In Silico Identification of the NLRP3 Inhibitors from Traditional Chinese Medicine
Abstract
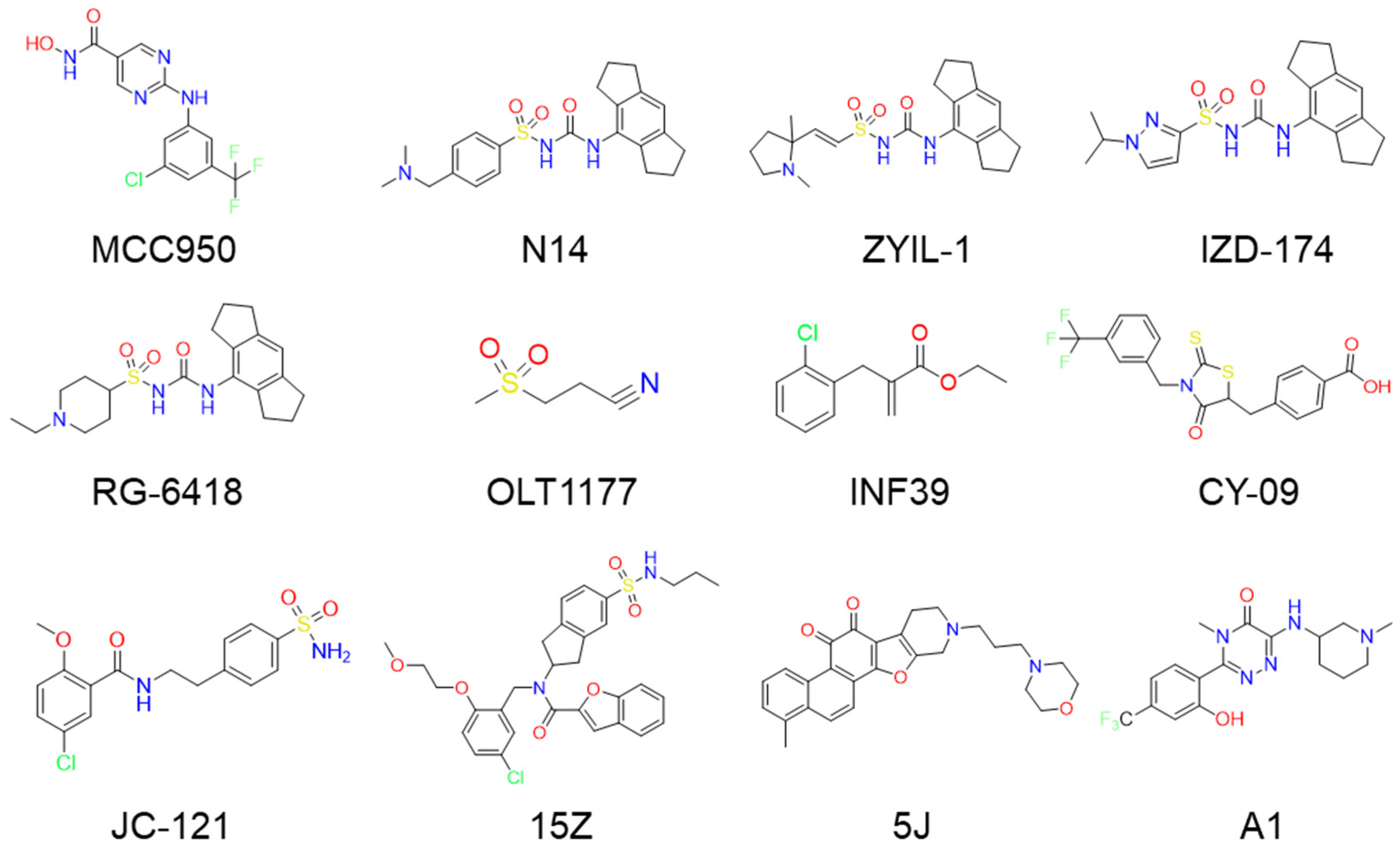
1. Introduction
2. Results
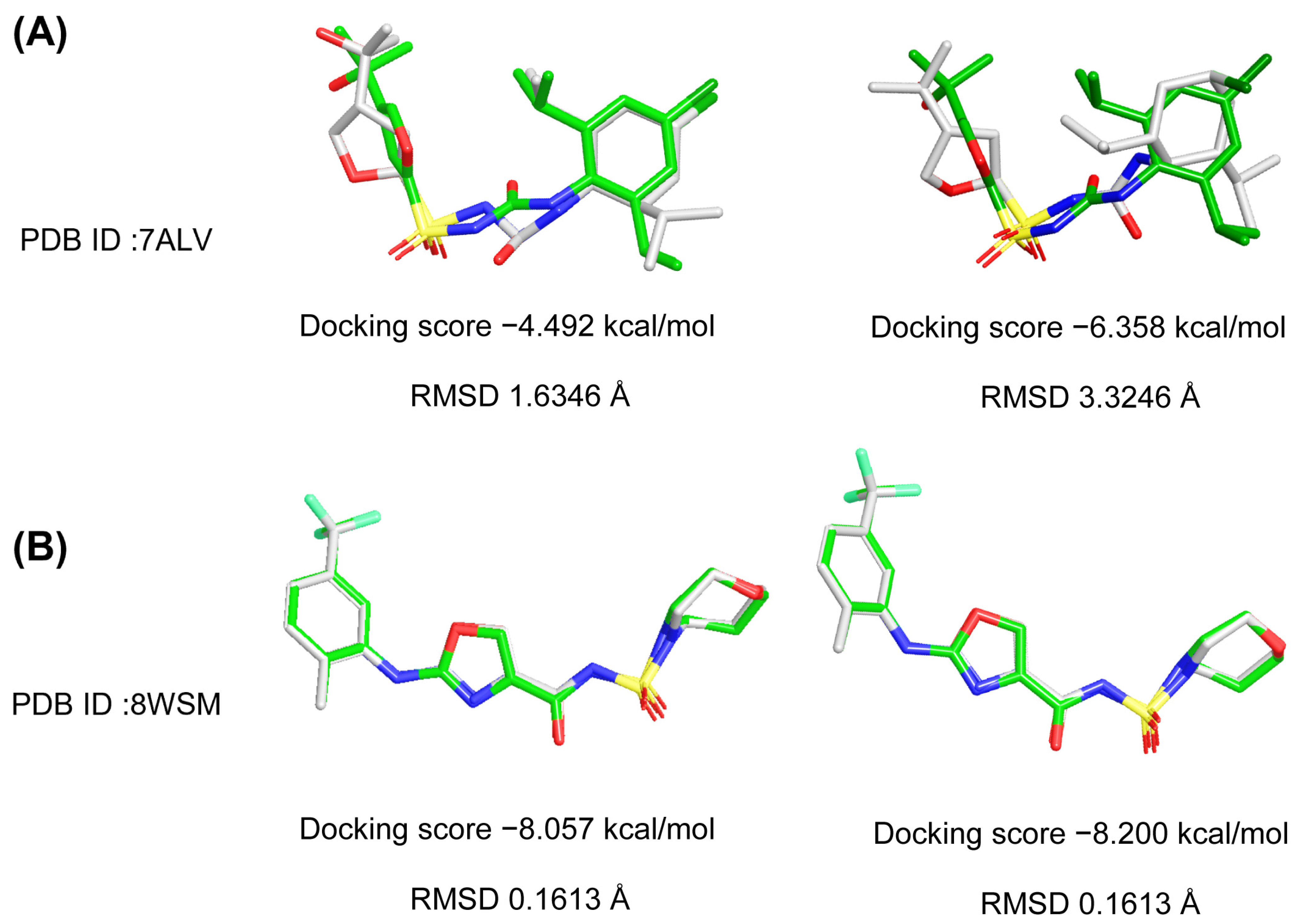
2.1. Glide Docking and Crystal Selection
2.2. Results of Docking-Based Virtual Screening
2.3. Results of Shape-Based Screening
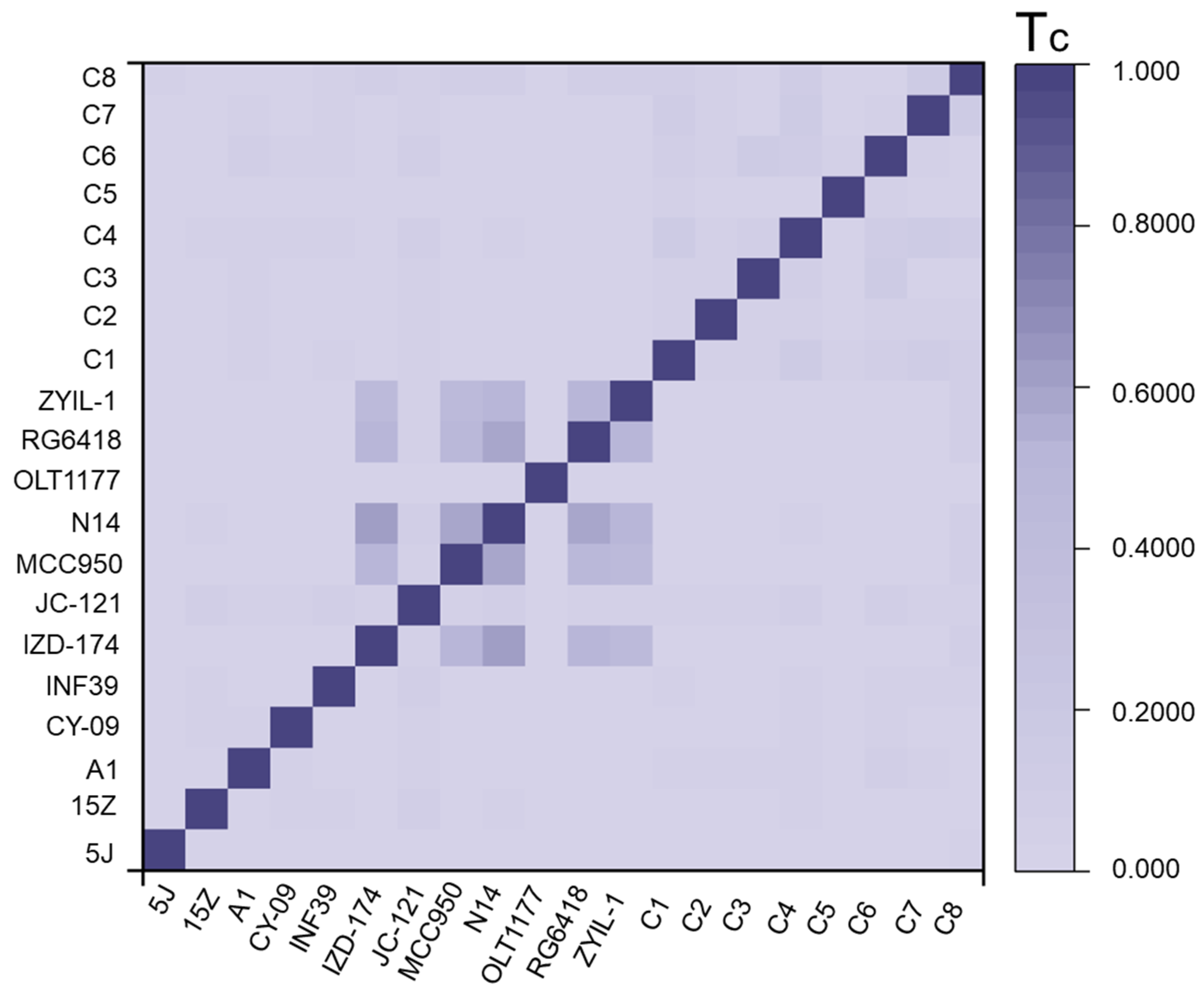
2.4. The Calculation of Molecular Similarity
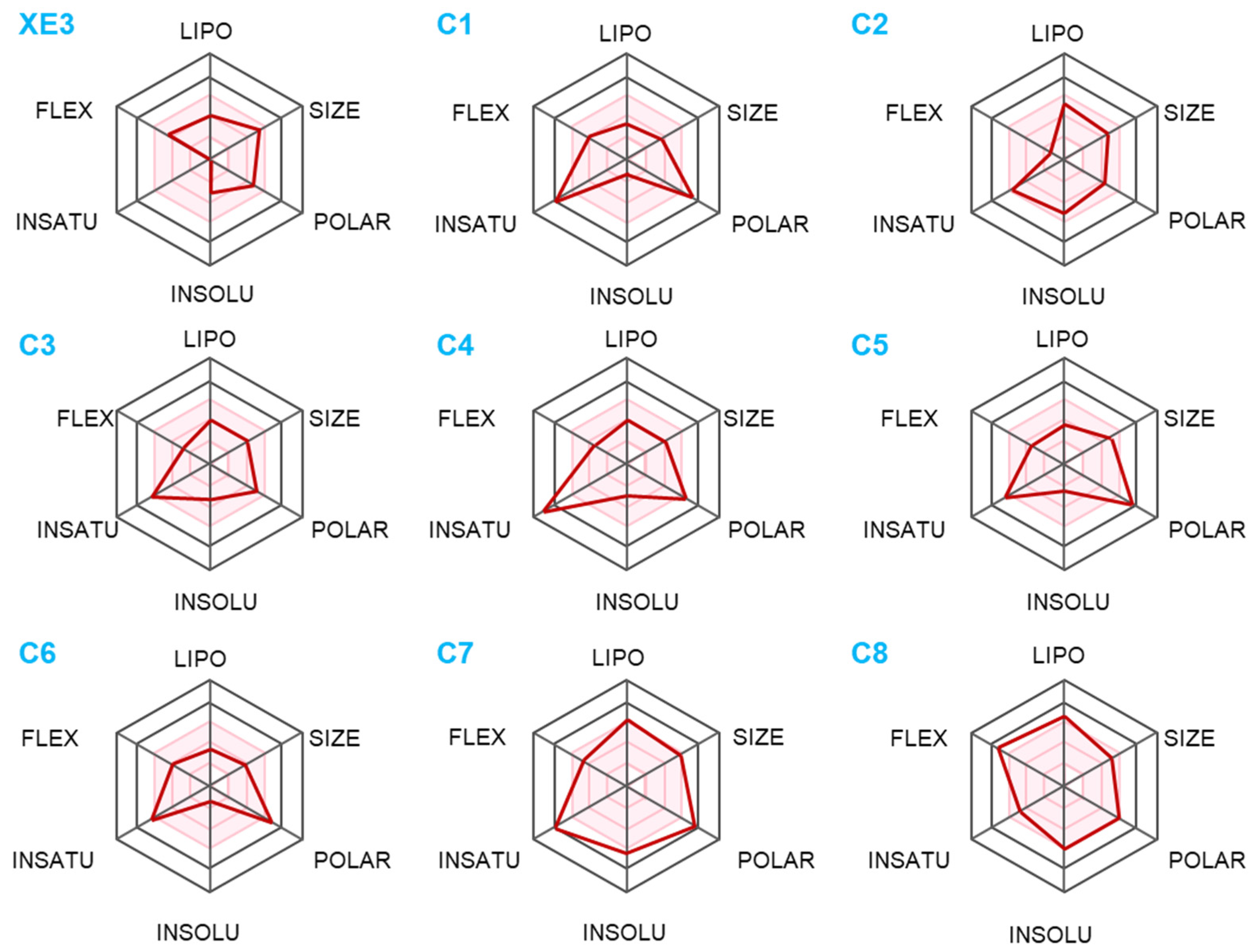
2.5. The Analysis of Pharmacokinetic Properties
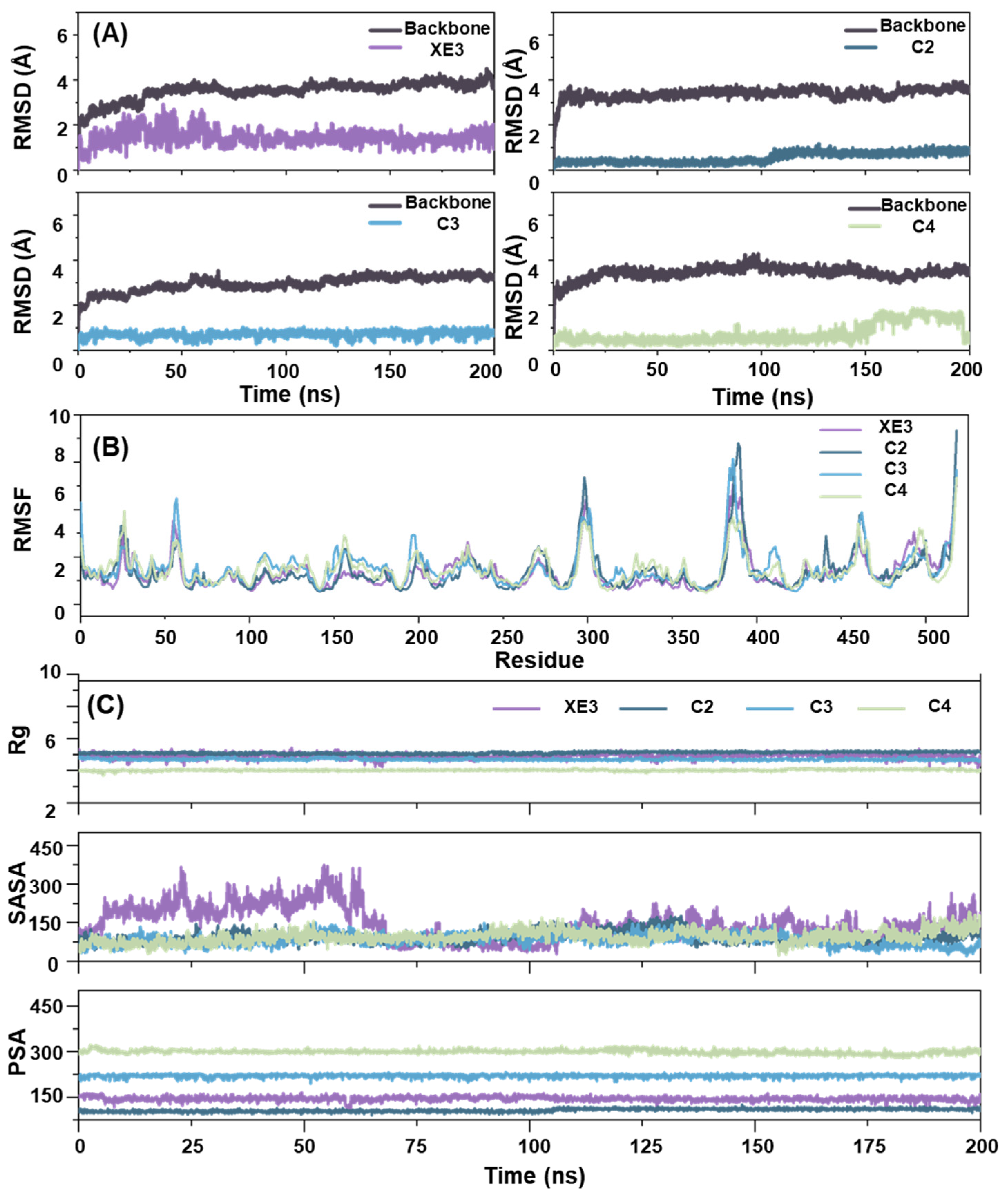
2.6. The Analysis of RMSD, RMSF, Rg, SASA and PSA
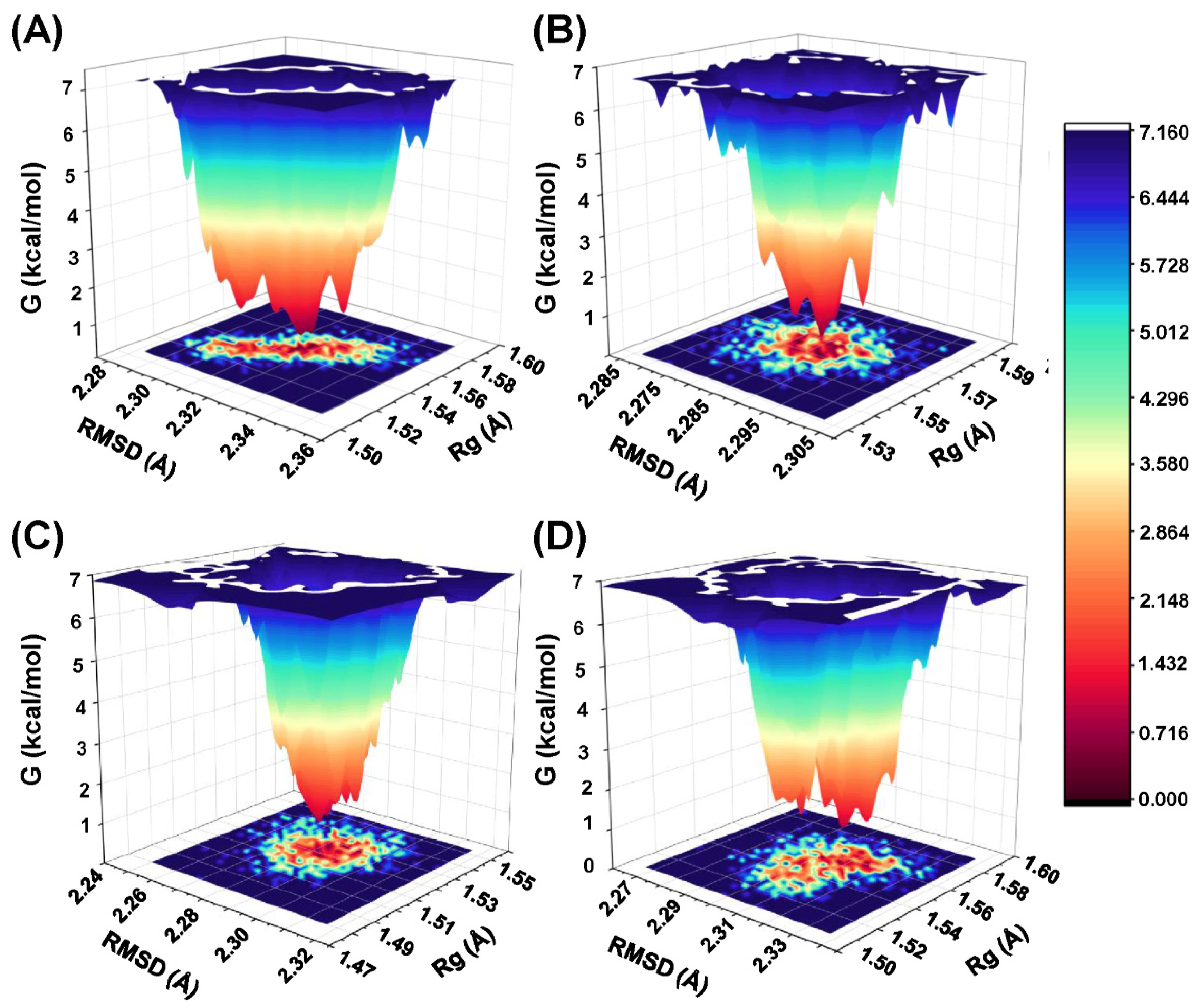
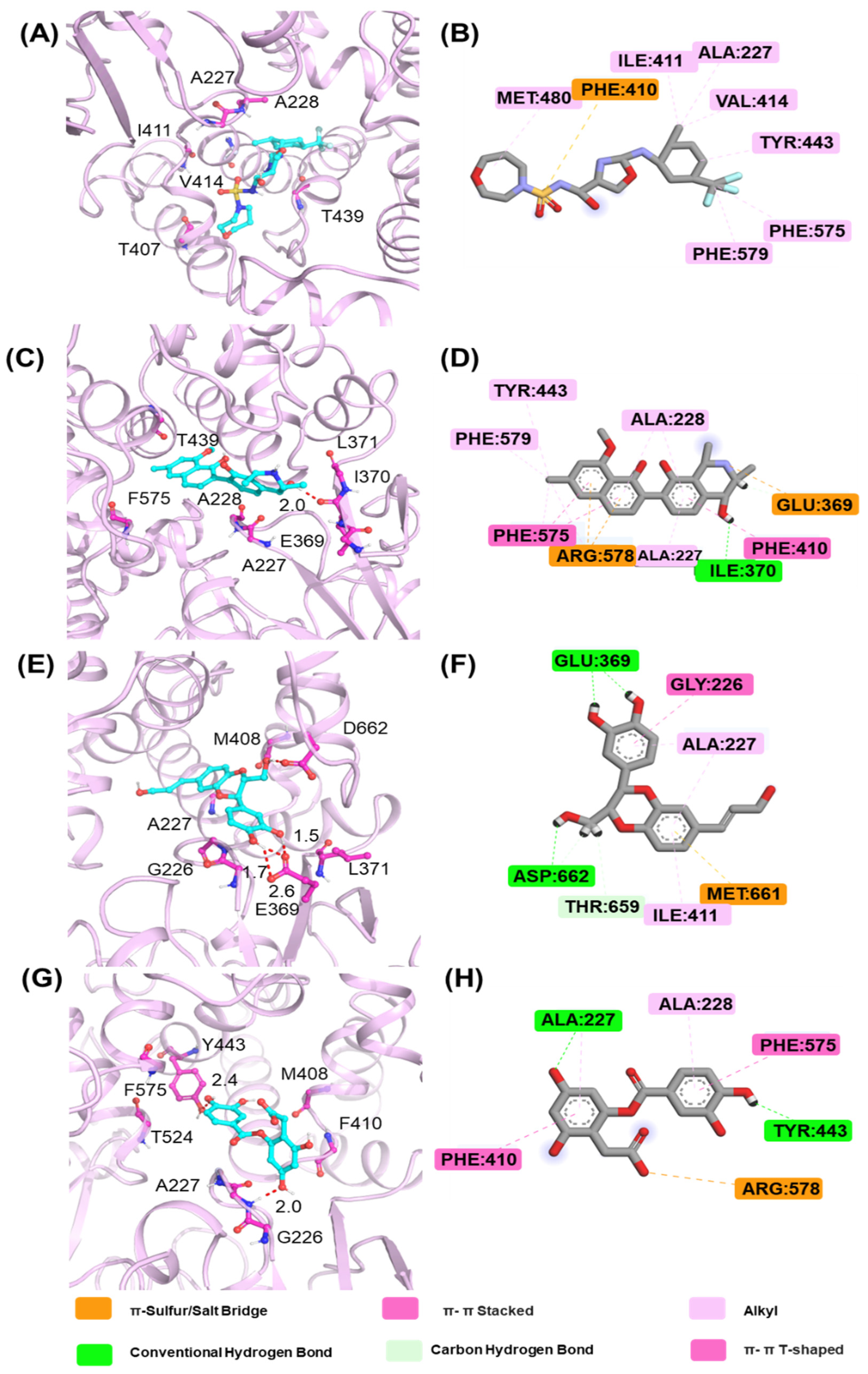
2.7. The Analysis of Free Energy Landscape and Binding Mode
2.8. The Calculation Binding Free Energy
3. Discussion
4. Materials and Methods
4.1. Database Integration
4.2. Ligand Preparation and Protein Selection
4.3. Docking-Based Virtual Screening
4.4. Shape-Based Screening
4.5. Similarity Calculations
4.6. The Identification of Pharmacokinetics Properties
4.7. Molecular Dynamics Simulation
4.8. MMGBSA
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wu, H. Structural Mechanisms of NLRP3 Inflammasome Assembly and Activation. Annu. Rev. Immunol. 2023, 41, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Seoane, P.I.; Lee, B.; Hoyle, C.; Yu, S.; Lopez-Castejon, G.; Lowe, M.; Brough, D. The NLRP3-inflammasome as a sensor of organelle dysfunction. J. Cell Biol. 2020, 219, e202006194. [Google Scholar] [CrossRef]
- Wang, Q.; Ou, Y.; Hu, G.; Wen, C.; Yue, S.; Chen, C.; Xu, L.; Xie, J.; Dai, H.; Xiao, H.; et al. Naringenin attenuates non-alcoholic fatty liver disease by down-regulating the NLRP3/NF-κB pathway in mice. Br. J. Pharmacol. 2020, 177, 1806–1821. [Google Scholar] [CrossRef]
- Zhao, C.; Zhao, W. NLRP3 Inflammasome-A Key Player in Antiviral Responses. Front. Immunol. 2020, 11, 211. [Google Scholar] [CrossRef]
- Ma, Q. Pharmacological Inhibition of the NLRP3 Inflammasome: Structure, Molecular Activation, and Inhibitor-NLRP3 Interaction. Pharmacol. Rev. 2023, 75, 487–520. [Google Scholar] [CrossRef]
- Li, Z.; Guo, J.; Bi, L. Role of the NLRP3 inflammasome in autoimmune diseases. Biomed. Pharmacother. 2020, 130, 110542. [Google Scholar] [CrossRef]
- Schmacke, N.A.; O’Duill, F.; Gaidt, M.M.; Szymanska, I.; Kamper, J.M.; Schmid-Burgk, J.L.; Mädler, S.C.; Mackens-Kiani, T.; Kozaki, T.; Chauhan, D.; et al. IKKβ primes inflammasome formation by recruiting NLRP3 to the trans-Golgi network. Immunity 2022, 55, 2271–2284.e2277. [Google Scholar] [CrossRef]
- Abdullaha, M.; Ali, M.; Kour, D.; Kumar, A.; Bharate, S.B. Discovery of benzo[cd]indol-2-one and benzylidene-thiazolidine-2,4-dione as new classes of NLRP3 inflammasome inhibitors via ER-β structure based virtual screening. Bioorganic Chem. 2020, 95, 103500. [Google Scholar] [CrossRef]
- Sánchez-Fernández, A.; Skouras, D.B.; Dinarello, C.A.; López-Vales, R. OLT1177 (Dapansutrile), a Selective NLRP3 Inflammasome Inhibitor, Ameliorates Experimental Autoimmune Encephalomyelitis Pathogenesis. Front. Immunol. 2019, 10, 2578. [Google Scholar] [CrossRef]
- Harrison, D.; Billinton, A.; Bock, M.G.; Doedens, J.R.; Gabel, C.A.; Holloway, M.K.; Porter, R.A.; Reader, V.; Scanlon, J.; Schooley, K.; et al. Discovery of Clinical Candidate NT-0796, a Brain-Penetrant and Highly Potent NLRP3 Inflammasome Inhibitor for Neuroinflammatory Disorders. J. Med. Chem. 2023, 66, 14897–14911. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, H.; Kouadir, M.; Song, H.; Shi, F. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 2019, 10, 128. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Niu, L.; Liu, Z.; Xu, X.; Shi, M.; Zhang, Y.; Deng, Y.; He, J.; Xu, Y.; Wan, W.; et al. Inhibition of the NLRP3 inflammasome with MCC950 prevents chronic social isolation-induced depression-like behavior in male mice. Neurosci. Lett. 2021, 765, 136290. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Jiang, H.; Chen, Y.; Wang, X.; Yang, Y.; Tao, J.; Deng, X.; Liang, G.; Zhang, H.; Jiang, W.; et al. Tranilast directly targets NLRP3 to treat inflammasome-driven diseases. EMBO Mol. Med. 2018, 10, e8689. [Google Scholar] [CrossRef] [PubMed]
- Youm, Y.H.; Nguyen, K.Y.; Grant, R.W.; Goldberg, E.L.; Bodogai, M.; Kim, D.; D’Agostino, D.; Planavsky, N.; Lupfer, C.; Kanneganti, T.D.; et al. The ketone metabolite β-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat. Med. 2015, 21, 263–269. [Google Scholar] [CrossRef]
- Ran, L.; Chen, M.; Ye, J.; Zhang, S.; Luo, Z.; Bai, T.; Qian, C.; Zhou, Q.; Shan, M.; Chu, Y.; et al. UK5099 Inhibits the NLRP3 Inflammasome Independently of its Long-Established Target Mitochondrial Pyruvate Carrier. Adv. Sci. 2024, 11, e2307224. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, J.; Jiang, Y.; Sun, S.; Wang, R.; Sun, J.; Ma, C.; Chen, Y.; Wang, W.; Hou, X.; et al. Sorbremnoids A and B: NLRP3 Inflammasome Inhibitors Discovered from Spatially Restricted Crosstalk of Biosynthetic Pathways. J. Am. Chem. Soc. 2024, 146, 18172–18183. [Google Scholar] [CrossRef]
- Shao, J.J.; Li, W.F.; Sun, J.F.; Zhuang, Z.S.; Min, J.L.; Long, X.H.; Wu, G.J.; Xu, H.W.; Liang, G. Britannin as a novel NLRP3 inhibitor, suppresses inflammasome activation in macrophages and alleviates NLRP3-related diseases in mice. Acta Pharmacol. Sin. 2024, 45, 803–814. [Google Scholar] [CrossRef]
- Michino, M.; Beautrait, A.; Boyles, N.A.; Nadupalli, A.; Dementiev, A.; Sun, S.; Ginn, J.; Baxt, L.; Suto, R.; Bryk, R.; et al. Shape-Based Virtual Screening of a Billion-Compound Library Identifies Mycobacterial Lipoamide Dehydrogenase Inhibitors. ACS Bio Med. Chem. Au 2023, 3, 507–515. [Google Scholar] [CrossRef]
- Potlitz, F.; Link, A.; Schulig, L. Advances in the discovery of new chemotypes through ultra-large library docking. Expert. Opin. Drug Discov. 2023, 18, 303–313. [Google Scholar] [CrossRef]
- Lanka, G.; Begum, D.; Banerjee, S.; Adhikari, N.; P, Y.; Ghosh, B. Pharmacophore-based virtual screening, 3D QSAR, Docking, ADMET, and MD simulation studies: An in silico perspective for the identification of new potential HDAC3 inhibitors. Comput. Biol. Med. 2023, 166, 107481. [Google Scholar] [CrossRef]
- Wang, Y.; Jia, S.; Wang, F.; Jiang, R.; Yin, X.; Wang, S.; Jin, R.; Guo, H.; Tang, Y.; Wang, Y. 3D-QSAR, Scaffold Hopping, Virtual Screening, and Molecular Dynamics Simulations of Pyridin-2-one as mIDH1 Inhibitors. Int. J. Mol. Sci. 2024, 25, 7434. [Google Scholar] [CrossRef]
- Sobhia, M.E.; Ghosh, K.; Sivangula, S.; Kumar, S.; Singh, H. Identification of potential SARS-CoV-2 M(pro) inhibitors integrating molecular docking and water thermodynamics. J. Biomol. Struct. Dyn. 2022, 40, 5079–5089. [Google Scholar] [CrossRef]
- Nahhas, A.F.; Webster, T.J. Applications of peptide-functionalized or unfunctionalized selenium nanoparticles for the passivation of SARS-CoV-2 variants and the respiratory syncytial virus (RSV). Colloids Surf. B Biointerfaces 2024, 233, 113638. [Google Scholar] [CrossRef]
- Yao, K.; Liu, H.; Liu, P.; Liu, W.; Yang, J.; Wei, Q.; Cao, P.; Lai, Y. Molecular modeling studies to discover novel mIDH2 inhibitors with high selectivity for the primary and secondary mutants. Comput. Biol. Chem. 2020, 86, 107261. [Google Scholar] [CrossRef]
- Matore, B.W.; Roy, P.P.; Singh, J. Discovery of novel VEGFR2-TK inhibitors by phthalimide pharmacophore based virtual screening, molecular docking, MD simulation and DFT. J. Biomol. Struct. Dyn. 2023, 41, 13056–13077. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lv, J.; Zhao, L.; Zhao, R.; Gao, T.; Xu, Q.; Liu, D.; Yu, Q.; Ma, F. Exploring the role of microbial proteins in controlling environmental pollutants based on molecular simulation. Sci. Total Environ. 2023, 905, 167028. [Google Scholar] [CrossRef] [PubMed]
- Durham, E.; Dorr, B.; Woetzel, N.; Staritzbichler, R.; Meiler, J. Solvent accessible surface area approximations for rapid and accurate protein structure prediction. J. Mol. Model. 2009, 15, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Singh, R.; Chatterjee, C.; Kumar, A.; Singh, S.K. Computational screening of coumarin derivatives as inhibitors of the NACHT domain of NLRP3 inflammasome for the treatment of Alzheimer’s disease. J. Biomol. Struct. Dyn. 2025, 43, 2187–2203. [Google Scholar] [CrossRef]
- Cabral, J.E.; Wu, A.; Zhou, H.; Pham, M.A.; Lin, S.; McNulty, R. Targeting the NLRP3 inflammasome for inflammatory disease therapy. Trends Pharmacol. Sci. 2025, 46, 503–519. [Google Scholar] [CrossRef]
- Alam, S.; Khan, F. Virtual screening, Docking, ADMET and System Pharmacology studies on Garcinia caged Xanthone derivatives for Anticancer activity. Sci. Rep. 2018, 8, 5524. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Kim, C.Y.; Lee, E.; Lee, C.; Lee, K.S.; Lee, J.; Park, H.; Choi, B.; Hwang, I.; Kim, J.; et al. Microglial NLRP3-gasdermin D activation impairs blood-brain barrier integrity through interleukin-1beta-independent neutrophil chemotaxis upon peripheral inflammation in mice. Nat. Commun. 2025, 16, 699. [Google Scholar] [CrossRef] [PubMed]
- Velcicky, J.; Janser, P.; Gommermann, N.; Brenneisen, S.; Ilic, S.; Vangrevelinghe, E.; Stiefl, N.; Boettcher, A.; Arnold, C.; Malinverni, C.; et al. Discovery of Potent, Orally Bioavailable, Tricyclic NLRP3 Inhibitors. J. Med. Chem. 2024, 67, 1544–1562. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, F.; Wang, Y.; Li, C.; Zhang, X.; Li, H.; Diao, L.; Gu, J.; Wang, W.; Li, D.; et al. BATMAN-TCM: A Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine. Sci. Rep. 2016, 6, 21146. [Google Scholar] [CrossRef]
- Xu, H.Y.; Zhang, Y.Q.; Liu, Z.M.; Chen, T.; Lv, C.Y.; Tang, S.H.; Zhang, X.B.; Zhang, W.; Li, Z.Y.; Zhou, R.R.; et al. ETCM: An encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019, 47, D976–D982. [Google Scholar] [CrossRef]
- Fang, S.; Dong, L.; Liu, L.; Guo, J.; Zhao, L.; Zhang, J.; Bu, D.; Liu, X.; Huo, P.; Cao, W.; et al. HERB: A high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 2021, 49, D1197–D1206. [Google Scholar] [CrossRef]
- Yan, D.; Zheng, G.; Wang, C.; Chen, Z.; Mao, T.; Gao, J.; Yan, Y.; Chen, X.; Ji, X.; Yu, J.; et al. HIT 2.0: An enhanced platform for Herbal Ingredients’ Targets. Nucleic Acids Res. 2022, 50, D1238–D1243. [Google Scholar] [CrossRef]
- Tian, S.; Zhang, J.; Yuan, S.; Wang, Q.; Lv, C.; Wang, J.; Fang, J.; Fu, L.; Yang, J.; Zu, X.; et al. Exploring pharmacological active ingredients of traditional Chinese medicine by pharmacotranscriptomic map in ITCM. Brief. Bioinform. 2023, 24, bbad027. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, F.; Yang, K.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Hu, H.; Gao, K.; Wang, W.; et al. SymMap: An integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res. 2019, 47, D1110–D1117. [Google Scholar] [CrossRef]
- Zhang, L.X.; Dong, J.; Wei, H.; Shi, S.H.; Lu, A.P.; Deng, G.M.; Cao, D.S. TCMSID: A simplified integrated database for drug discovery from traditional chinese medicine. J. Cheminform. 2022, 14, 89. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chao, H.; Chen, L.; Craig, P.A.; Crichlow, G.V.; Dalenberg, K.; Duarte, J.M.; et al. RCSB Protein Data bank: Tools for visualizing and understanding biological macromolecules in 3D. Protein Sci. 2022, 31, e4482. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput.-Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R.; et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. [Google Scholar] [CrossRef]
- Qin, P.; Niu, Y.; Duan, J.; Lin, P. Computational study on the mechanism of small molecules inhibiting NLRP3 with ensemble docking and molecular dynamic simulations. BMC Pharmacol. Toxicol. 2025, 26, 49. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Kunimoto, R.; Bajorath, J. Combining Similarity Searching and Network Analysis for the Identification of Active Compounds. ACS Omega 2018, 3, 3768–3777. [Google Scholar] [CrossRef]
- Mendolia, I.; Contino, S.; De Simone, G.; Perricone, U.; Pirrone, R. EMBER-Embedding Multiple Molecular Fingerprints for Virtual Screening. Int. J. Mol. Sci. 2022, 23, 2156. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Luo, H.; Li, S.; Wu, F.X.; Wang, J. Drug-drug similarity measure and its applications. Brief. Bioinform. 2021, 22, bbaa265. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Yoon, H.J.; Kundu, S.; Wu, S. Molecular Dynamics Simulation Study of the Selective Inhibition of Coagulation Factor IXa over Factor Xa. Molecules 2023, 28, 6909. [Google Scholar] [CrossRef]
- Botelho, F.D.; Nepovimova, E.; Kamil, K.; Franca, T.C.C. Virtual screening and molecular dynamic study of potential new binders to mTOR. J. Mol. Model. 2022, 28, 315. [Google Scholar] [CrossRef] [PubMed]
- Benny, S.; Rajappan Krishnendu, P.; Kumar, S.; Bhaskar, V.; Manisha, D.S.; Abdelgawad, M.A.; Ghoneim, M.M.; Naguib, I.A.; Pappachen, L.K.; Mary Zachariah, S.; et al. A computational investigation of thymidylate synthase inhibitors through a combined approach of 3D-QSAR and pharmacophore modelling. J. Biomol. Struct. Dyn. 2024, 42, 8473–8492. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zeng, Z.; Zhang, J.; Wu, D.; Li, H.; Geng, F. Molecular dynamics simulation of the interaction of food proteins with small molecules. Food Chem. 2023, 405, 134824. [Google Scholar] [CrossRef] [PubMed]
- Chaieb, K.; Kouidhi, B.; Hosawi, S.B.; Baothman, O.A.S.; Zamzami, M.A.; Altayeb, H.N. Computational screening of natural compounds as putative quorum sensing inhibitors targeting drug resistance bacteria: Molecular docking and molecular dynamics simulations. Comput. Biol. Med. 2022, 145, 105517. [Google Scholar] [CrossRef]
- Proj, M.; Zidar, M.; Lebar, B.; Strašek, N.; Miličić, G.; Žula, A.; Gobec, S. Discovery of compounds with viscosity-reducing effects on biopharmaceutical formulations with monoclonal antibodies. Comput. Struct. Biotechnol. J. 2022, 20, 5420–5429. [Google Scholar] [CrossRef]
- Bai, H.; Cheng, Y.; Jia, S.; Wang, X.; Jin, R.; Guo, H.; Tang, Y.; Wang, Y. Machine learning-based QSAR and structure-based virtual screening guided discovery of novel mIDH1 inhibitors from natural products. J. Comput. Aided Mol. Des. 2025, 39, 44. [Google Scholar] [CrossRef]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef]
| ID | PubChem ID | Name | Chemical Structure | Docking Scores (Kcal/mol) | Shape Sim | Source (English/Latin Name) | Database |
|---|---|---|---|---|---|---|---|
| XE3 | NA | NA | 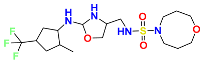 | −8.057 | 1 | NA | NA |
| C1 | 101010963 | l-malicacid 2-o-gallate |  | −10.522 | 0.328 | Phyllanthus emblica L. | iTCM |
| C2 | 443775 | Dioncophyllinol B | 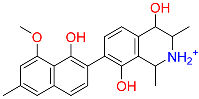 | −10.802 | 0.305 | Akebia trifoliata | HERB |
| C3 | 6444016 | Isoamericanol A | 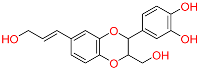 | −10.119 | 0.280 | Phytolacca americana L | HERB |
| C4 | NA | NA | 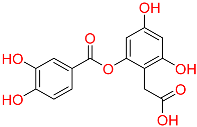 | −10.137 | 0.251 | NA | HERB |
| C5 | 135612764 | Isobetanidin | 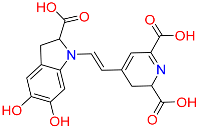 | −10.129 | 0.247 | Portulaca grandiflora Hook/Cichorium intybus L/Portulaca oleracea L/Portulaca pilosa L | HERB |
| C6 | 129716404 | Monocaffeyltartaric acid | 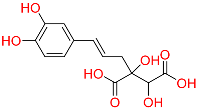 | −11.277 | 0.229 | Cichorium intybus L/Monarda didyma L/Salvia coccinea Buc’hoz ex Etl | HERB BATMAN-TCM iTCM SymMap V2.0 |
| C7 | 135728 | Gyrophoric acid | 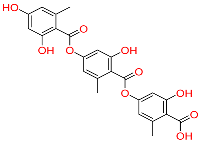 | −10.669 | 0.226 | Gypsophila pacifica | HERB |
| C8 | 71435823 | Paludosicacid | 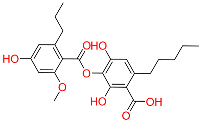 | −10.161 | 0.217 | Silene jenisseensis/Parmelia Lichen | HERB |
| ID | PlogPo/w (−2.0–6.5) | PlogS (−6.5–0.5) | PlogHERG (<−5) | PPCaco (<25 Poor) (>500 Great) | PlogBB (−3.0–1.2) |
|---|---|---|---|---|---|
| XE3 | 1.948 | −2.398 | −6.186 | 61.217 | 0.107 |
| C1 | −0.581 | −1.752 | −0.75 | 0.236 | −3.256 |
| C2 | 3.007 | −4.01 | −6.167 | 270.285 | −0.349 |
| C3 | 1.48 | −3.58 | −5.528 | 114.108 | −1.87 |
| C4 | 0.17 | −2.393 | −3.152 | 2.603 | −2.845 |
| C5 | 1.113 | −3.674 | 0.443 | 0.024 | −3.905 |
| C6 | 0.863 | −2.055 | −0.982 | 0.463 | −3.089 |
| C7 | 2.819 | −4.59 | −3.118 | 1.446 | −3.127 |
| C8 | 3.854 | −5.275 | −3.653 | 22.402 | −2.559 |
| No. | ΔG Bind (kcal/mol) | ΔG Bind Coulomb | ΔG Bind H Bond | ΔG Bind Lipo | ΔG Bind vdW |
|---|---|---|---|---|---|
| XE3 | −42.31 ± 5.31 | −6.57 ± 5.45 | −0.98 ± 0.66 | −17.37 ± 1.55 | −43.41 ± 3.72 |
| C2 | −34.17 ± 5.79 | −81.64 ± 10.54 | −1.20 ± 1.32 | −19.64 ± 1.23 | −41.06 ± 2.79 |
| C3 | −48.81 ± 3.89 | −29.45 ± 5.13 | −3.04 ± 0.56 | −19.61 ± 1.76 | −34.60 ± 2.77 |
| C4 | −33.50 ± 5.25 | −3.64 ± 17.94 | −2.21 ± 0.87 | −13.45 ± 1.75 | −29.05 ± 2.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, S.; Lai, H.; Chen, X.; Lu, J.; Ding, W.; Cui, D.; Zhao, P.; Zhang, Q.; Wang, Y.; Cheng, C. In Silico Identification of the NLRP3 Inhibitors from Traditional Chinese Medicine. Int. J. Mol. Sci. 2025, 26, 10569. https://doi.org/10.3390/ijms262110569
Jia S, Lai H, Chen X, Lu J, Ding W, Cui D, Zhao P, Zhang Q, Wang Y, Cheng C. In Silico Identification of the NLRP3 Inhibitors from Traditional Chinese Medicine. International Journal of Molecular Sciences. 2025; 26(21):10569. https://doi.org/10.3390/ijms262110569
Chicago/Turabian StyleJia, Shunjiang, Huanling Lai, Xinyu Chen, Jiajie Lu, Wei Ding, Dongxiao Cui, Peng Zhao, Qiao Zhang, Yuwei Wang, and Chunsong Cheng. 2025. "In Silico Identification of the NLRP3 Inhibitors from Traditional Chinese Medicine" International Journal of Molecular Sciences 26, no. 21: 10569. https://doi.org/10.3390/ijms262110569
APA StyleJia, S., Lai, H., Chen, X., Lu, J., Ding, W., Cui, D., Zhao, P., Zhang, Q., Wang, Y., & Cheng, C. (2025). In Silico Identification of the NLRP3 Inhibitors from Traditional Chinese Medicine. International Journal of Molecular Sciences, 26(21), 10569. https://doi.org/10.3390/ijms262110569







