Optineurin Shapes Basal and LPS-Induced Transcriptomes in BV2 Microglia
Abstract
1. Introduction
2. Results
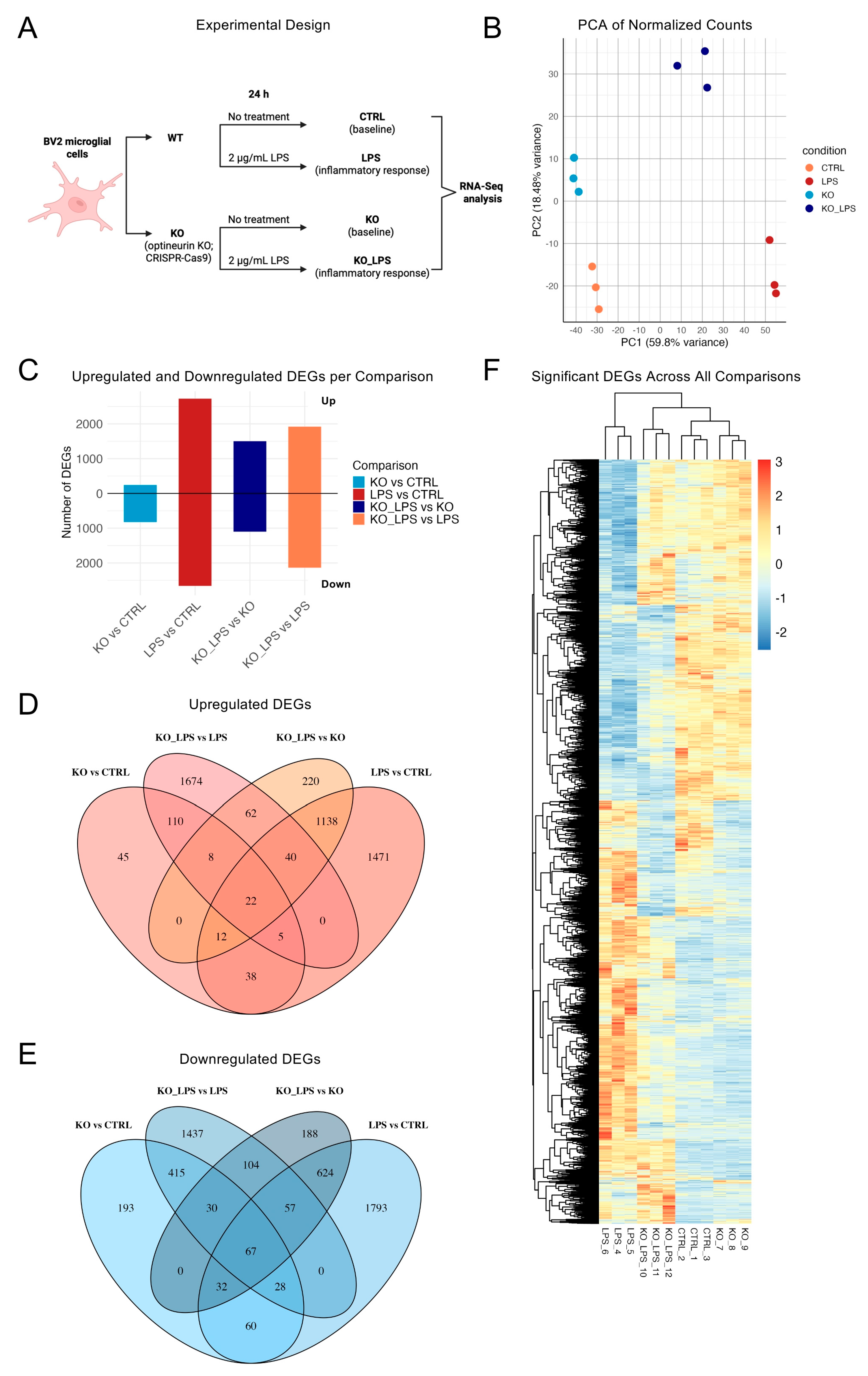
2.1. Optineurin Loss and LPS Drove Distinct Transcriptional Programs in BV2 Microglia
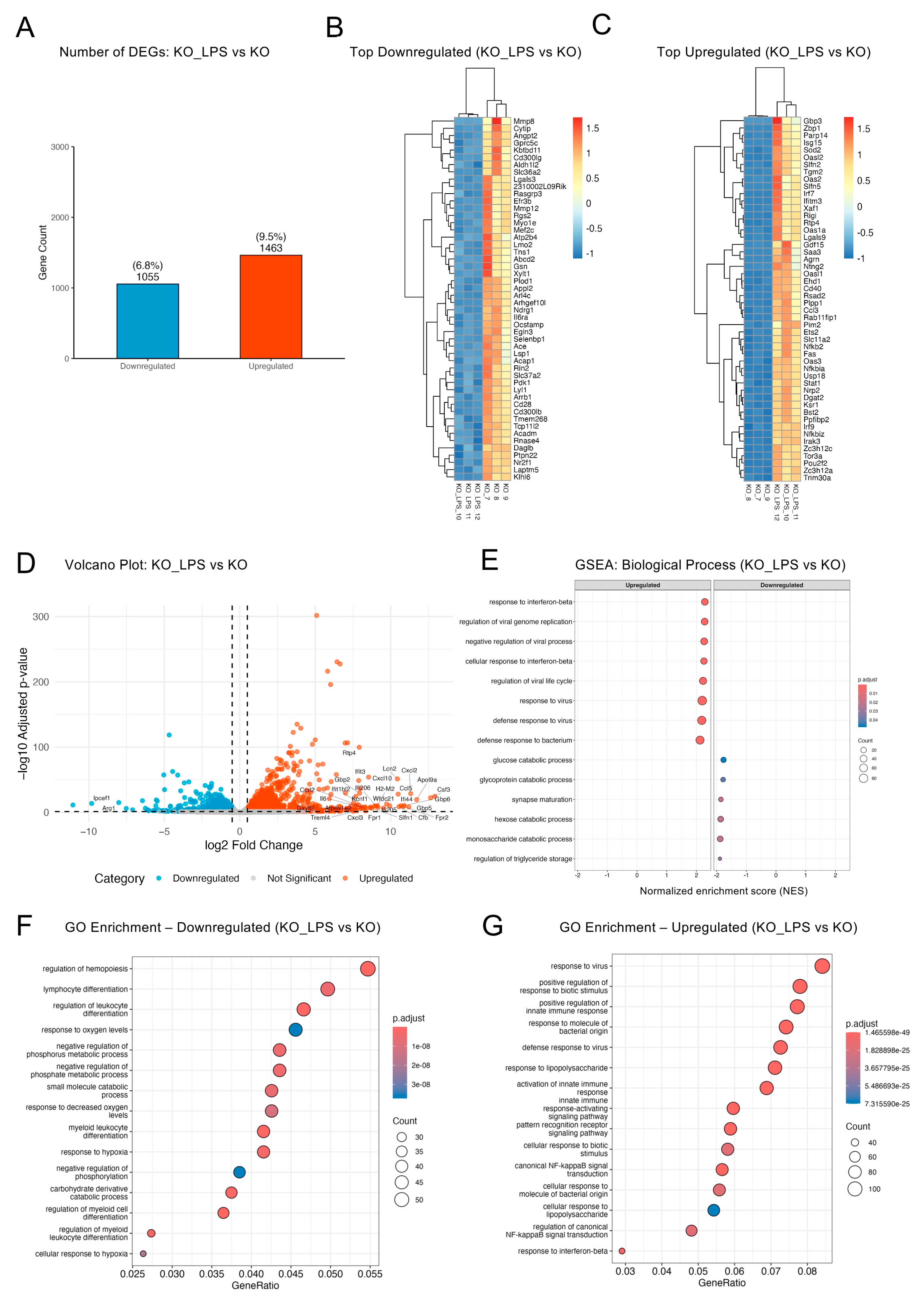
2.2. Optineurin Loss Reshaped the Basal BV2 Transcriptome
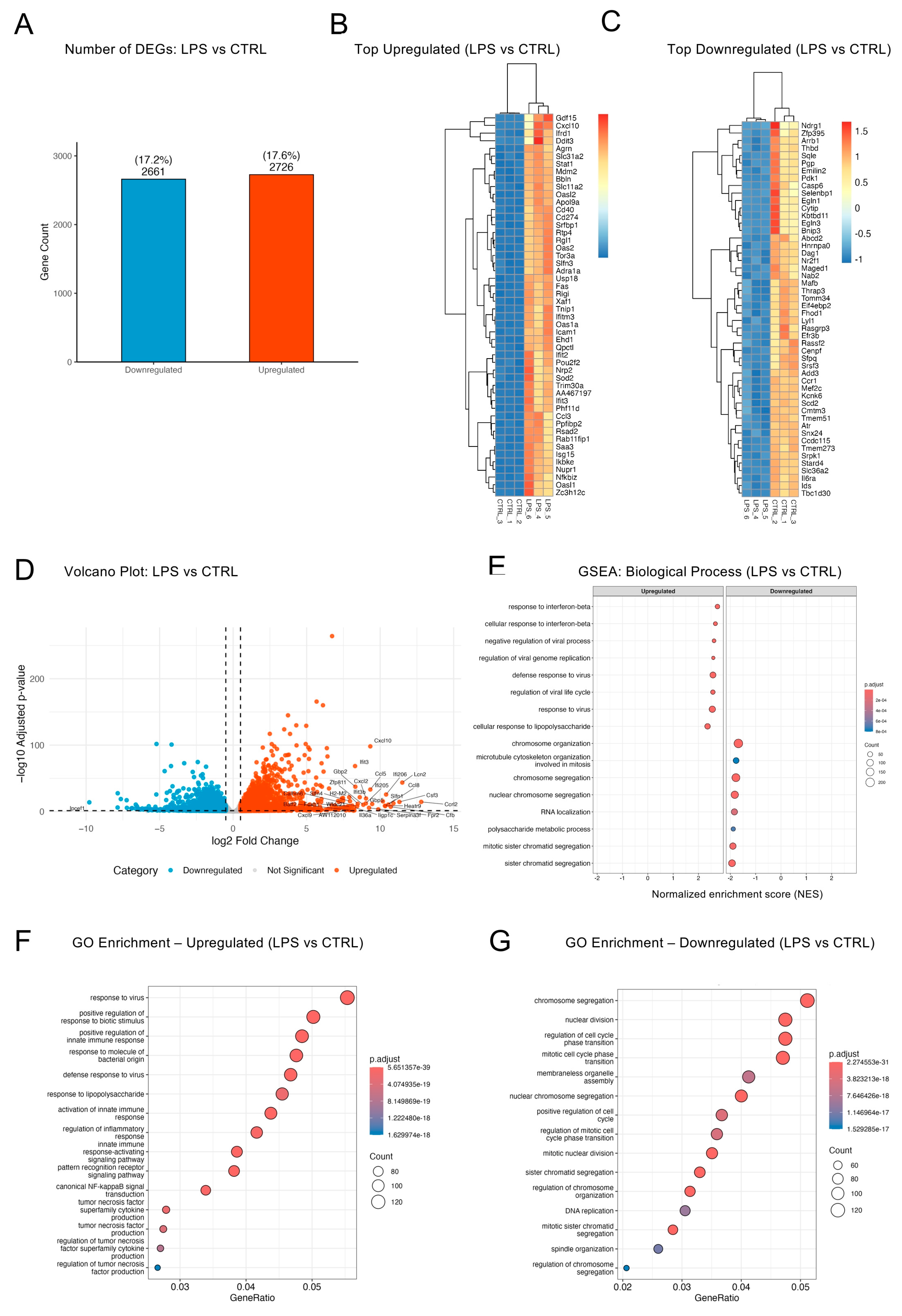
2.3. LPS Triggered a Broad Inflammatory Transcriptome in BV2 Cells
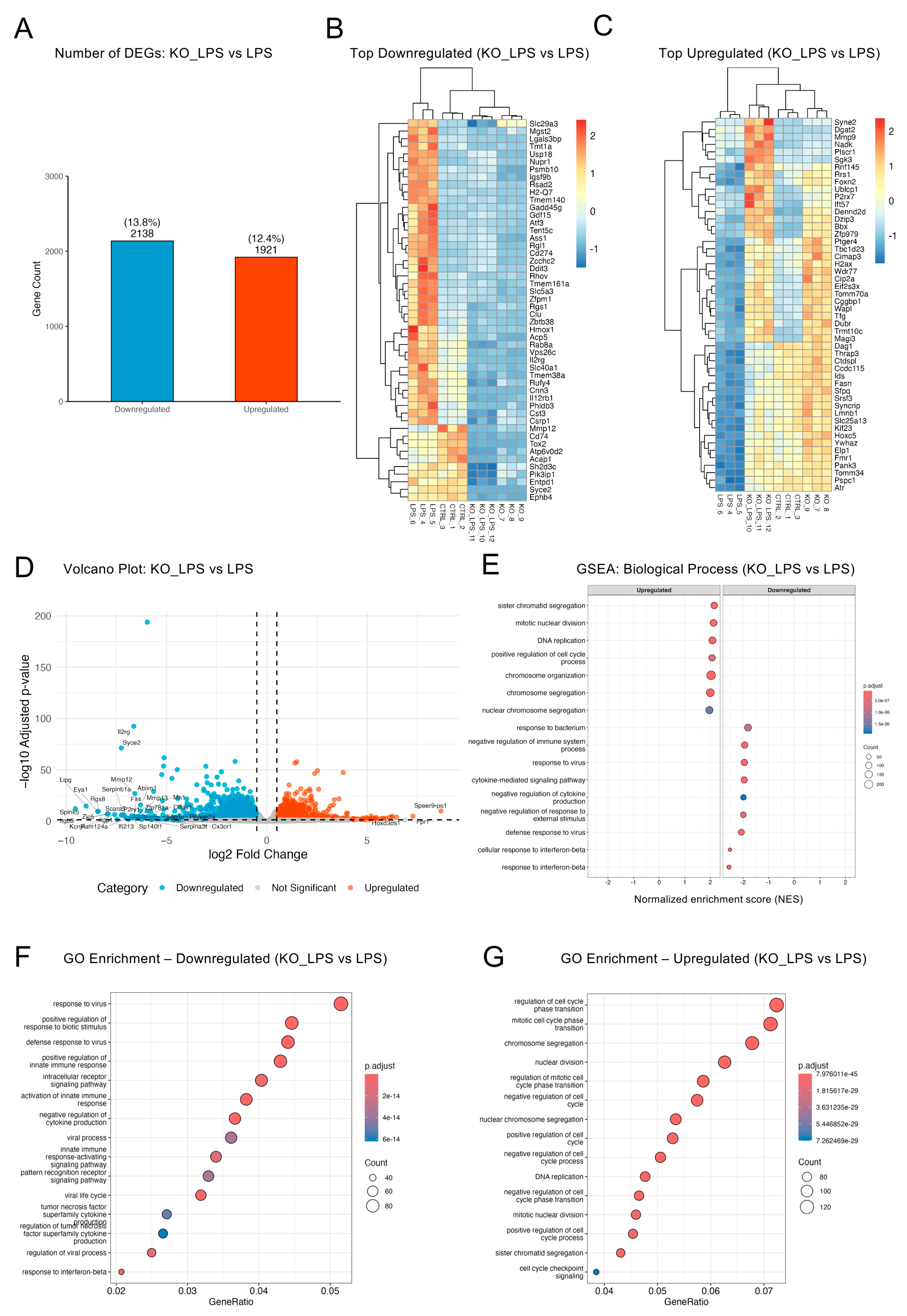
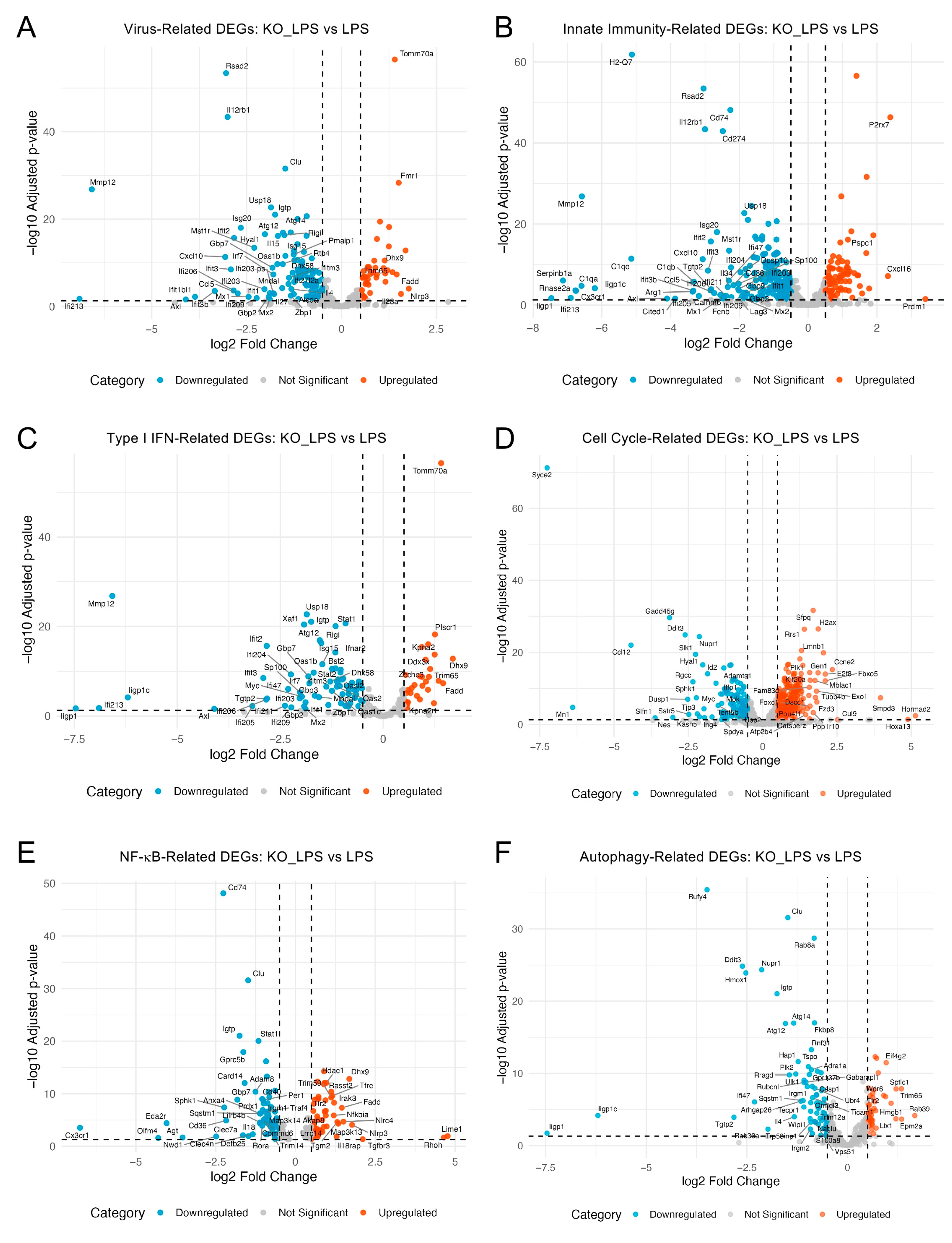
2.4. Optineurin Deficiency Attenuated LPS-Induced Inflammatory Transcriptome
2.5. Optineurin Influenced Inflammatory and Cell Cycle Gene Regulation in Response to LPS
2.6. Targeted Literature Mining for Potential Modulators of Optineurin-Mediated Pathways
3. Discussion
4. Materials and Methods
4.1. Generation of Optineurin Knockout Cells, Cell Culture and Treatments
4.2. RNA Isolation, Sequencing, and Transcriptomic Analysis
4.3. Literature-Based Identification of Potential Modulators
4.4. Data Availability
4.5. Use of Generative AI
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALS | Amyotrophic Lateral Sclerosis |
| ATCC | American Type Culture Collection |
| BV2 | BV2 Microglial Cell Line |
| CNS | Central Nervous System |
| Cas9 | CRISPR-associated Protein 9 |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CTRL | Control (Wild Type, Untreated) |
| DEG | Differentially Expressed Gene |
| DESeq2 | Differential Expression analysis using Sequencing, version 2 |
| DMEM | Dulbecco’s Modified Eagle Medium |
| FBS | Fetal Bovine Serum |
| FTD | Frontotemporal Dementia |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| GSEA | Gene Set Enrichment Analysis |
| IFN | Interferon |
| IFNAR | Interferon α/β receptor |
| IRF | Interferon Regulatory Factor |
| ISG | Interferon-Stimulated Gene |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KO | Knockout |
| KO_LPS | Optineurin Knockout + LPS |
| LPS | Lipopolysaccharide |
| NFκB | Nuclear Factor Kappa Light Chain Enhancer of Activated B Cells |
| NRF2 | Nuclear factor erythroid 2-related factor 2 |
| PCA | Principal Component Analysis |
| RNA-Seq | RNA Sequencing |
| TBK1 | TANK binding kinase 1 |
| TF | Transcription Factor (used in Section 2.5 and Section 2.6). |
| TLR4 | Toll-Like Receptor 4 |
| WT | Wild Type |
Appendix A
Curated GO Terms for Targeted Pathway Analysis
References
- Kettenmann, H.; Hanisch, U.K.; Noda, M.; Verkhratsky, A. Physiology of microglia. Physiol. Rev. 2011, 91, 461–553. [Google Scholar] [CrossRef]
- Paolicelli, R.C.; Sierra, A.; Stevens, B.; Tremblay, M.E.; Aguzzi, A.; Ajami, B.; Amit, I.; Audinat, E.; Bechmann, I.; Bennett, M.; et al. Microglia states and nomenclature: A field at its crossroads. Neuron 2022, 110, 3458–3483. [Google Scholar] [CrossRef]
- Ransohoff, R.M. How neuroinflammation contributes to neurodegeneration. Science 2016, 353, 777–783. [Google Scholar] [CrossRef]
- Deczkowska, A.; Keren-Shaul, H.; Weiner, A.; Colonna, M.; Schwartz, M.; Amit, I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 2018, 173, 1073–1081. [Google Scholar] [CrossRef]
- Béland, L.C.; Markovinovic, A.; Jakovac, H.; De Marchi, F.; Bilic, E.; Mazzini, L.; Kriz, J.; Munitic, I. Immunity in amyotrophic lateral sclerosis: Blurred lines between excessive inflammation and inefficient immune responses. Brain Commun. 2020, 2, fcaa124. [Google Scholar] [CrossRef]
- De Marchi, F.; Franjkic, T.; Schito, P.; Russo, T.; Nimac, J.; Chami, A.A.; Mele, A.; Vidatic, L.; Kriz, J.; Julien, J.P.; et al. Emerging Trends in the Field of Inflammation and Proteinopathy in ALS/FTD Spectrum Disorder. Biomedicines 2023, 11, 1599. [Google Scholar] [CrossRef]
- Buratti, E. Functional Significance of TDP-43 Mutations in Disease. Adv. Genet. 2015, 91, 1–53. [Google Scholar] [CrossRef]
- Peters, O.M.; Ghasemi, M.; Brown, R.H. Emerging mechanisms of molecular pathology in ALS. J. Clin. Investig. 2015, 125, 1767–1779, Erratum in J. Clin. Investig. 2015, 125, 2548. [Google Scholar] [CrossRef]
- Feldman, E.L.; Goutman, S.A.; Petri, S.; Mazzini, L.; Savelieff, M.G.; Shaw, P.J.; Sobue, G. Amyotrophic lateral sclerosis. Lancet 2022, 400, 1363–1380. [Google Scholar] [CrossRef]
- Liscic, R.M.; Alberici, A.; Cairns, N.J.; Romano, M.; Buratti, E. From basic research to the clinic: Innovative therapies for ALS and FTD in the pipeline. Mol. Neurodegener. 2020, 15, 31. [Google Scholar] [CrossRef]
- Rezaie, T.; Child, A.; Hitchings, R.; Brice, G.; Miller, L.; Coca-Prados, M.; Heon, E.; Krupin, T.; Ritch, R.; Kreutzer, D.; et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science 2002, 295, 1077–1079. [Google Scholar] [CrossRef]
- Maruyama, H.; Morino, H.; Ito, H.; Izumi, Y.; Kato, H.; Watanabe, Y.; Kinoshita, Y.; Kamada, M.; Nodera, H.; Suzuki, H.; et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 2010, 465, 223–226. [Google Scholar] [CrossRef]
- Slowicka, K.; Vereecke, L.; van Loo, G. Cellular Functions of Optineurin in Health and Disease. Trends Immunol. 2016, 37, 621–633. [Google Scholar] [CrossRef]
- Markovinovic, A.; Cimbro, R.; Ljutic, T.; Kriz, J.; Rogelj, B.; Munitic, I. Optineurin in amyotrophic lateral sclerosis: Multifunctional adaptor protein at the crossroads of different neuroprotective mechanisms. Prog. Neurobiol. 2017, 154, 1–20. [Google Scholar] [CrossRef]
- Xiao, Y.; Tan, Y.; Li, C.; Wei, Q.; Jiang, Q.; Wang, S.; Yang, T.; Lin, J.; Zhang, L.; Shang, H. Genetic and clinical analysis of OPTN in amyotrophic lateral sclerosis. J. Med. Genet. 2025, 62, 242–248. [Google Scholar] [CrossRef]
- Ying, H.; Yue, B.Y. Cellular and molecular biology of optineurin. Int. Rev. Cell Mol. Biol. 2012, 294, 223–258. [Google Scholar] [CrossRef]
- Prtenjaca, N.; Dominovic, M.; Peradinovic, J.; Šajn, R.; Markovinovic, A.; Munitic, I. Optineurin dysfunction in amyotrophic lateral sclerosis: Why so puzzling. Period. Biol. 2020, 121, 23–34. [Google Scholar] [CrossRef]
- Ito, Y.; Ofengeim, D.; Najafov, A.; Das, S.; Saberi, S.; Li, Y.; Hitomi, J.; Zhu, H.; Chen, H.; Mayo, L.; et al. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science 2016, 353, 603–608. [Google Scholar] [CrossRef]
- Liu, D.; Webber, H.C.; Bian, F.; Xu, Y.; Prakash, M.; Feng, X.; Yang, M.; Yang, H.; You, I.J.; Li, L.; et al. Optineurin-facilitated axonal mitochondria delivery promotes neuroprotection and axon regeneration. Nat. Commun. 2025, 16, 1789. [Google Scholar] [CrossRef]
- Mohovic, N.; Peradinovic, J.; Markovinovic, A.; Cimbro, R.; Minic, Z.; Dominovic, M.; Jakovac, H.; Nimac, J.; Rogelj, B.; Munitic, I. Neuroimmune characterization of optineurin insufficiency mouse model during ageing. Sci. Rep. 2023, 13, 11840. [Google Scholar] [CrossRef]
- Slowicka, K.; Vereecke, L.; Mc Guire, C.; Sze, M.; Maelfait, J.; Kolpe, A.; Saelens, X.; Beyaert, R.; van Loo, G. Optineurin deficiency in mice is associated with increased sensitivity to Salmonella but does not affect proinflammatory NF-kappaB signaling. Eur. J. Immunol. 2016, 46, 971–980. [Google Scholar] [CrossRef]
- Kurashige, T.; Kuramochi, M.; Ohsawa, R.; Yamashita, Y.; Shioi, G.; Morino, H.; Kamada, M.; Ayaki, T.; Ito, H.; Sotomaru, Y.; et al. Optineurin defects cause TDP43-pathology with autophagic vacuolar formation. Neurobiol. Dis. 2020, 148, 105215. [Google Scholar] [CrossRef]
- McCall, A.L.; Dhindsa, J.S.; Pucci, L.A.; Kahn, A.F.; Fusco, A.F.; Biswas, D.D.; Strickland, L.M.; Tseng, H.C.; ElMallah, M.K. Respiratory pathology in the Optn−/− mouse model of Amyotrophic Lateral Sclerosis. Respir. Physiol. Neurobiol. 2020, 282, 103525. [Google Scholar] [CrossRef]
- Moharir, S.C.; Swarup, G. Optineurin deficiency induces patchy hair loss but it is not sufficient to cause amyotrophic lateral sclerosis in mice. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166470. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; McWhirter, S.M.; Faia, K.L.; Rowe, D.C.; Latz, E.; Golenbock, D.T.; Coyle, A.J.; Liao, S.M.; Maniatis, T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003, 4, 491–496. [Google Scholar] [CrossRef]
- Qin, L.; Wu, X.; Block, M.L.; Liu, Y.; Breese, G.R.; Hong, J.S.; Knapp, D.J.; Crews, F.T. Systemic LPS causes chronic neuroinflammation and progressive neurodegeneration. Glia 2007, 55, 453–462. [Google Scholar] [CrossRef]
- Swarup, V.; Phaneuf, D.; Dupre, N.; Petri, S.; Strong, M.; Kriz, J.; Julien, J.P. Deregulation of TDP-43 in amyotrophic lateral sclerosis triggers nuclear factor kappaB-mediated pathogenic pathways. J. Exp. Med. 2011, 208, 2429–2447. [Google Scholar] [CrossRef]
- Peradinovic, J.; Mohovic, N.; Bulic, K.; Markovinovic, A.; Cimbro, R.; Munitic, I. Ageing-Induced Decline in Primary Myeloid Cell Phagocytosis Is Unaffected by Optineurin Insufficiency. Biology 2023, 12, 240. [Google Scholar] [CrossRef]
- Heneka, M.T.; Kummer, M.P.; Latz, E. Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol. 2014, 14, 463–477. [Google Scholar] [CrossRef]
- Gao, C.; Jiang, J.; Tan, Y.; Chen, S. Microglia in neurodegenerative diseases: Mechanism and potential therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 359. [Google Scholar] [CrossRef]
- McCauley, M.E.; Baloh, R.H. Inflammation in ALS/FTD pathogenesis. Acta Neuropathol. 2019, 137, 715–730. [Google Scholar] [CrossRef]
- Beers, D.R.; Appel, S.H. Immune dysregulation in amyotrophic lateral sclerosis: Mechanisms and emerging therapies. Lancet Neurol. 2019, 18, 211–220. [Google Scholar] [CrossRef]
- Clarke, B.E.; Patani, R. The microglial component of amyotrophic lateral sclerosis. Brain 2020, 143, 3526–3539. [Google Scholar] [CrossRef]
- Markovinovic, A.; Ljutic, T.; Beland, L.C.; Munitic, I. Optineurin Insufficiency Disbalances Proinflammatory and Anti-inflammatory Factors by Reducing Microglial IFN-beta Responses. Neuroscience 2018, 388, 139–151. [Google Scholar] [CrossRef]
- Gleason, C.E.; Ordureau, A.; Gourlay, R.; Arthur, J.S.; Cohen, P. Polyubiquitin binding to optineurin is required for optimal activation of TANK-binding kinase 1 and production of interferon β. J. Biol. Chem. 2011, 286, 35663–35674. [Google Scholar] [CrossRef]
- Munitic, I.; Giardino Torchia, M.L.; Meena, N.P.; Zhu, G.; Li, C.C.; Ashwell, J.D. Optineurin insufficiency impairs IRF3 but not NF-κB activation in immune cells. J. Immunol. 2013, 191, 6231–6240. [Google Scholar] [CrossRef]
- Meena, N.P.; Zhu, G.; Mittelstadt, P.R.; Giardino Torchia, M.L.; Pourcelot, M.; Arnoult, D.; Ashwell, J.D.; Munitic, I. The TBK1-binding domain of optineurin promotes type I interferon responses. FEBS Lett. 2016, 590, 1498–1508. [Google Scholar] [CrossRef]
- Prtenjaca, N.; Rob, M.; Alam, M.S.; Markovinovic, A.; Stuani, C.; Buratti, E.; Munitic, I. Optineurin Deficiency and Insufficiency Lead to Higher Microglial TDP-43 Protein Levels. Int. J. Mol. Sci. 2022, 23, 6829. [Google Scholar] [CrossRef]
- Sudhakar, C.; Nagabhushana, A.; Jain, N.; Swarup, G. NF-kappaB mediates tumor necrosis factor alpha-induced expression of optineurin, a negative regulator of NF-kappaB. PLoS ONE 2009, 4, e5114. [Google Scholar] [CrossRef]
- Okun, E.; Griffioen, K.J.; Mattson, M.P. Toll-like receptor signaling in neural plasticity and disease. Trends Neurosci. 2011, 34, 269–281. [Google Scholar] [CrossRef]
- Owens, R.; Grabert, K.; Davies, C.L.; Alfieri, A.; Antel, J.P.; Healy, L.M.; McColl, B.W. Divergent Neuroinflammatory Regulation of Microglial TREM Expression and Involvement of NF-κB. Front. Cell Neurosci. 2017, 11, 56. [Google Scholar] [CrossRef]
- Toth, R.P.; Atkin, J.D. Dysfunction of Optineurin in Amyotrophic Lateral Sclerosis and Glaucoma. Front. Immunol. 2018, 9, 1017. [Google Scholar] [CrossRef]
- Li, F.; Xu, D.; Wang, Y.; Zhou, Z.; Liu, J.; Hu, S.; Gong, Y.; Yuan, J.; Pan, L. Structural insights into the ubiquitin recognition by OPTN (optineurin) and its regulation by TBK1-mediated phosphorylation. Autophagy 2018, 14, 66–79. [Google Scholar] [CrossRef]
- Brenner, D.; Sieverding, K.; Bruno, C.; Lüningschrör, P.; Buck, E.; Mungwa, S.; Fischer, L.; Brockmann, S.J.; Ulmer, J.; Bliederhäuser, C.; et al. Heterozygous Tbk1 loss has opposing effects in early and late stages of ALS in mice. J. Exp. Med. 2019, 216, 267–278. [Google Scholar] [CrossRef]
- Wild, P.; Farhan, H.; McEwan, D.G.; Wagner, S.; Rogov, V.V.; Brady, N.R.; Richter, B.; Korac, J.; Waidmann, O.; Choudhary, C.; et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011, 333, 228–233. [Google Scholar] [CrossRef]
- Wong, Y.C.; Holzbaur, E.L. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. USA 2014, 111, E4439–E4448. [Google Scholar] [CrossRef]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef]
- Chew, T.S.; O’Shea, N.R.; Sewell, G.W.; Oehlers, S.H.; Mulvey, C.M.; Crosier, P.S.; Godovac-Zimmermann, J.; Bloom, S.L.; Smith, A.M.; Segal, A.W. Optineurin deficiency contributes to impaired cytokine secretion and neutrophil recruitment in bacteria driven colitis. Dis. Model. Mech. 2015, 8, 817–829. [Google Scholar] [CrossRef]
- Pourcelot, M.; Zemirli, N.; Silva Da Costa, L.; Loyant, R.; Garcin, D.; Vitour, D.; Munitic, I.; Vazquez, A.; Arnoult, D. The Golgi apparatus acts as a platform for TBK1 activation after viral RNA sensing. BMC Biol. 2016, 14, 69. [Google Scholar] [CrossRef]
- Wang, J.; Hong, W.; Zhang, L.; Song, L.; Shi, Q.; Shao, Y.; Hao, G.; Fang, C.; Qiu, Y.; Yang, L.; et al. Optineurin modulates the maturation of dendritic cells to regulate autoimmunity through JAK2-STAT3 signaling. Nat. Commun. 2021, 12, 6198. [Google Scholar] [CrossRef]
- Hu, X.; Wong, S.W.; Liang, K.; Wu, T.H.; Wang, S.; Wang, L.; Liu, J.; Yamauchi, M.; Foster, B.L.; Ting, J.P.; et al. Optineurin regulates NRF2-mediated antioxidant response in a mouse model of Paget’s disease of bone. Sci. Adv. 2023, 9, eade6998. [Google Scholar] [CrossRef]
- Génin, P.; Cuvelier, F.; Lambin, S.; Côrte-Real Filipe, J.; Autrusseau, E.; Laurent, C.; Laplantine, E.; Weil, R. Optineurin regulates the interferon response in a cell cycle-dependent manner. PLoS Pathog. 2015, 11, e1004877, Erratum in PLoS Pathog. 2015, 11, e1004971. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- He, Y.; Yao, X.; Taylor, N.; Bai, Y.; Lovenberg, T.; Bhattacharya, A. RNA sequencing analysis reveals quiescent microglia isolation methods from postnatal mouse brains and limitations of BV2 cells. J. Neuroinflamm. 2018, 15, 153. [Google Scholar] [CrossRef]
- Kaneko, Y.S.; Ota, A.; Nakashima, A.; Nagasaki, H.; Kodani, Y.; Mori, K.; Nagatsu, T. Lipopolysaccharide treatment arrests the cell cycle of BV-2 microglial cells in G1 phase and protects them from UV light-induced apoptosis. J. Neural. Transm. 2015, 122, 187–199. [Google Scholar] [CrossRef]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e17. [Google Scholar] [CrossRef]
- Brenner, D.; Sieverding, K.; Srinidhi, J.; Zellner, S.; Secker, C.; Yilmaz, R.; Dyckow, J.; Amr, S.; Ponomarenko, A.; Tunaboylu, E.; et al. A TBK1 variant causes autophagolysosomal and motoneuron pathology without neuroinflammation in mice. J. Exp. Med. 2024, 221, e20221190. [Google Scholar] [CrossRef]
- Lenoel, I.; Ribon, M.; Lorenc, F.; Diebold, A.; Philibert, C.E.; Robaldo, D.; Badsi, M.; Perronnet, J.; Lameth, J.; Berriat, F.; et al. ALS/FTD-linked TBK1 deficiency in microglia induces an aged-like microglial signature and drives social recognition deficits in mice. Nat. Commun. 2025, 16, 7951. [Google Scholar] [CrossRef]
- Dosanjh, A.; Won, C.Y. Amlexanox: A Novel Therapeutic for Atopic, Metabolic, and Inflammatory Disease. Yale J. Biol. Med. 2020, 93, 759–763. [Google Scholar]
- Duan, Q.Q.; Wang, H.; Su, W.M.; Gu, X.J.; Shen, X.F.; Jiang, Z.; Ren, Y.L.; Cao, B.; Li, G.B.; Wang, Y.; et al. TBK1, a prioritized drug repurposing target for amyotrophic lateral sclerosis: Evidence from druggable genome Mendelian randomization and pharmacological verification in vitro. BMC Med. 2024, 22, 96. [Google Scholar] [CrossRef]
- Freischmidt, A.; Wieland, T.; Richter, B.; Ruf, W.; Schaeffer, V.; Muller, K.; Marroquin, N.; Nordin, F.; Hubers, A.; Weydt, P.; et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 2015, 18, 631–636. [Google Scholar] [CrossRef]
- Cirulli, E.T.; Lasseigne, B.N.; Petrovski, S.; Sapp, P.C.; Dion, P.A.; Leblond, C.S.; Couthouis, J.; Lu, Y.F.; Wang, Q.; Krueger, B.J.; et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 2015, 347, 1436–1441. [Google Scholar] [CrossRef]
- Boutej, H.; Rahimian, R.; Thammisetty, S.S.; Béland, L.C.; Lalancette-Hébert, M.; Kriz, J. Diverging mRNA and Protein Networks in Activated Microglia Reveal SRSF3 Suppresses Translation of Highly Upregulated Innate Immune Transcripts. Cell Rep. 2017, 21, 3220–3233. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Shippy, D.C.; Ulland, T.K. Lipid metabolism transcriptomics of murine microglia in Alzheimer’s disease and neuroinflammation. Sci. Rep. 2023, 13, 14800. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Alexander, S.P.H.; Fabbro, D.; Kelly, E.; Mathie, A.A.; Peters, J.A.; Veale, E.L.; Armstrong, J.F.; Faccenda, E.; Harding, S.D.; Davies, J.A.; et al. The Concise Guide to PHARMACOLOGY 2023/24: Enzymes. Br. J. Pharmacol. 2023, 180 (Suppl. 2), S289–S373. [Google Scholar] [CrossRef]
- Apic, G. SysWiz database and software tool providing an insight into mechanism of action of molecules associated with ciliopathies. Cilia 2012, 1 (Suppl. 1), P12. [Google Scholar] [CrossRef]
- Kühnel, M.P.; Cosovic, B.; Medic, G.; Russell, R.B.; Apic, G. Unraveling Mechanisms of Toxicity with the Power of Pathways: ToxWiz Tool as an Illustrative Example. In Pathway Analysis for Drug Discovery; Wiley: Hoboken, NJ, USA, 2008; pp. 195–217. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cappelli, S.; Peradinovic, J.; Mohovic, N.; Mandal, P.; Stuani, C.; Longo, A.; Cannon, J.R.; Baloni, P.; Leoni, B.; Krsmanovic, T.; et al. Optineurin Shapes Basal and LPS-Induced Transcriptomes in BV2 Microglia. Int. J. Mol. Sci. 2025, 26, 10453. https://doi.org/10.3390/ijms262110453
Cappelli S, Peradinovic J, Mohovic N, Mandal P, Stuani C, Longo A, Cannon JR, Baloni P, Leoni B, Krsmanovic T, et al. Optineurin Shapes Basal and LPS-Induced Transcriptomes in BV2 Microglia. International Journal of Molecular Sciences. 2025; 26(21):10453. https://doi.org/10.3390/ijms262110453
Chicago/Turabian StyleCappelli, Sara, Josip Peradinovic, Nikolina Mohovic, Purba Mandal, Cristiana Stuani, Alessandra Longo, Jason R. Cannon, Priyanka Baloni, Benedetta Leoni, Tamara Krsmanovic, and et al. 2025. "Optineurin Shapes Basal and LPS-Induced Transcriptomes in BV2 Microglia" International Journal of Molecular Sciences 26, no. 21: 10453. https://doi.org/10.3390/ijms262110453
APA StyleCappelli, S., Peradinovic, J., Mohovic, N., Mandal, P., Stuani, C., Longo, A., Cannon, J. R., Baloni, P., Leoni, B., Krsmanovic, T., Stojanov, K., Apic, G., Russell, R. B., Romano, M., Buratti, E., & Munitic, I. (2025). Optineurin Shapes Basal and LPS-Induced Transcriptomes in BV2 Microglia. International Journal of Molecular Sciences, 26(21), 10453. https://doi.org/10.3390/ijms262110453








