A Novel Germline Frameshift Variant in the Tumor Suppressor Gene OBSCN in a Melanoma Patient
Abstract
1. Introduction
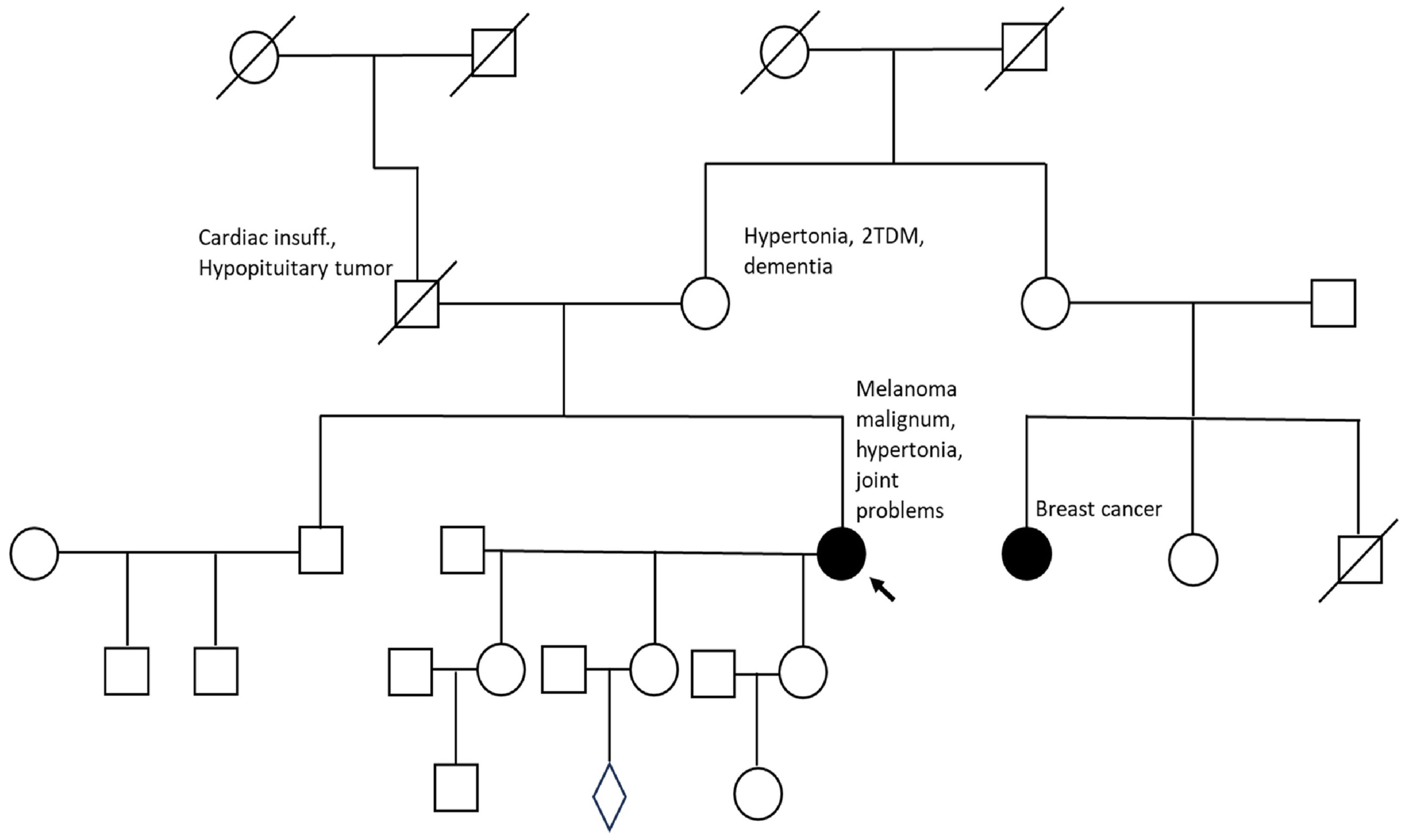
2. Results
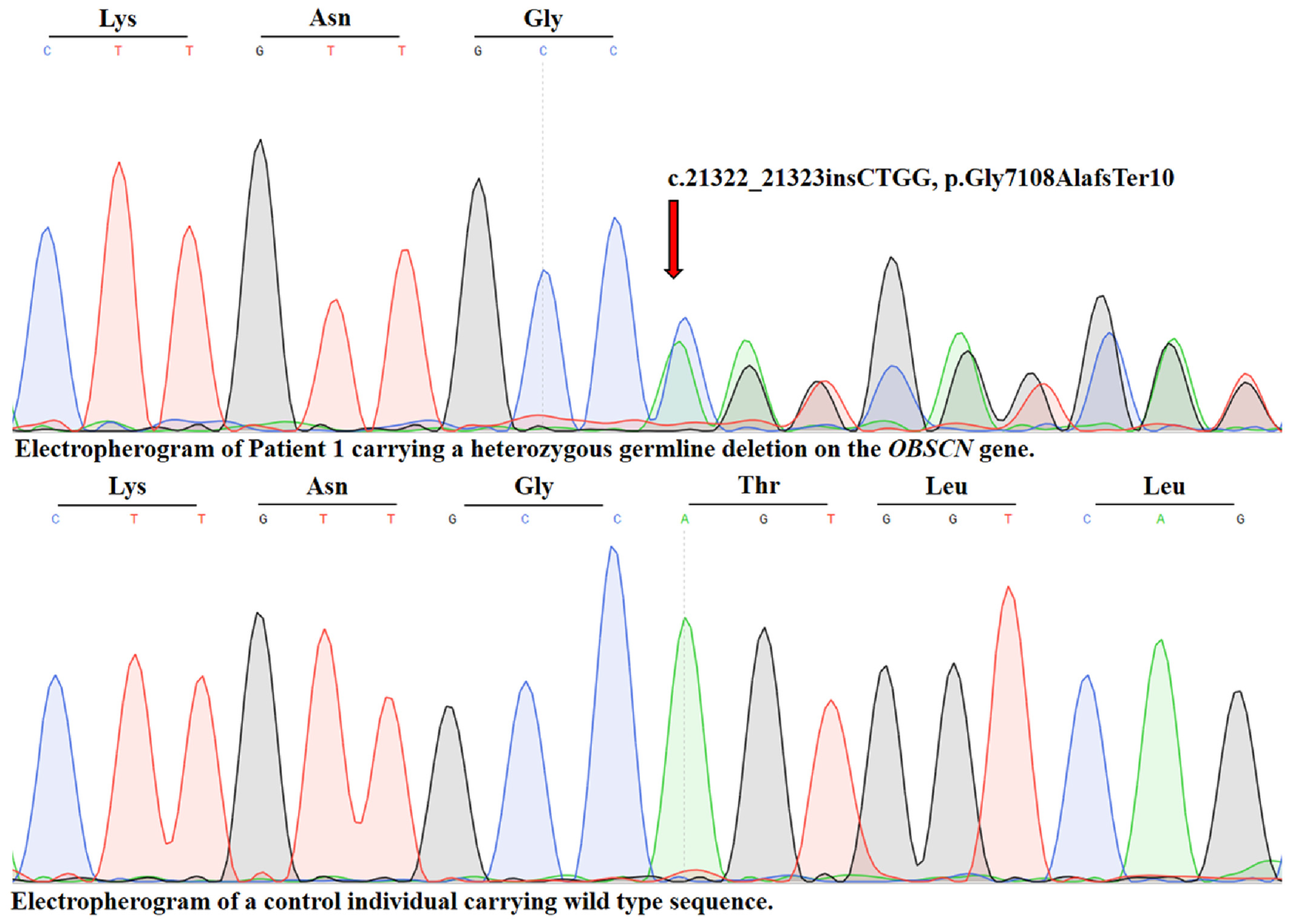
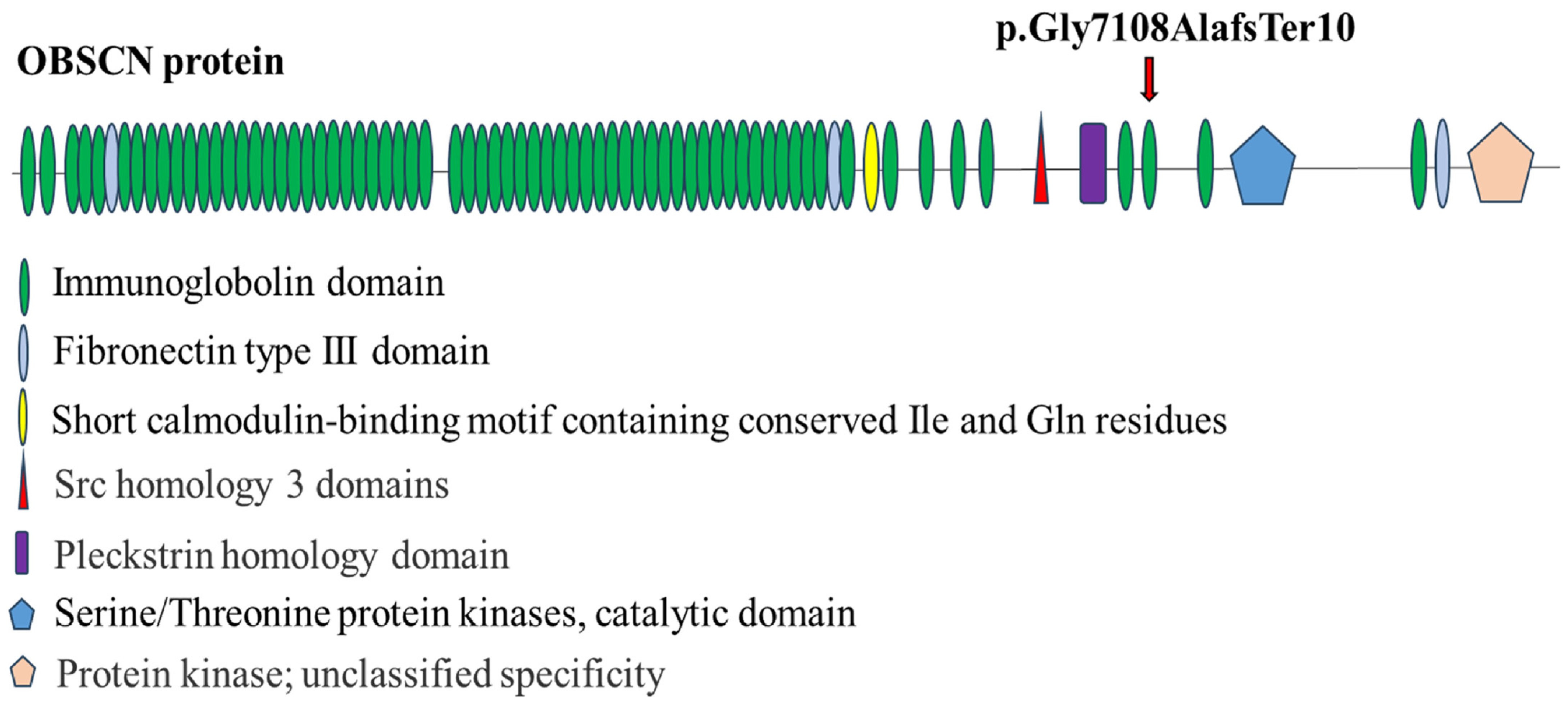
2.1. Identification of a Novel OBSCN Variant
2.2. Figures, Tables and Schemes
- PVS1 (very strong evidence): A null variant (frameshift/nonsense) in a gene where loss of function is a known disease mechanism.
- PM2 (moderate evidence): The variant is absent, or extremely rare, in large population databases such as gnomAD, suggesting it is not a common benign polymorphism.
3. Discussion
- Genetic counseling: Long-term dermatologic and oncologic follow-up is warranted for both the proband and her mother, considering the germline nature of the variant and its potential association with multi-tumor risk.
- Surveillance: Given the involvement of OBSCN in breast, gastrointestinal, and gynecologic cancers, carriers might benefit from comprehensive surveillance protocols including annual dermatologic examinations, mammography or breast MRI starting at age 40, and colonoscopic screening per standard population guidelines.
- Research directions: Future research should focus on both predictive and functional validation. Immediate steps include comprehensive in silico modeling and gene network analyses to identify pathways potentially affected by OBSCN loss. Definitive insights, however, will require in vitro studies assessing cytoskeletal organization and cell adhesion in melanoma cells expressing truncated OBSCN.
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACMG | American College of Medical Genetics and Genomics |
| CDKN2A | Cyclin Dependent Kinase Inhibitor 2A |
| CDK4 | Cyclin Dependent Kinase 4 |
| TERT | Telomerase Reverse Transcriptase |
| POT1 | Protection of Telomeres 1 |
| ACD | Adrenocortical Dysplasia protein homolog (also known as TPP1) |
| TERF2IP | Telomeric Repeat Binding Factor 2 Interacting Protein |
| BAP1 | BRCA1 Associated Protein 1 |
| BRCA1/2 | Breast Cancer Gene 1/2 |
| TP53 | Tumor Protein p53 |
| MC1R | Melanocortin 1 Receptor |
| TYR | Tyrosinase |
| OCA2 | Oculocutaneous Albinism II |
| SLC45A2 | Solute Carrier Family 45 Member 2 |
| NGS | Next Generation Sequencing |
| LOH | Loss of Heterozygosity |
| OBSCN | Obscurin |
| gnomAD | Genome Aggregation Database |
| Ig-like | Immunoglobulin like domain |
| FnIII | Fibronectin type III domain |
References
- Wang, M.; Gao, X.; Zhang, L. Recent global patterns in skin cancer incidence, mortality, and prevalence. Chin. Med. J. 2025, 138, 185–192. Available online: https://journals.lww.com/cmj/fulltext/2025/01200/recent_global_patterns_in_skin_cancer_incidence,.6.aspx (accessed on 18 October 2025). [CrossRef] [PubMed]
- Funchain, P.; Ni, Y.; Heald, B.; Bungo, B.; Arbesman, M.; Behera, T.R.; McCormick, S.; Song, J.M.; Kennedy, L.B.; Nielsen, S.M.; et al. Germline cancer susceptibility in individuals with melanoma. J. Am. Acad. Dermatol. 2024, 91, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Read, J.; Wadt, K.A.; Hayward, N.K. Melanoma genetics. J. Med. Genet. 2016, 53, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Manganelli, M.; Stabile, G.; Scharf, C.; Podo Brunetti, A.; Paolino, G.; Giuffrida, R.; Bigotto, G.D.; Damiano, G.; Mercuri, S.R.; Sallustio, F.; et al. Skin Photodamage and Melanomagenesis: A Comprehensive Review. Cancers 2025, 17, 1784. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, L.P.; Milne, R.L.; Pita, G.; Floristan, U.; Sendagorta, E.; Feito, M.; Avilés, J.A.; Martin-Gonzalez, M.; Lázaro, P.; Benítez, J.; et al. Pigmentation-related genes and their implication in malignant melanoma susceptibility. Exp. Dermatol. 2009, 18, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Bokor, B.A.; Abdolreza, A.; Kaptás, F.; Pál, M.; Battyani, Z.; Széll, M.; Nagy, N. Novel variants in medium and low penetrance predisposing genes in a Hungarian malignant melanoma cohort with increased risk. Pigment. Cell Melanoma Res. 2025, 38, e13214. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, A.; Bleeker, F.E.; Lamba, S.; Rodolfo, M.; Daniotti, M.; Scarpa, A.; Van Tilborg, A.A.; Leenstra, S.; Zanon, C.; Bardelli, A. Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res. 2007, 67, 3545–3550. [Google Scholar] [CrossRef] [PubMed]
- Young, P.; Ehler, E.; Gautel, M. Obscurin, a giant sarcomeric rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J. Cell Biol. 2001, 154, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Kontrogianni-Konstantopoulos, A.; Ackermann, M.A.; Bowman, A.L.; Yap, S.V.; Bloch, R.J. Muscle giants: Molecular scaffolds in sarcomerogenesis. Physiol. Rev. 2009, 89, 1217–1267. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.A.; King, B.; Lieberman, N.A.; Bobbili, P.J.; Rudloff, M.; Berndsen, C.E.; Wright, N.T.; Hecker, P.A.; Kontrogianni-Konstantopoulos, A. Novel obscurins mediate cardiomyocyte adhesion and size via the pi3k/akt/mtor signaling pathway. J. Mol. Cell. Cardiol. 2017, 111, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Mendiratta, G.; Ke, E.; Aziz, M.; Liarakos, D.; Tong, M.; Stites, E.C. Cancer gene mutation frequencies for the U.S. population. Nat. Commun. 2021, 12, 5961. Available online: https://pubmed.ncbi.nlm.nih.gov/34645806 (accessed on 18 October 2025). [CrossRef]
- Guardia, T.; Eason, M.; Kontrogianni-Konstantopoulos, A. Obscurin: A multitasking giant in the fight against cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188567. [Google Scholar] [CrossRef] [PubMed]
- Sjöblom, T.; Jones, S.; Wood, L.D.; Parsons, D.W.; Lin, J.; Barber, T.D.; Mandelker, D.; Leary, R.J.; Ptak, J.; Silliman, N.; et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006, 314, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Babur, Ö.; Gönen, M.; Aksoy, B.A.; Schultz, N.; Ciriello, G.; Sander, C.; Demir, E. Systematic identification of cancer driving signaling pathways based on mutual exclusivity of genomic alterations. Genome Biol. 2015, 16, 45. Available online: https://genomebiology.biomedcentral.com/articles/10.1186/s13059-015-0612-6 (accessed on 18 October 2025). [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bokor, B.A.; Abdolreza, A.; Pál, M.; Battyani, Z.; Széll, M.; Nagy, N. A Novel Germline Frameshift Variant in the Tumor Suppressor Gene OBSCN in a Melanoma Patient. Int. J. Mol. Sci. 2025, 26, 10553. https://doi.org/10.3390/ijms262110553
Bokor BA, Abdolreza A, Pál M, Battyani Z, Széll M, Nagy N. A Novel Germline Frameshift Variant in the Tumor Suppressor Gene OBSCN in a Melanoma Patient. International Journal of Molecular Sciences. 2025; 26(21):10553. https://doi.org/10.3390/ijms262110553
Chicago/Turabian StyleBokor, Barbara Anna, Aliasgari Abdolreza, Margit Pál, Zita Battyani, Márta Széll, and Nikoletta Nagy. 2025. "A Novel Germline Frameshift Variant in the Tumor Suppressor Gene OBSCN in a Melanoma Patient" International Journal of Molecular Sciences 26, no. 21: 10553. https://doi.org/10.3390/ijms262110553
APA StyleBokor, B. A., Abdolreza, A., Pál, M., Battyani, Z., Széll, M., & Nagy, N. (2025). A Novel Germline Frameshift Variant in the Tumor Suppressor Gene OBSCN in a Melanoma Patient. International Journal of Molecular Sciences, 26(21), 10553. https://doi.org/10.3390/ijms262110553







