| trnH | N | 7976–7911 | 74 | GUG | ||
| ND4 | N | 7986–9296 | 13 | ATA | TAA | |
| ND4l | N | 9310–9588 | 166 | ATA | TAA | |
| trnP | N | 9755–9824 | 17 | UGG | ||
| trnT | N | 9842–9906 | 16 | UGU | ||
| ND6 | J | 9923–10,456 | −1 | ATG | TAA | |
| CYTB | J | 10,456–11,604 | 4 | ATG | TAG | |
| trnS2 | J | 11,609–11,676 | 1 | UGA | ||
| ND1 | N | 11,678–12,607 | −3 | ATA | TAA | |
| trnL1 | N | 12,605–12,673 | −23 | UAG | ||
| rrnL | N | 12,651–14,004 | 30 | |||
| trnV | J | 14,035–14,103 | −4 | UAC | ||
| rrnS | N | 14,100–14,907 | 13 | |||
| trnQ | N | 14,921–14,987 | 0 | UUG | ||
| D-loop | - | 14,988–15,458 | 0 |
| Length (bp) | A (bp) | C (bp) | G (bp) | T (bp) | A (%) | C (%) | G (%) | T (%) | A + T (%) | AT-Skew | GC-Skew | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Whole genome | 15,458 | 6408 | 1405 | 1092 | 6553 | 41.5 | 9.1 | 7.1 | 42.4 | 83.9 | −0.011 | −0.125 |
| PCGs 2 | 11,121 | 4538 | 1012 | 843 | 4728 | 40.8 | 9.1 | 7.6 | 42.5 | 83.3 | −0.020 | −0.091 |
| tRNAs | 1268 | 570 | 66 | 90 | 542 | 44.9 | 5.2 | 7.1 | 42.7 | 87.6 | 0.025 | 0.154 |
| rRNAs | 2162 | 876 | 160 | 256 | 870 | 40.5 | 7.4 | 11.8 | 40.2 | 80.7 | 0.003 | 0.231 |
| rrnL (N-strand) | 1354 | 542 | 103 | 150 | 559 | 40.0 | 7.6 | 11.1 | 41.3 | 81.3 | −0.015 | 0.186 |
| rrnS (N-strand) | 808 | 334 | 57 | 106 | 311 | 41.3 | 7.1 | 13.1 | 38.5 | 79.8 | 0.036 | 0.301 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.-J.; Wu, J.; Wubie, A.J.; Wang, C.-Y. The Complete Mitochondrial Genome of the Stingless Bee Meliplebeia beccarii (Hymenoptera: Apidae: Meliponini) and Insights into Unusual Gene Rearrangement. Int. J. Mol. Sci. 2025, 26, 10588. https://doi.org/10.3390/ijms262110588
Wang S-J, Wu J, Wubie AJ, Wang C-Y. The Complete Mitochondrial Genome of the Stingless Bee Meliplebeia beccarii (Hymenoptera: Apidae: Meliponini) and Insights into Unusual Gene Rearrangement. International Journal of Molecular Sciences. 2025; 26(21):10588. https://doi.org/10.3390/ijms262110588
Chicago/Turabian StyleWang, Shi-Jie, Jiao Wu, Abebe Jenberie Wubie, and Cheng-Ye Wang. 2025. "The Complete Mitochondrial Genome of the Stingless Bee Meliplebeia beccarii (Hymenoptera: Apidae: Meliponini) and Insights into Unusual Gene Rearrangement" International Journal of Molecular Sciences 26, no. 21: 10588. https://doi.org/10.3390/ijms262110588
APA StyleWang, S.-J., Wu, J., Wubie, A. J., & Wang, C.-Y. (2025). The Complete Mitochondrial Genome of the Stingless Bee Meliplebeia beccarii (Hymenoptera: Apidae: Meliponini) and Insights into Unusual Gene Rearrangement. International Journal of Molecular Sciences, 26(21), 10588. https://doi.org/10.3390/ijms262110588






