Trans-Omic Analysis Identifies the ‘PRMT1–STAT3–Integrin αVβ6 Axis’ as a Novel Therapeutic Target in Tacrolimus-Induced Chronic Nephrotoxicity
Abstract
1. Introduction
2. Results
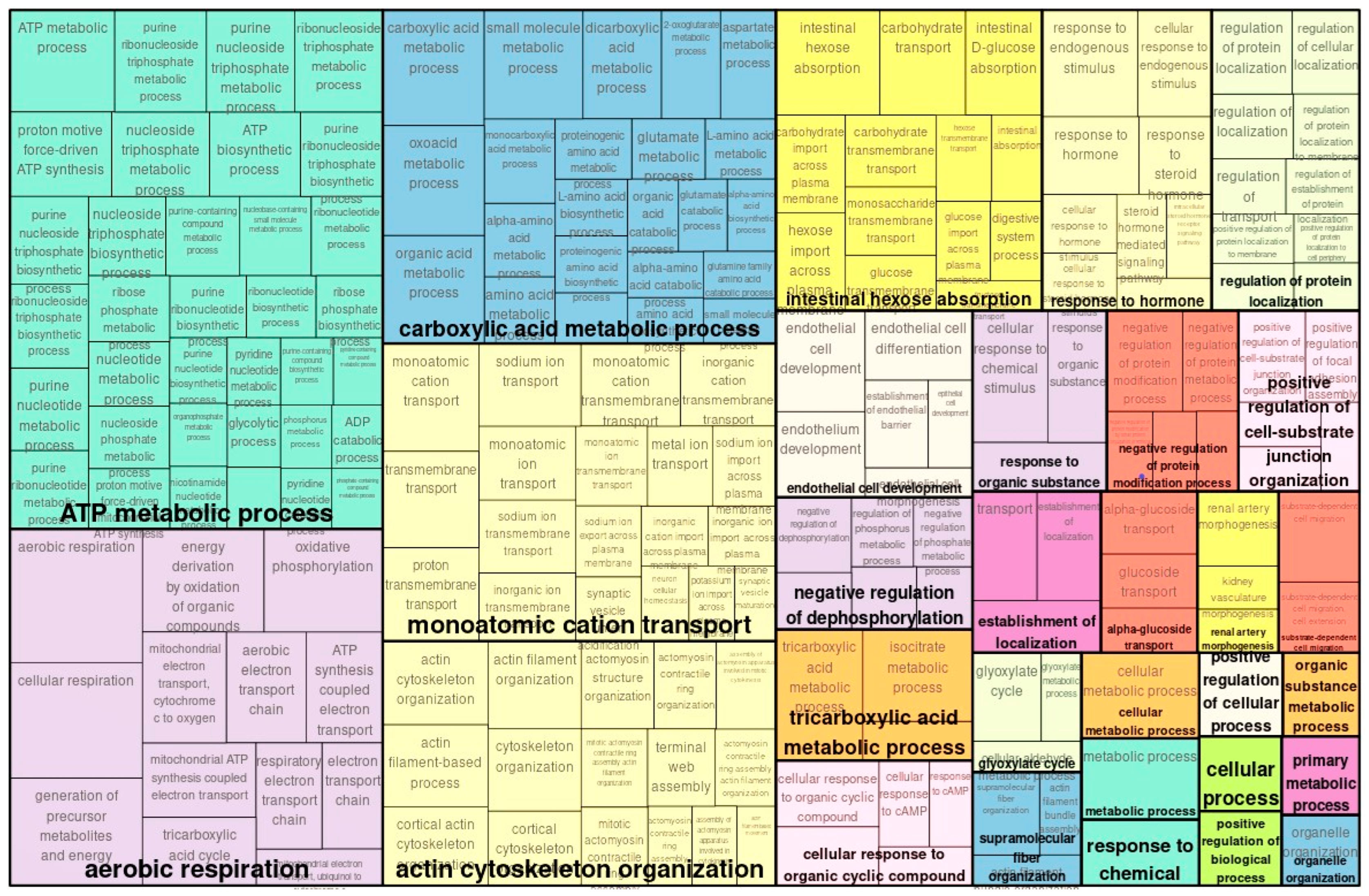
2.1. Transcriptome Analysis and GO Analysis Functional Classification of Genes
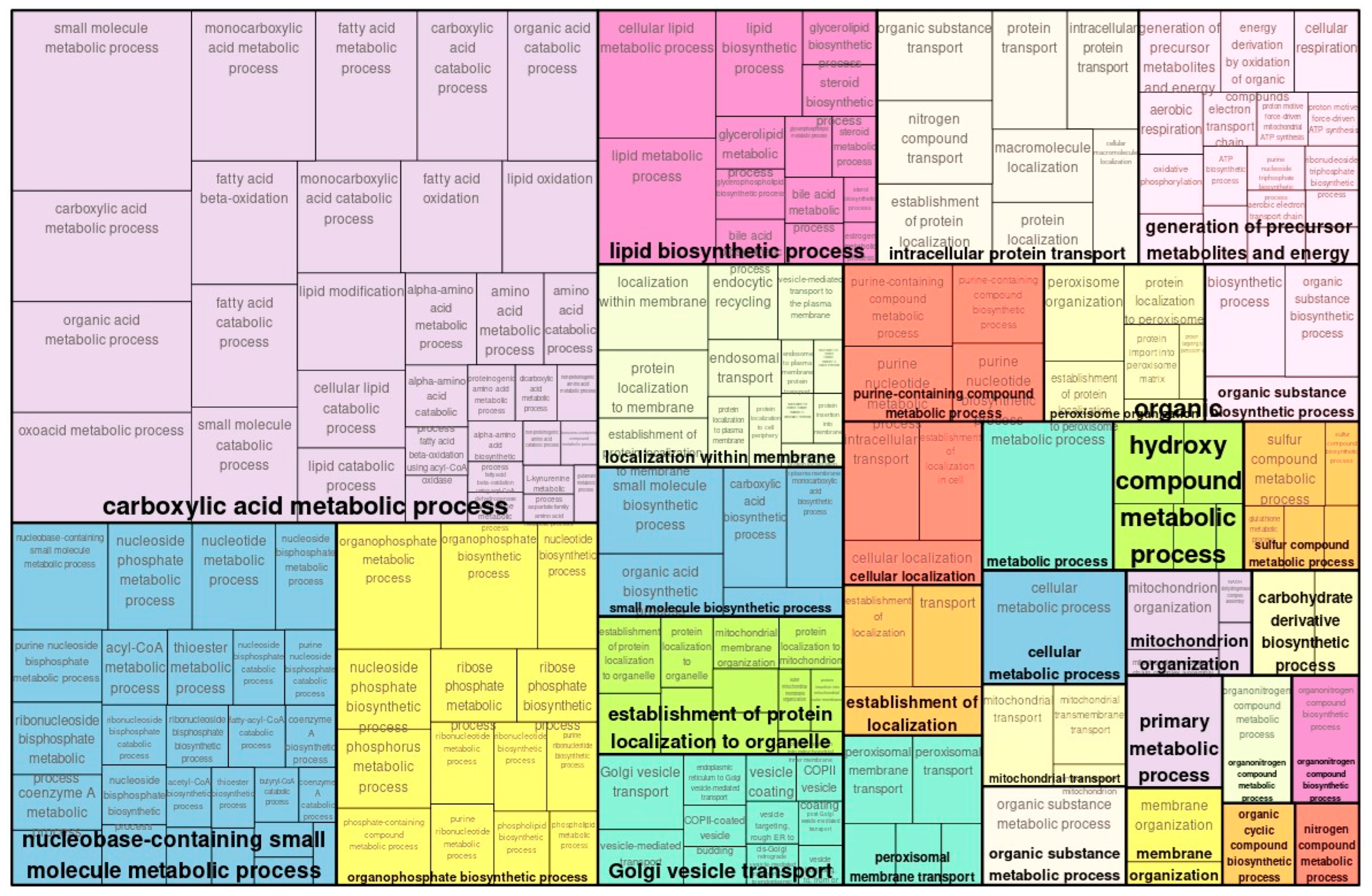
2.2. Proteome Analysis and GO Analysis of Proteins
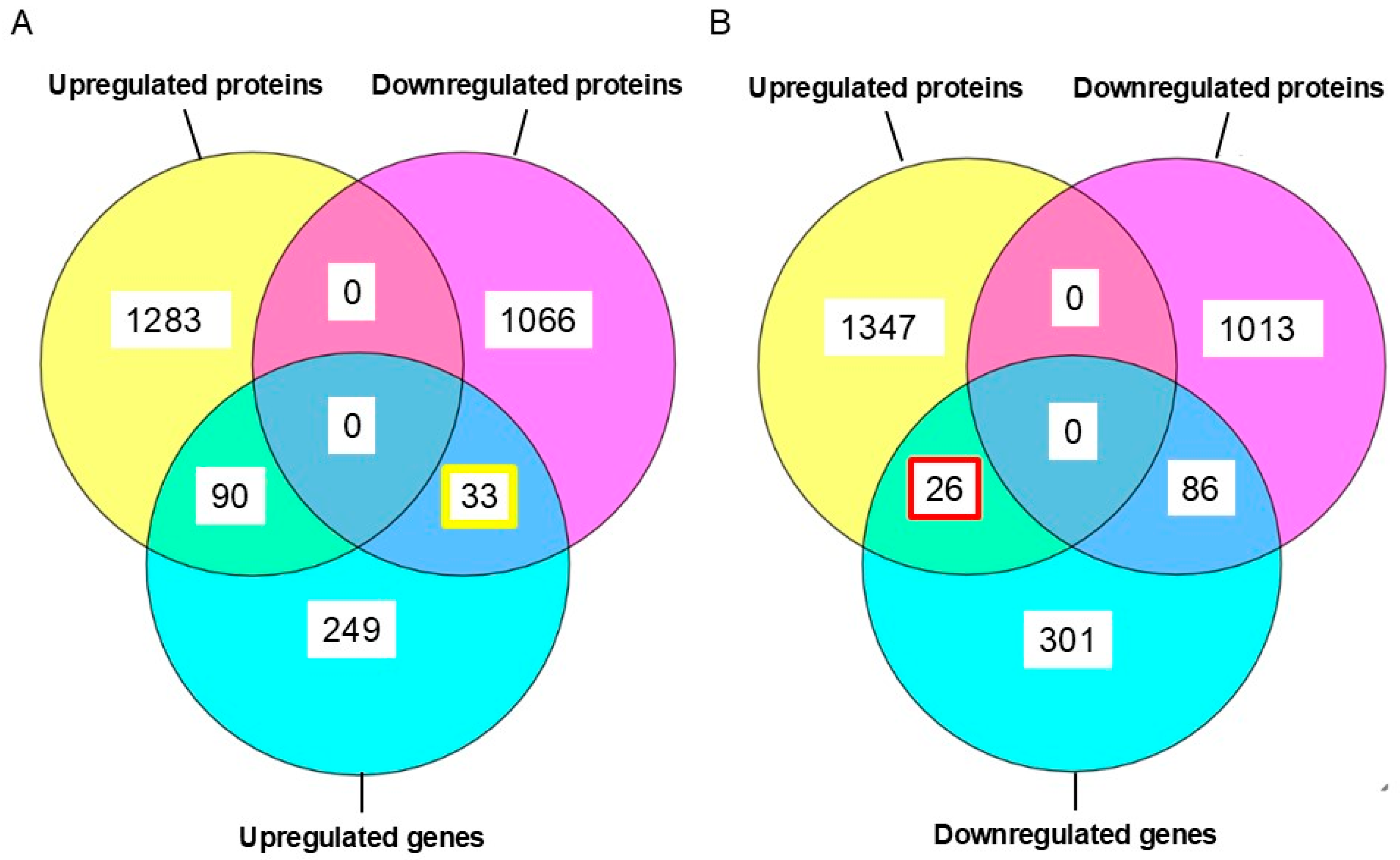
2.3. Comparison of Transcriptomic and Proteomic Expression
2.4. Detecting Transcription Factors
2.5. Identification of Master Regulators Through Upstream Network Analysis
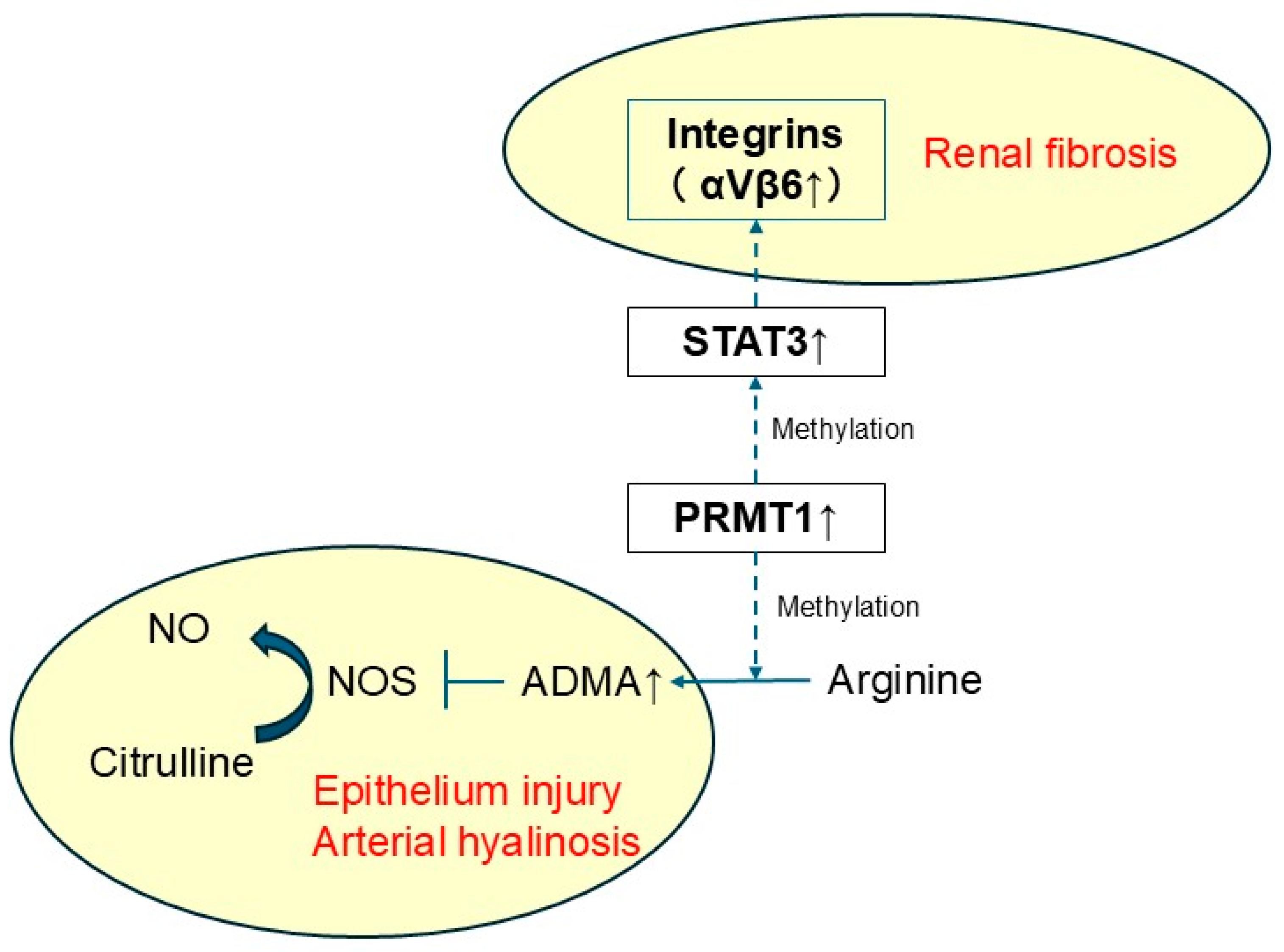
2.6. Identification of Therapeutic Target Molecules in Master Regulators
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Sample Preparation for Transcriptome Analysis
4.3. Sample Preparation for Proteome Analysis
4.4. LC-MS/MS
| Condition | MS 1 | MS 2 |
| m/z | 495–745 | More than 200 |
| Mass resolution | 30,000 | 30,000 |
| Auto gain control target | 3 × 106 | 3 × 106 |
| Maximum injection time | 55 ms | Auto |
| Fixed normalized collision energy | - | 23% |
4.5. Data Processing
4.6. Data Analysis Using Genome Enhancer
4.6.1. Databases Used in the Study
4.6.2. Methods for Analysis of Enriched Transcription Factor Binding Sites and Composite Modules
4.6.3. Methods for Finding Master Regulators in Networks
4.6.4. Methods for Analysis of Pharmaceutical Compounds
4.6.5. Methods for Analysis of Known Pharmaceutical Compounds
- (i)
- Ranking by “Target activity score” (T-scorePSD);
- (ii)
- Ranking by “Disease activity score” (D-scorePSD);
- (iii)
- Ranking by “Clinical validity score”.
4.6.6. Method for Prediction of Pharmaceutical Compounds
- (i)
- Toxicity below a chosen toxicity threshold (defined as Pa, probability to be active as a toxic substance).
- (ii)
- For all predicted pharmacological effects that correspond to a set of user-selected disease(s), Pa is greater than a chosen effect threshold.
- (iii)
- There are at least 2 targets (corresponding to the predicted activity mechanisms) with predicted Pa greater than a chosen target threshold.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADMA | Asymmetric dimethyl arginine |
| CCND1 | Cyclin D1 |
| CDK4 | Cyclin-dependent kinase 4 |
| CKD | Chronic kidney disease |
| CMA | Composite Module Analyst |
| CSF1R | Colony-stimulating factor 1 receptor |
| DUSP6 | Dual-specificity phosphatase 6 |
| EN1 | Engrailed homeobox 1 |
| EOMES | Eomesodermin |
| FDRs | False discovery rates |
| FIGLA | Folliculogenesis-specific bHLH transcription factor |
| FLI1 | Fli-1 proto-oncogene, ETS transcription factor |
| HIC1 | HIC ZBTB transcriptional repressor 1 |
| HNF4A | Hepatocyte nuclear factor 4 alpha |
| HSF1 | Heat shock transcription factor 1 |
| HSPA1A | Heat shock protein family A(Hsp70) member 1A |
| KLF4 | KLF transcription factor 4 |
| LEF1 | Lymphoid enhancer-binding factor 1 |
| MEIS1 | Meis homebox 1 |
| MZF1 | Myeloid zinc finger 1 |
| NAD+ | Nicotinamide adenine dinucleotide |
| NF2 | NF2, moesin–ezrin–radixin-like (MERLIN) tumor suppressor |
| NFATC1 | Nuclear factor of activated T cells 1 |
| NFIC | Nuclear factor I C |
| NKX2-5 | NK2 homeobox 5 |
| NO | Nitric oxide |
| NOS | Nitric oxide synthase |
| NR2F1 | Nuclear receptor subfamily 2 group F member 1 |
| PAX2 | Paired box 2 |
| PRMT1 | Protein arginine methyltransferase-1 |
| PTPRD | Protein tyrosine phosphatasereceptor type D |
| PWMs | Position weight matrices |
| SAE1, | SUMO1-activating enzyme subunit 1 |
| SIAH1 | Siah E3 ubiquitin proteinligase 1 |
| SIRT2 | Sirtuin 2 |
| SOX9 | SRY-box transcription factor 9 |
| STAT3 | Signal transducer and activator of transcription 3 |
| SUMO3, | Small ubiquitin-like modifier 3, |
| TAC | Tacrolimus |
| TACN | Tacrolimus-induced chronic nephrotoxicity |
| TFs | Transcription factors |
| TFBSs | Transcription factor binding sites |
| TFCP2 | Transcription factor CP2 |
| UBA2 | Ubiquitin-like modifier-activating enzyme 2 |
| UBE2J1 | Ubiquitin-conjugating enzyme E2 J1 |
| VDR | Vitamin D receptor |
References
- Aizawa, K.; Kimura, N.; Goda, T.; Nishida, S.; Sakuma, Y.; Iwami, D.; Nagai, R. Analytical Validation of an LC-MS/MS Method for Simultaneous Quantification of Multiple Immunosuppressants in Microvolume Whole Blood. Int. J. Mol. Sci. 2025, 26, 6358. [Google Scholar] [CrossRef]
- Dheer, D.; Jyoti; Gupta, P.N.; Shankar, R. Tacrolimus: An updated review on delivering strategies for multifarious diseases. Eur. J. Pharm. Sci. 2018, 114, 217–227. [Google Scholar] [CrossRef]
- Baumgart, D.C.; Pintoffl, J.P.; Sturm, A.; Wiedenmann, B.; Dignass, A.U. Tacrolimus is safe and effective in patients with severe steroid-refractory or steroid-dependent inflammatory bowel disease—A long-term follow-up. Am. J. Gastroenterol. 2006, 101, 1048–1056. [Google Scholar] [CrossRef]
- Ekberg, H.; Tedesco-Silva, H.; Demirbas, A.; Vítko, Š.; Nashan, B.; Gürkan, A.; Margreiter, R.; Hugo, C.; Grinyó, J.M.; Frei, U.; et al. Reduced Exposure to Calcineurin Inhibitors in Renal Transplantation. N. Engl. J. Med. 2007, 357, 2562–2575. [Google Scholar] [CrossRef]
- Lentine, K.L.; Smith, J.M.; Lyden, G.R.; Miller, J.M.; Booker, S.E.; Dolan, T.G.; Temple, K.R.; Weiss, S.; Handarova, D.; Israni, A.K.; et al. OPTN/SRTR 2023 Annual Data Report: Kidney. Am. J. Transplant. 2025, 25, S22–S137. [Google Scholar] [CrossRef]
- Nishida, S.; Ishima, T.; Kimura, N.; Iwami, D.; Nagai, R.; Imai, Y.; Aizawa, K. Metabolomic Profiling of Mice with Tacrolimus-Induced Nephrotoxicity: Carnitine Deficiency in Renal Tissue. Biomedicines 2024, 12, 521. [Google Scholar] [CrossRef] [PubMed]
- Bentata, Y. Tacrolimus: 20 years of use in adult kidney transplantation. What we should know about its nephrotoxicity. Artif. Organs 2020, 44, 140–152. [Google Scholar] [CrossRef] [PubMed]
- Farouk, S.S.; Rein, J.L. The Many Faces of Calcineurin Inhibitor Toxicity—What the FK? Adv. Chronic Kidney Dis. 2020, 27, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Randhawa, M.B.B.S.; Shapiro, R.; Jordan, M.L.; Starzl, T.E.; Demetris, A.J. The Histopathological Changes Associated with Allograft Rejection and Drug Toxicity in Renal Transplant Recipients Maintained on FK506. Clinical significance and comparison with cyclosporine. Am. J. Surg. Pathol. 1993, 17, 60–68. [Google Scholar] [CrossRef]
- Abbiss, H.; Maker, G.L.; Trengove, R.D. Metabolomics Approaches for the Diagnosis and Understanding of Kidney Diseases. Metabolites 2019, 9, 34. [Google Scholar] [CrossRef]
- Hanifa, M.A.; Skott, M.; Maltesen, R.G.; Rasmussen, B.S.; Nielsen, S.; Frøkiær, J.; Ring, T.; Wimmer, R. Tissue, urine and blood metabolite signatures of chronic kidney disease in the 5/6 nephrectomy rat model. Metabolomics 2019, 15, 112. [Google Scholar] [CrossRef]
- Khattri, R.B.; Thome, T.; Ryan, T.E. Tissue-Specific 1H-NMR Metabolomic Profiling in Mice with Adenine-Induced Chronic Kidney Disease. Metabolites 2021, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Si, S.; Liu, H.; Xu, L.; Zhan, S. Identification of novel therapeutic targets for chronic kidney disease and kidney function by integrating multi-omics proteome with transcriptome. Genome Med. 2024, 16, 84. [Google Scholar] [CrossRef]
- Dubin, R.F.; Deo, R.; Ren, Y.; Wang, J.; Zheng, Z.; Shou, H.; Go, A.S.; Parsa, A.; Lash, J.P.; Rahman, M.; et al. Proteomics of CKD progression in the chronic renal insufficiency cohort. Nat. Commun. 2023, 14, 6340. [Google Scholar] [CrossRef]
- Checa-Ros, A.; Locascio, A.; Steib, N.; Okojie, O.J.; Malte-Weier, T.; Bermúdez, V.; D’Marco, L. In silico medicine and -omics strategies in nephrology: Contributions and relevance to the diagnosis and prevention of chronic kidney disease. Kidney Res. Clin. Pract. 2025, 44, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Nishida, S.; Ishima, T.; Iwami, D.; Nagai, R.; Aizawa, K. Whole Blood Metabolomic Profiling of Mice with Tacrolimus-Induced Chronic Nephrotoxicity: NAD+ Depletion with Salvage Pathway Impairment. Antioxidants 2025, 14, 62. [Google Scholar] [CrossRef] [PubMed]
- Koshiba, S.; Motoike, I.N.; Saigusa, D.; Inoue, J.; Aoki, Y.; Tadaka, S.; Shirota, M.; Katsuoka, F.; Tamiya, G.; Minegishi, N.; et al. Identification of critical genetic variants associated with metabolic phenotypes of the Japanese population. Commun. Biol. 2020, 3, 662. [Google Scholar] [CrossRef]
- Lemke, O.; Heineike, B.M.; Viknander, S.; Cohen, N.; Li, F.; Steenwyk, J.L.; Spranger, L.; Agostini, F.; Lee, C.T.; Aulakh, S.K.; et al. The role of metabolism in shaping enzyme structures over 400 million years. Nature 2025, 644, 280–289. [Google Scholar] [CrossRef]
- Noda-Garcia, L.; Liebermeister, W.; Tawfik, D.S. Metabolite-Enzyme Coevolution: From Single Enzymes to Metabolic Pathways and Networks. Annu. Rev. Biochem. 2018, 87, 187–216. [Google Scholar] [CrossRef]
- Demirci, H.; Popovic, S.; Dittmayer, C.; Yilmaz, D.E.; El-Shimy, I.A.; Mülleder, M.; Hinze, C.; Su, M.; Mertins, P.; Kirchner, M.; et al. Immunosuppression with cyclosporine versus tacrolimus shows distinctive nephrotoxicity profiles within renal compartments. Acta Physiol. 2024, 240, e14190. [Google Scholar] [CrossRef]
- Yen, N.T.H.; Phat, N.K.; Oh, J.H.; Park, S.M.; Moon, K.S.; Thu, V.T.A.; Cho, Y.S.; Shin, J.G.; Long, N.P.; Kim, D.H. Pathway-level multi-omics analysis of the molecular mechanisms underlying the toxicity of long-term tacrolimus exposure. Toxicol. Appl. Pharmacol. 2023, 473, 116597. [Google Scholar] [CrossRef]
- Waleev, T.; Shtokalo, D.; Konovalova, T.; Voss, N.; Cheremushkin, E.; Stegmaier, P.; Kel-Margoulis, O.; Wingender, E.; Kel, A. Composite Module Analyst: Identification of transcription factor binding site combinations using genetic algorithm. Nucleic Acids Res. 2006, 34, W541–W545. [Google Scholar] [CrossRef] [PubMed]
- Kel, A.E.; Stegmaier, P.; Valeev, T.; Koschmann, J.; Poroikov, V.; Kel-Margoulis, O.V.; Wingender, E. Multi-omics “upstream analysis” of regulatory genomic regions helps identifying targets against methotrexate resistance of colon cancer. EuPA Open Proteom. 2016, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Michael, H.; Hogan, J.; Kel, A.; Kel-Margoulis, O.; Schacherer, F.; Voss, N.; Wingender, E. Building a knowledge base for systems pathology. Brief. Bioinform. 2008, 9, 518–531. [Google Scholar] [CrossRef] [PubMed]
- Filimonov, D.; Poroikov, V.; Borodina, Y.; Gloriozova, T. ChemInform Abstract: Chemical Similarity Assessment Through Multilevel Neighborhoods of Atoms: Definition and Comparison with the Other Descriptors. ChemInform. 1999, 39, 666–670. [Google Scholar] [CrossRef]
- Bock, F.; Li, S.; Pozzi, A.; Zent, R. Integrins in the kidney—Beyond the matrix. Nat. Rev. Nephrol. 2025, 21, 157–174. [Google Scholar] [CrossRef]
- Hahm, K.; Lukashev, M.E.; Luo, Y.; Yang, W.J.; Dolinski, B.M.; Weinreb, P.H.; Simon, K.J.; Chun Wang, L.; Leone, D.R.; Lobb, R.R.; et al. Alphav beta6 integrin regulates renal fibrosis and inflammation in Alport mouse. Am. J. Pathol. 2007, 170, 110–125. [Google Scholar] [CrossRef]
- Trevillian, P.; Paul, H.; Millar, E.; Hibberd, A.; Agrez, M.V. alpha(v)beta(6) Integrin expression in diseased and transplanted kidneys. Kidney Int. 2004, 66, 1423–1433. [Google Scholar] [CrossRef]
- Zambruno, G.; Marchisio, P.C.; Marconi, A.; Vaschieri, C.; Melchiori, A.; Giannetti, A.; De Luca, M. Transforming growth factor-beta 1 modulates beta 1 and beta 5 integrin receptors and induces the de novo expression of the alpha v beta 6 heterodimer in normal human keratinocytes: Implications for wound healing. J. Cell Biol. 1995, 129, 853–865. [Google Scholar] [CrossRef]
- Ma, L.J.; Yang, H.; Gaspert, A.; Carlesso, G.; Barty, M.M.; Davidson, J.M.; Sheppard, D.; Fogo, A.B. Transforming growth factor-beta-dependent and -independent pathways of induction of tubulointerstitial fibrosis in beta6(-/-) mice. Am. J. Pathol. 2003, 163, 1261–1273. [Google Scholar] [CrossRef]
- Henderson, N.C.; Arnold, T.D.; Katamura, Y.; Giacomini, M.M.; Rodriguez, J.D.; McCarty, J.H.; Pellicoro, A.; Raschperger, E.; Betsholtz, C.; Ruminski, P.G.; et al. Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat. Med. 2013, 19, 1617–1624. [Google Scholar] [CrossRef]
- Zhu, C.; Zheng, R.; Han, X.; Tang, Z.; Li, F.; Hu, X.; Lin, R.; Shen, J.; Pei, Q.; Wang, R.; et al. Knockout of integrin αvβ6 protects against renal inflammation in chronic kidney disease by reduction of pro-inflammatory macrophages. Cell Death Dis. 2024, 15, 397. [Google Scholar] [CrossRef] [PubMed]
- Tain, Y.L.; Hsu, C.N. Toxic Dimethylarginines: Asymmetric Dimethylarginine (ADMA) and Symmetric Dimethylarginine (SDMA). Toxins 2017, 9, 92. [Google Scholar] [CrossRef]
- Oliva-Damaso, E.; Oliva-Damaso, N.; Rodriguez-Esparragon, F.; Payan, J.; Baamonde-Laborda, E.; Gonzalez-Cabrera, F.; Santana-Estupiñan, R.; Rodriguez-Perez, J.C. Asymmetric (ADMA) and Symmetric (SDMA) Dimethylarginines in Chronic Kidney Disease: A Clinical Approach. Int. J. Mol. Sci. 2019, 20, 3668. [Google Scholar] [CrossRef]
- Vallance, P.; Leone, A.; Calver, A.; Collier, J.; Moncada, S. Accumulation of an endogenous inhibitor of nitric oxide synthesis in chronic renal failure. Lancet 1992, 339, 572–575. [Google Scholar] [CrossRef]
- Zoccali, C.; Bode-Böger, S.; Mallamaci, F.; Benedetto, F.; Tripepi, G.; Malatino, L.; Cataliotti, A.; Bellanuova, I.; Fermo, I.; Frölich, J.; et al. Plasma concentration of asymmetrical dimethylarginine and mortality in patients with end-stage renal disease: A prospective study. Lancet 2001, 358, 2113–2117. [Google Scholar] [CrossRef]
- Jayachandran, I.; Sundararajan, S.; Venkatesan, S.; Paadukaana, S.; Balasubramanyam, M.; Mohan, V.; Manickam, N. Asymmetric dimethylarginine (ADMA) accelerates renal cell fibrosis under high glucose condition through NOX4/ROS/ERK signaling pathway. Sci. Rep. 2020, 10, 16005. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Wu, N.; Lv, C.; Wang, Y. P4HB regulates the TGFβ/SMAD3 signaling pathway through PRMT1 to participate in high glucose-induced epithelial-mesenchymal transition and fibrosis of renal tubular epithelial cells. BMC Nephrol. 2024, 25, 297. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yu, C.; Zhuang, S. Protein arginine methyltransferase 1 mediates renal fibroblast activation and fibrogenesis through activation of Smad3 signaling. Am. J. Physiol. Renal Physiol. 2020, 318, F375–F387. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Chen, H.; Su, B.; Zhang, L.; Hu, J.; Wang, Q.; Zhuang, S. PRMT1-mediated BRD4 arginine methylation and phosphorylation promote partial epithelial-mesenchymal transformation and renal fibrosis. FASEB J. 2025, 39, e70293. [Google Scholar] [CrossRef]
- Honda, M.; Nakashima, K.; Katada, S. PRMT1 regulates astrocytic differentiation of embryonic neural stem/precursor cells. J. Neurochem. 2017, 142, 901–907. [Google Scholar] [CrossRef]
- Xu, M.; Chen, X.; Yin, H.; Yin, L.; Liu, F.; Fu, Y.; Yao, J.; Deng, X. Cloning and characterization of the human integrin β6 gene promoter. PLoS ONE 2015, 10, e0121439. [Google Scholar] [CrossRef]
- Meecham, A.; Marshall, J.F. The ITGB6 gene: Its role in experimental and clinical biology. Gene X 2020, 5, 100023. [Google Scholar] [CrossRef]
- Solez, K.; Colvin, R.B.; Racusen, L.C.; Haas, M.; Sis, B.; Mengel, M.; Halloran, P.F.; Baldwin, W.; Banfi, G.; Collins, A.B.; et al. Banff 07 classification of renal allograft pathology: Updates and future directions. Am. J. Transplant. 2008, 8, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Liu, J.; Wu, Q.; Wang, B.; Hu, T.; Li, Y.; Yan, X.; Ma, L.; Tan, Z. A dual αvβ1/αvβ6 integrin inhibitor Bexotegrast (PLN-74809) ameliorates organ injury and fibrogenesis in fibrotic kidney disease. Eur. J. Pharmacol. 2024, 983, 176983. [Google Scholar] [CrossRef]
- Wu, J.; Li, D.; Wang, L. Overview of PRMT1 modulators: Inhibitors and degraders. Eur. J. Med. Chem. 2024, 279, 116887. [Google Scholar] [CrossRef]
- Dominici, C.; Sgarioto, N.; Yu, Z.; Sesma-Sanz, L.; Masson, J.Y.; Richard, S.; Raynal, N.J. Synergistic effects of type I PRMT and PARP inhibitors against non-small cell lung cancer cells. Clin. Epigenet. 2021, 13, 54. [Google Scholar] [CrossRef]
- Fedoriw, A.; Rajapurkar, S.R.; O’Brien, S.; Gerhart, S.V.; Mitchell, L.H.; Adams, N.D.; Rioux, N.; Lingaraj, T.; Ribich, S.A.; Pappalardi, M.B.; et al. Anti-tumor Activity of the Type I PRMT Inhibitor, GSK3368715, Synergizes with PRMT5 Inhibition through MTAP Loss. Cancer Cell 2019, 36, 100–114.e25. [Google Scholar] [CrossRef] [PubMed]
- Yuk, N.; Jung, H.J. Inhibition of PRMT1 Suppresses the Growth of U87MG-Derived Glioblastoma Stem Cells by Blocking the STAT3 Signaling Pathway. Int. J. Mol. Sci. 2024, 25, 2950. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, C.; Van der Hauwaert, C.; Dewaeles, E.; Franczak, J.; Hennino, M.F.; Gnemmi, V.; Savary, G.; Tavernier, Q.; Nottet, N.; Paquet, A.; et al. Tacrolimus-induced nephrotoxicity in mice is associated with microRNA deregulation. Arch. Toxicol. 2018, 92, 1539–1550. [Google Scholar] [CrossRef]
- Isogai, N.; Shiono, Y.; Kuramoto, T.; Yoshioka, K.; Ishihama, H.; Funao, H.; Nakamura, M.; Matsumoto, M.; Ishii, K. Potential osteomyelitis biomarkers identified by plasma metabolome analysis in mice. Sci. Rep. 2020, 10, 839. [Google Scholar] [CrossRef]
- Zhou, Q.; Kerbl-Knapp, J.; Zhang, F.; Korbelius, M.; Kuentzel, K.B.; Vujić, N.; Akhmetshina, A.; Hörl, G.; Paar, M.; Steyrer, E.; et al. Metabolomic Profiles of Mouse Tissues Reveal an Interplay between Aging and Energy Metabolism. Metabolites 2021, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, Y.; Okada, F.; Takamiya, R.; Kibe, N.; Tsunoda, S.; Nakajima, O.; Toyoda, K.; Nagae, R.; Suematsu, M.; Soga, T.; et al. Rescue of anaemia and autoimmune responses in SOD1-deficient mice by transgenic expression of human SOD1 in erythrocytes. Biochem. J. 2009, 422, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Kel, A.E.; Gössling, E.; Reuter, I.; Cheremushkin, E.; Kel-Margoulis, O.V.; Wingender, E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003, 31, 3576–3579. [Google Scholar] [CrossRef] [PubMed]
- Koschmann, J.; Bhar, A.; Stegmaier, P.; Kel, A.E.; Wingender, E. “Upstream Analysis”: An Integrated Promoter-Pathway Analysis Approach to Causal Interpretation of Microarray Data. Microarrays 2015, 4, 270–286. [Google Scholar] [CrossRef]
- Krull, M.; Pistor, S.; Voss, N.; Kel, A.; Reuter, I.; Kronenberg, D.; Michael, H.; Schwarzer, K.; Potapov, A.; Choi, C.; et al. TRANSPATH: An information resource for storing and visualizing signaling pathways and their pathological aberrations. Nucleic Acids Res. 2006, 34, D546–D551. [Google Scholar] [CrossRef]


| Gene Symbol | Gene Description | Regulatory Score 1 | Yes–No Ratio 2 |
|---|---|---|---|
| HNF4A | Hepatocyte nuclear factor 4 alpha | 3.75 | 1.51 |
| STAT3 | Signal transducer and activator of transcription 3 | 3.33 | 1.87 |
| LEF1 | Lymphoid enhancer-binding factor 1 | 2.43 | 1.67 |
| SOX9 | SRY-box transcription factor 9 | 2.36 | 1.29 |
| NFATC1 | Nuclear factor of activated T cells 1 | 2.21 | 3.61 |
| NR2F1 | Nuclear receptor subfamily 2 group F member 1 | 2.17 | 5 |
| EOMES | Eomesodermin | 1.9 | 5.83 |
| TFCP2 | Transcription factor CP2 | 1.73 | 2.27 |
| EN1 | Engrailed homeobox 1 | 1.25 | 2.31 |
| MZF1 | Myeloid zinc finger 1 | 0.33 | 2.42 |
| MEIS1 | Meis homebox 1 | 0 | 8.33 |
| Gene Symbol | Gene Description | Regulatory Score 1 | Yes–No Ratio 2 |
|---|---|---|---|
| HSF1 | Heat shock transcription factor 1 | 2.57 | 2.75 |
| SMAD3 | SMAD family member 3 | 2.51 | 1.2 |
| SMAD2 | SMAD family member 2 | 2.36 | 1.4 |
| SMAD5 | SMAD family member 5 | 2.28 | 1.26 |
| SMAD1 | SMAD family member 1 | 2.21 | 1.2 |
| SOX9 | SRY-box transcription factor 9 | 2.1 | 3.34 |
| SMAD4 | SMAD family member 4 | 2.08 | 1.2 |
| SMAD9 | SMAD family member 9 | 1.98 | 1.2 |
| SMAD7 | SMAD family member 7 | 1.94 | 1.15 |
| SMAD6 | SMAD family member 6 | 1.78 | 1.2 |
| FLI1 | Fli-1 proto-oncogene, ETS transcription factor | 1.67 | 2.63 |
| HIC1 | HIC ZBTB transcriptional repressor 1 | 1.53 | 1.17 |
| VDR | Vitamin D receptor | 1.41 | 2.32 |
| LEF1 | Lymphoid enhancer-binding factor 1 | 1.41 | 10 |
| KLF4 | KLF transcription factor 4 | 1.35 | 1.89 |
| PAX2 | Paired box 2 | 1.25 | 1.52 |
| NKX2-5 | NK2 homeobox 5 | 0.97 | 1.63 |
| NFIC | Nuclear factor I C | 0.55 | 1.47 |
| FIGLA | Folliculogenesis-specific bHLH transcription factor | 0 | 1.15 |
| Master Molecule Name | Gene Symbol | Gene Description | Total Rank 1 | Log FC (Transcriptome) | Log FC (Proteome) |
|---|---|---|---|---|---|
| HRMT1L2(h) | PRMT1 | Protein arginine methyltransferase 1 | 94 | 0.43 | |
| Integrins | ITGA1, ITGA2B, ITGA3, ITGA4, ITGA5, ITGA6, ITGA8, ITGA9, ITGAL, ITGAV, ITGB1, ITGB2, ITGB3, ITGB4 | Integrin subunit alpha 1, integrin subunit alpha 2b, integrin subunit alpha 3 | 95 | 1.57 | 0.43 |
| PTPRD(h) | PTPRD | Protein tyrosine phosphatase receptor type D | 154 | 0.4 | |
| STAT3(h) | STAT3 | Signal transducer and activator of transcription 3 | 167 | 0.25 | 0.34 |
| Cdk4-isoform1(h): cyclinD1a(h) | CCND1, CDK4 | Cyclin D1, Cyclin-dependent kinase 4 | 176 | 0.47 | 0.38 |
| HRMT1L2-isoform2(h) | PRMT1 | Protein arginine methyltransferase 1 | 182 | 0.43 | |
| HRMT1L2-isoform4(h) | PRMT1 | Protein arginine methyltransferase 1 | 182 | 0.43 | |
| HRMT1L2-isoform1(h) | PRMT1 | Protein arginine methyltransferase 1 | 184 | 0.43 | |
| HRMT1L2-isoform3(h) | PRMT1 | Protein arginine methyltransferase 1 | 184 | 0.43 | |
| SIRT2(h) | SIRT2 | Sirtuin 2 | 198 | 0.27 |
| Master Molecule Name | Gene Symbol | Gene Description | Total Rank 1 | Log FC (Transcriptome) | Log FC (Proteome) |
|---|---|---|---|---|---|
| Ubc9(h)sumo3C93: sumo3(h){clCG92,93} | SUMO3, UBE2I | Small ubiquitin-like modifier 3, Ubiquitin-conjugating enzyme E2 I | 55 | −0.62 | |
| Ubc9{sumo3C93}: sumo3{clCG92,93} | SUMO3, UBE2I | Small ubiquitin-like modifier 3, Ubiquitin-conjugating enzyme E2 I | 81 | −0.62 | |
| M-CSF-1-R(h) | CSF1R | Colony-stimulating factor 1receptor | 99 | −0.48 | |
| M-CSF-1-R-isoform2(h) | CSF1R | Colony-stimulating factor 1receptor | 109 | −0.48 | |
| Aos1(h): SAE2(h)sumo3C173: sumo3(h){clCG92,173} | SAE1, SUMO3, UBA2 | SUMO1-activating enzyme subunit 1, small ubiquitin-like modifier 3, ubiquitin-like modifier-activating enzyme 2 | 140 | −0.62 | |
| MKP3(h) | DUSP6 | Dual-specificity phosphatase 6 | 168 | −0.27 | |
| Siah1(h) | SIAH1 | Siah E3 ubiquitin protein ligase 1 | 169 | −0.44 | |
| Merlin(h) | NF2 | NF2, moesin–ezrin–radixin-like (MERLIN) tumor suppressor | 181 | −0.34 | |
| Ubc6(h) | UBE2J1 | Ubiquitin-conjugating enzyme E2 J1 | 184 | −0.47 | |
| Hsp70-1(h) | HSPA1A | Heat shock protein family A(Hsp70) member 1A | 197 | −2.96 |
| Gene Symbol | Gene Description | Druggability Score | Total Rank 1 | Log FC (Transcriptome) | Log FC (Proteome) |
|---|---|---|---|---|---|
| PRMT1 | Protein arginine methyltransferase 1 | 2 | 94 | 0.43 | |
| ITGAL | Integrin subunit alpha L | 14 | 95 | 1.57 | 0.43 |
| ITGAV | Integrin subunit alpha V | 3 | 95 | 1.57 | 0.43 |
| STAT3 | Signal transducer and activator of transcription 3 | 33 | 167 | 0.25 | 0.34 |
| CCND1 | cyclin D1 | 45 | 176 | 0.47 | 0.38 |
| SIRT2 | Sirtuin 2 | 5 | 198 | 0.27 |
| Gene Symbol | Gene Description | Druggability Score | Total Rank 1 | Log FC (Transcriptome) | Log FC (Proteome) |
|---|---|---|---|---|---|
| PRMT1 | Protein arginine methyltransferase 1 | 0.65 | 94 | 0.43 | |
| ITGAL | Integrin subunit alpha L | 7.77 | 95 | 1.57 | 0.43 |
| ITGB6 | Integrin subunit beta 6 | 5.31 | 95 | 1.57 | 0.43 |
| ITGAV | Integrin subunit alpha V | 5.31 | 95 | 1.57 | 0.43 |
| ITGA1 | Integrin subunit alpha 1 | 5.31 | 95 | 1.57 | 0.43 |
| PTPRD | Protein tyrosine phosphatase receptor type D | 17.02 | 154 | 0.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishida, S.; Ishima, T.; Iwami, D.; Nagai, R.; Aizawa, K. Trans-Omic Analysis Identifies the ‘PRMT1–STAT3–Integrin αVβ6 Axis’ as a Novel Therapeutic Target in Tacrolimus-Induced Chronic Nephrotoxicity. Int. J. Mol. Sci. 2025, 26, 10282. https://doi.org/10.3390/ijms262110282
Nishida S, Ishima T, Iwami D, Nagai R, Aizawa K. Trans-Omic Analysis Identifies the ‘PRMT1–STAT3–Integrin αVβ6 Axis’ as a Novel Therapeutic Target in Tacrolimus-Induced Chronic Nephrotoxicity. International Journal of Molecular Sciences. 2025; 26(21):10282. https://doi.org/10.3390/ijms262110282
Chicago/Turabian StyleNishida, Sho, Tamaki Ishima, Daiki Iwami, Ryozo Nagai, and Kenichi Aizawa. 2025. "Trans-Omic Analysis Identifies the ‘PRMT1–STAT3–Integrin αVβ6 Axis’ as a Novel Therapeutic Target in Tacrolimus-Induced Chronic Nephrotoxicity" International Journal of Molecular Sciences 26, no. 21: 10282. https://doi.org/10.3390/ijms262110282
APA StyleNishida, S., Ishima, T., Iwami, D., Nagai, R., & Aizawa, K. (2025). Trans-Omic Analysis Identifies the ‘PRMT1–STAT3–Integrin αVβ6 Axis’ as a Novel Therapeutic Target in Tacrolimus-Induced Chronic Nephrotoxicity. International Journal of Molecular Sciences, 26(21), 10282. https://doi.org/10.3390/ijms262110282






