GC Content and Thermal Stability of Double-Stranded RNA: Fragments of Microsporidia Vairimorpha ceranae and Nosema bombycis AT-Rich Genes Are Sensitive to Standard Heat Treatment
Abstract
1. Introduction
2. Results
2.1. GC Content in CDSs of Microsporidia V. ceranae and N. bombycis Genes Encoding DNA Replication Enzymes
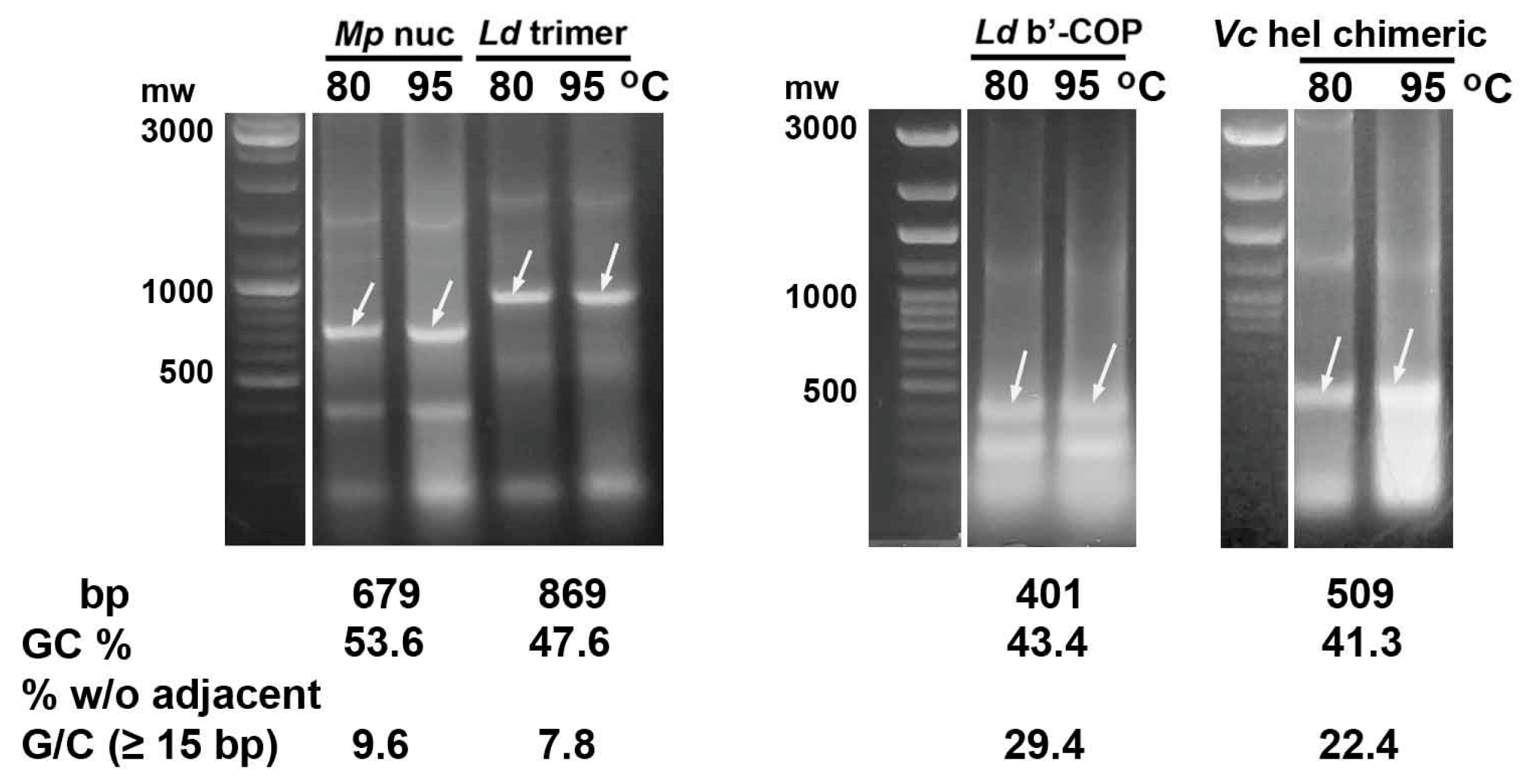
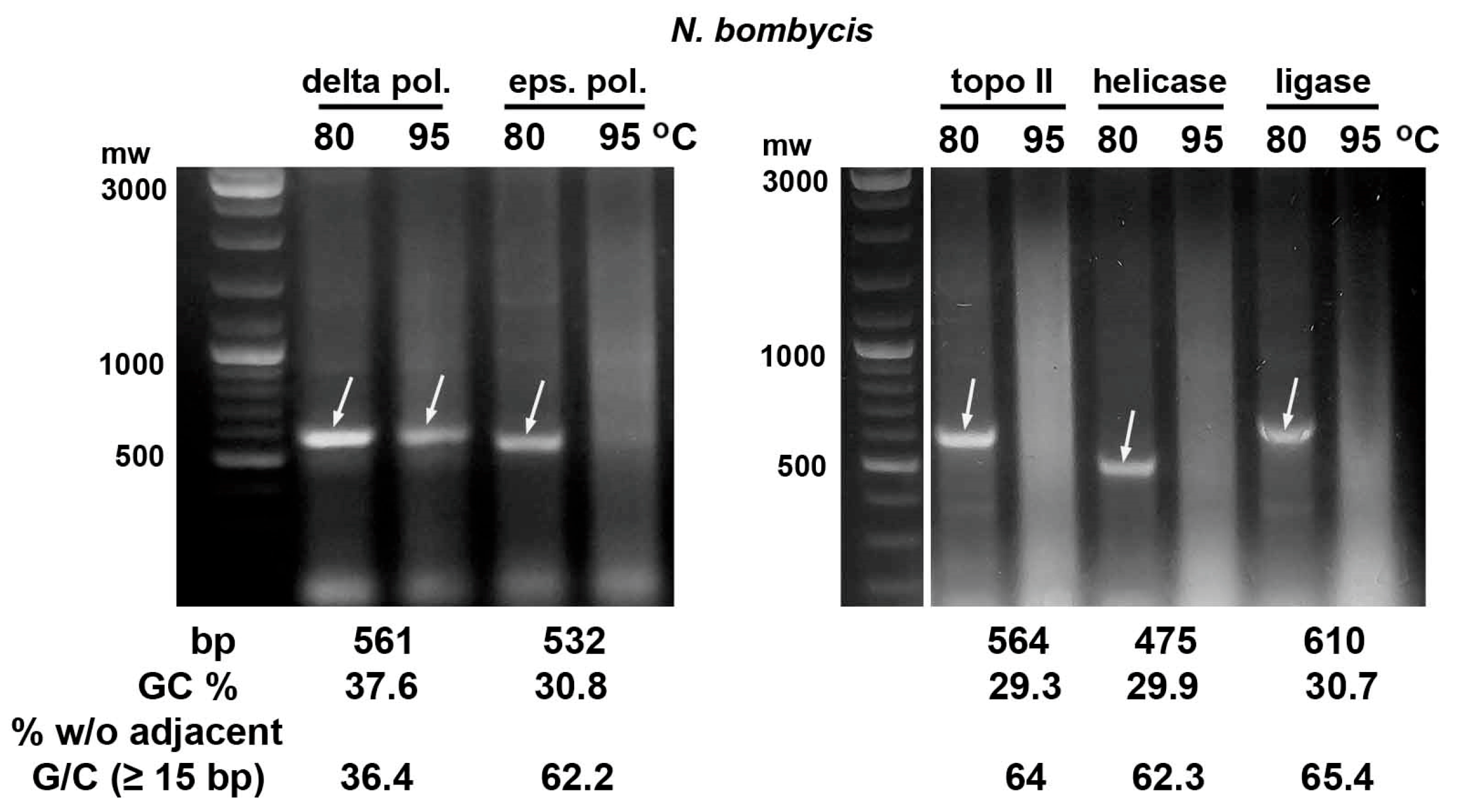
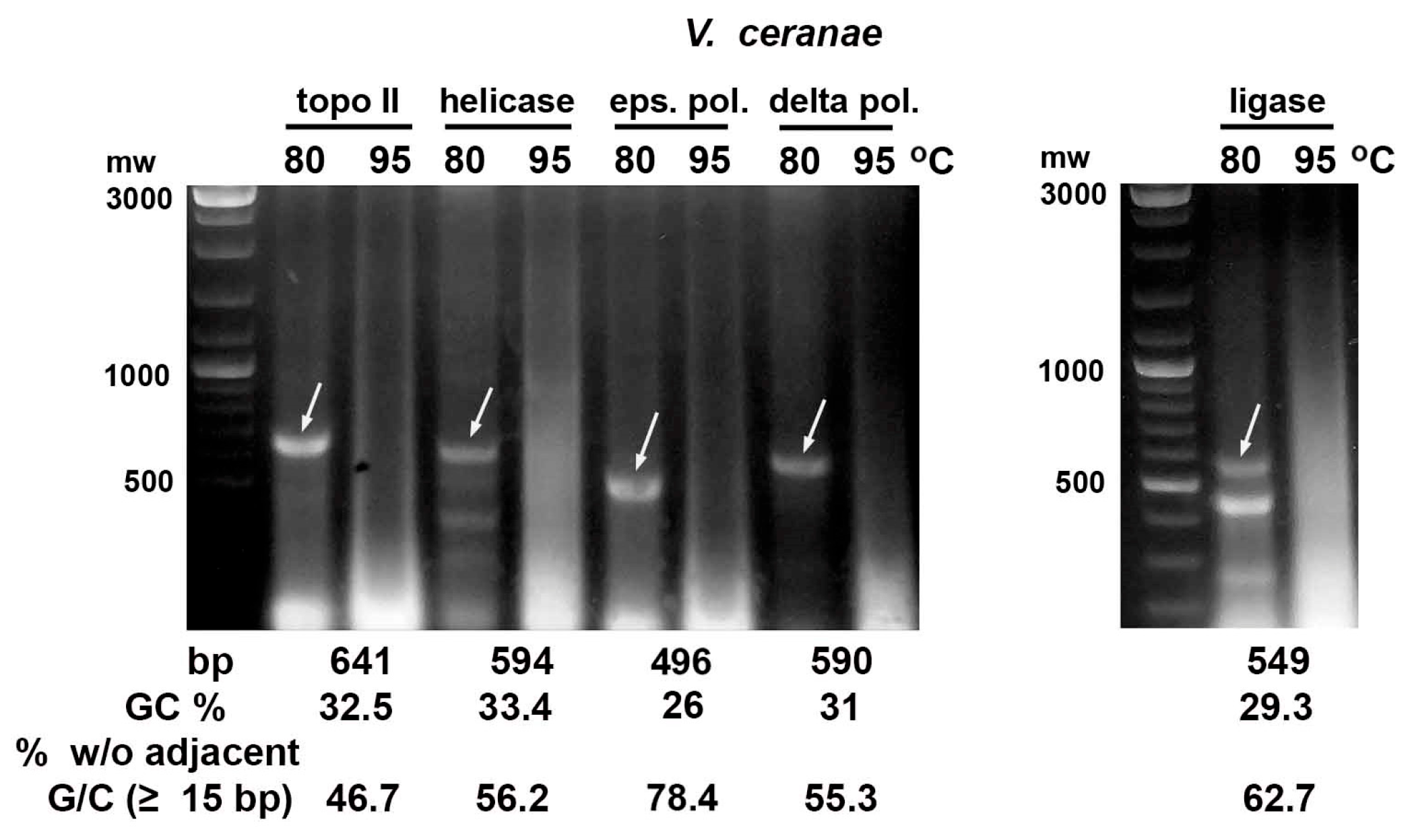
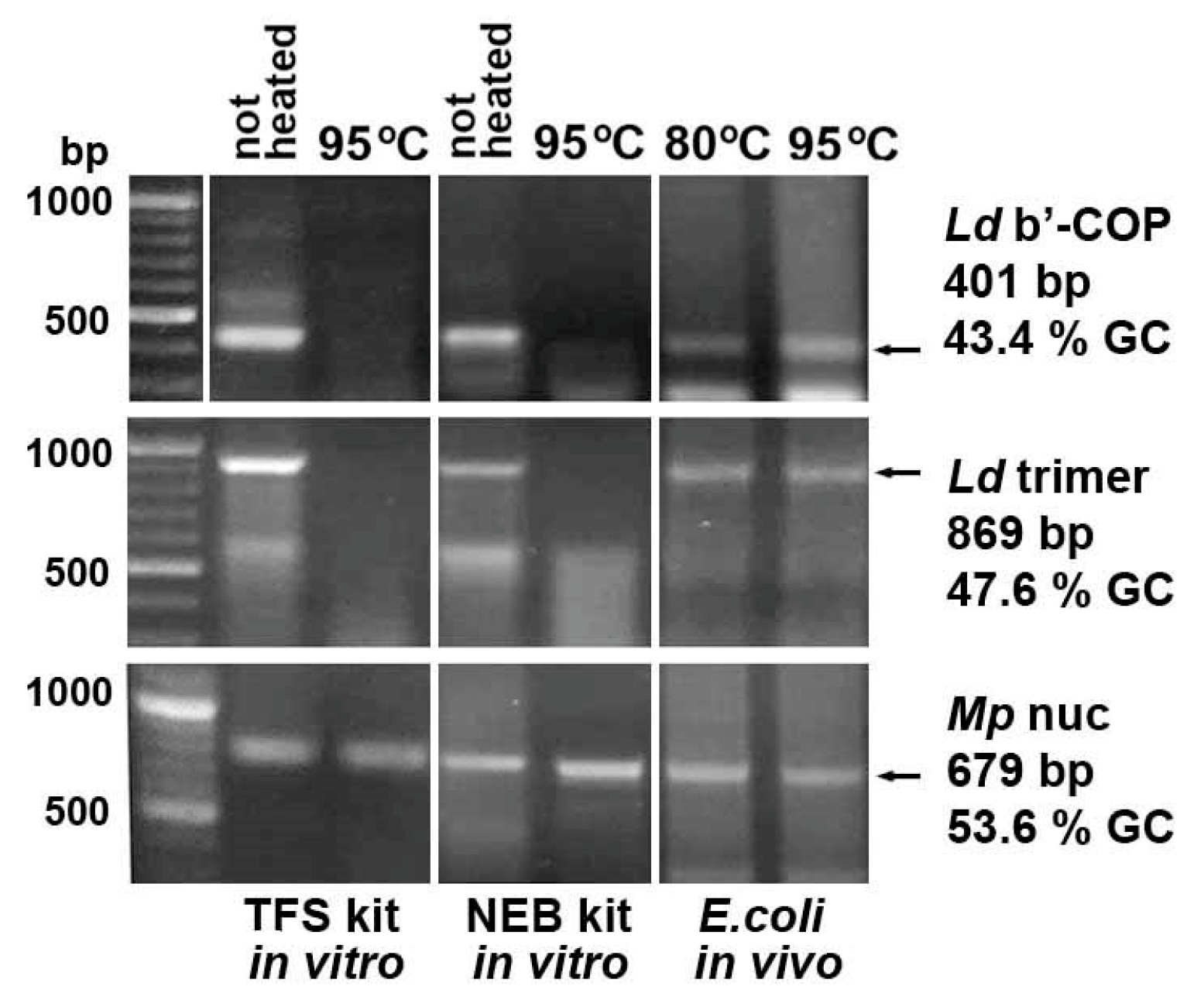
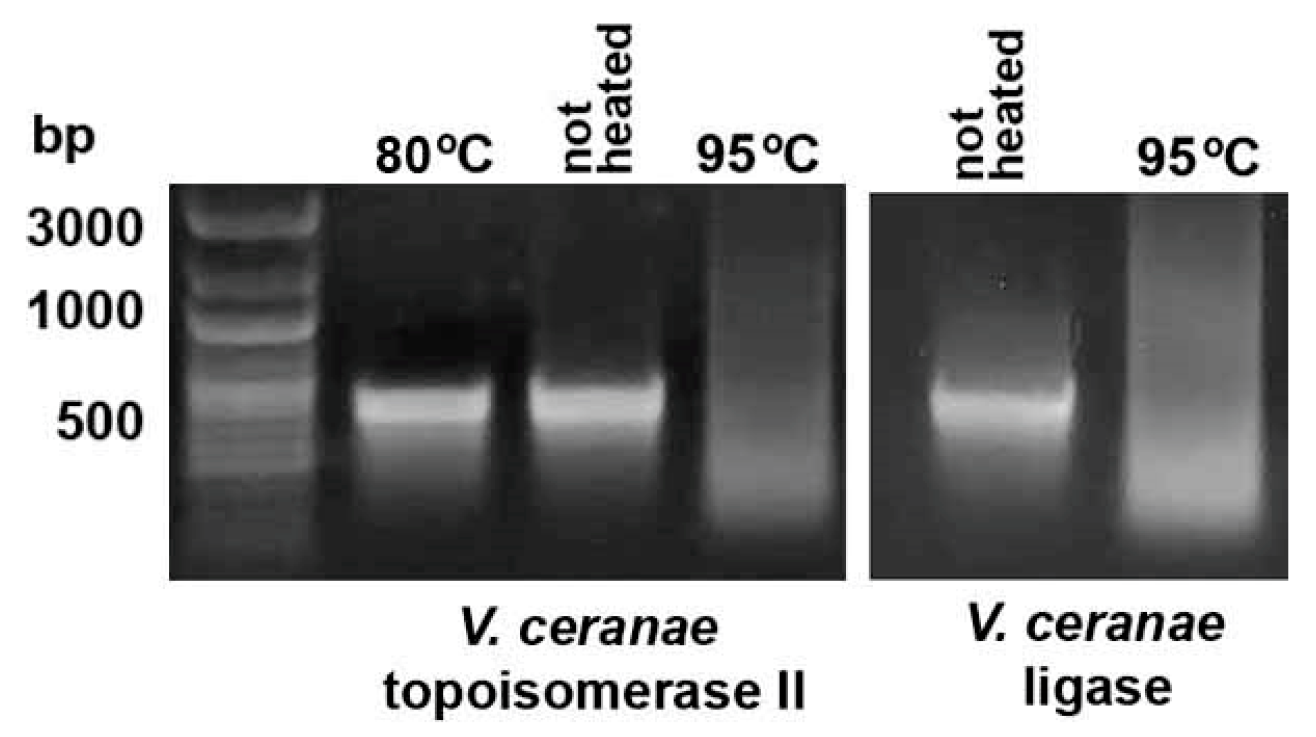
2.2. Heating E. coli HT115 (DE3) Cells Expressing dsRNA Fragments of AT-Enriched Microsporidia Genes at 95 °C Resulted in Their Destruction
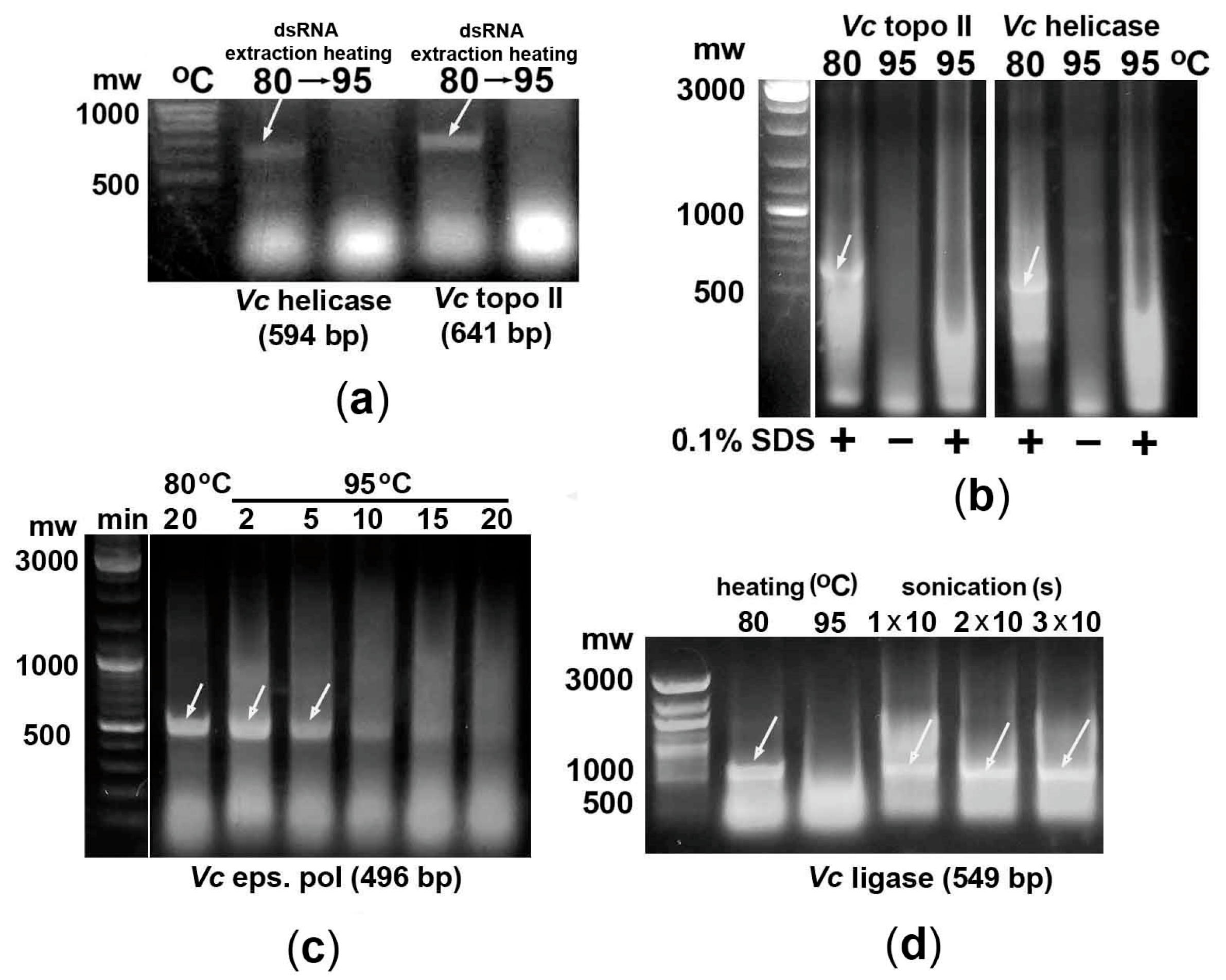
2.3. Effect of Bacteria Heating Time on the Degradation of the dsRNA Fragment of the AT-Rich Gene
2.4. Bacteria Sonication May Be Used to Efficiently Extract dsRNA Fragments of AT-Rich Genes
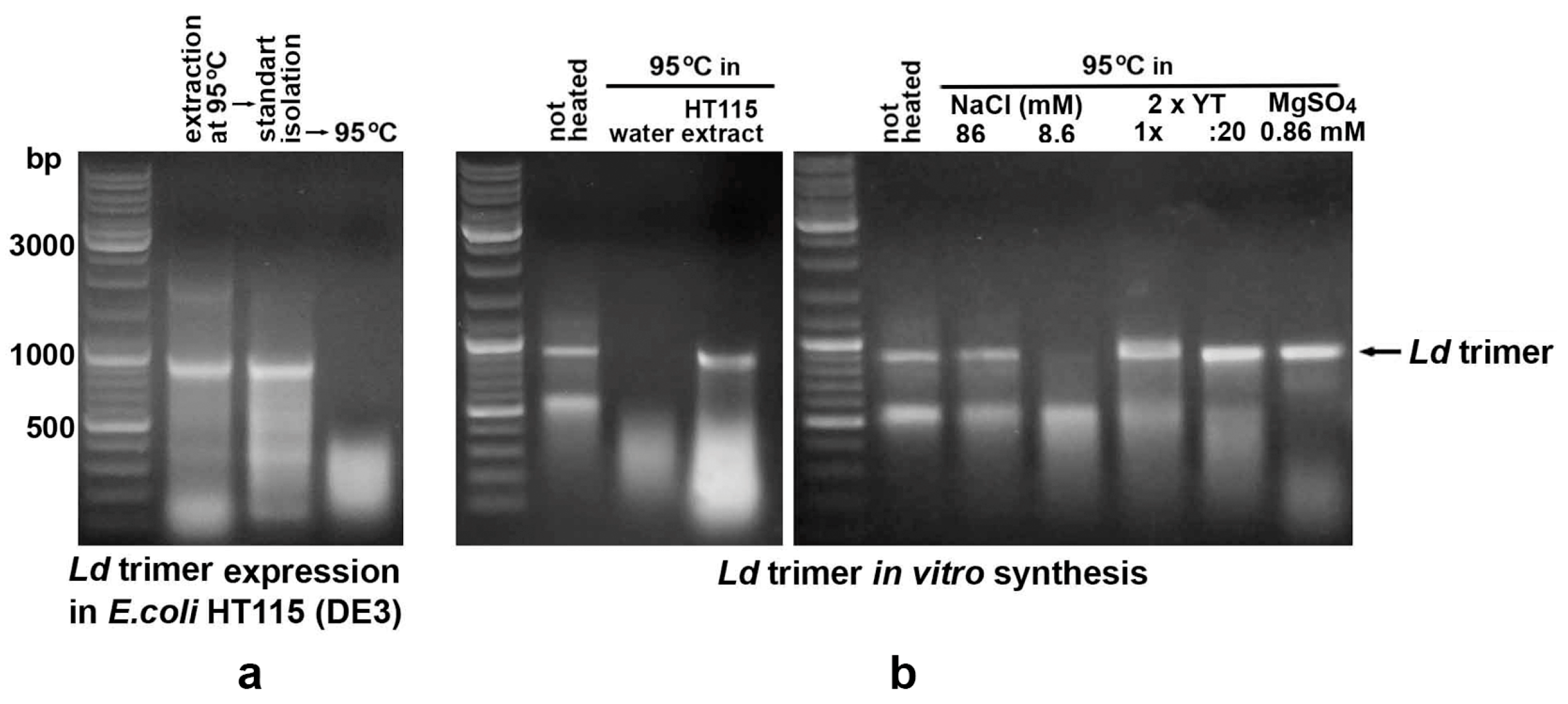
2.5. In Vitro Synthesized dsRNA Fragments Are More Sensitive to High-Temperature Treatment than E. coli-Derived Ones
2.6. Mono- and Divalent Cations Present in the Bacterial Culture Medium Play an Important Role in Stabilizing Heated dsRNA
2.7. Thermal Destruction of In Vitro-Synthesized Ld Trimer dsRNA Containing Fragments of Three L. decemlineata Genes Was Accompanied by a Decrease in Pest-Suppressing Activity
3. Discussion
4. Materials and Methods
4.1. Analysis of GC Content in the Studied Sequences
4.2. Plasmid Construction
4.3. Synthesis of dsRNA in Bacteria E. coli HT115 (DE3)
4.4. In Vitro Synthesis of dsRNA
4.5. Treatment of E. coli Cells and dsRNA
4.6. Analysis of dsRNA in Agarose Gel
4.7. Feeding of L. decemlineata Larvae with dsRNA, Statistical Analysis and Data Visualization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| dsRNA | Double-stranded RNA |
| ssRNA | Single-stranded RNA |
| CDS | Coding sequence |
| SDS | Sodium dodecyl sulfate |
| RISC | RNA-induced repression complex |
| siRNA | Small interfering RNA |
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.B.; Cai, W.J.; Wang, J.W.; Hong, G.J.; Tao, X.Y.; Wang, L.J.; Huang, Y.P.; Chen, X.Y. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007, 25, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Petek, M.; Coll, A.; Ferenc, R.; Razinger, J.; Gruden, K. Validating the potential of double-stranded RNA targeting Colorado potato beetle mesh gene in laboratory and field trial. Front. Plant Sci. 2020, 11, 1250. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Höfle, L.; Werner, B.T.; Imani, J.; Schmidt, A.; Jelonek, L.; Kogel, K.H. SIGS vs HIGS: A study on the efficacy of two dsRNA delivery strategies to silence fusarium FgCYP51 genes in infected host and non-host plants. Mol. Plant Pathol. 2019, 20, 1636–1644. [Google Scholar] [CrossRef]
- Vetukuri, R.R.; Dubey, M.; Kalyandurg, P.B.; Carlsson, A.S.; Whisson, S.C.; Ortiz, R. Spray-induced gene silencing: An innovative strategy for plant trait improvement and disease control. Crop Breed. Appl. Biotechnol. 2021, 21, e387921S11. [Google Scholar] [CrossRef]
- Tenllado, F.; Martínez-García, B.; Vargas, M.; Díaz-Ruíz, J.R. Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol. 2003, 3, 3. [Google Scholar] [CrossRef]
- Akbar, S.; Wei, Y.; Zhang, M.Q. RNA interference: Promising approach to combat plant viruses. Int. J. Mol. Sci. 2022, 23, 5312. [Google Scholar] [CrossRef]
- Desai, S.D.; Eu, Y.J.; Whyard, S.; Currie, R.W. Reduction in deformed wing virus infection in larval and adult honey bees (Apis mellifera L.) by double-stranded RNA ingestion. Insect. Mol. Biol. 2012, 21, 446–455. [Google Scholar] [CrossRef]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.W.; Broeck, J.V. RNA interference in insects: Protecting beneficials and controlling pests. Front. Physiol. 2019, 9, 1912. [Google Scholar] [CrossRef]
- McGruddy, R.A.; Smeele, Z.E.; Manley, B.; Masucci, J.D.; Haywood, J.; Lester, P.J. RNA interference as a next-generation control method for suppressing Varroa destructor reproduction in honey bee (Apis mellifera) hives. Pest. Manag. Sci. 2024, 80, 4770–4778. [Google Scholar] [CrossRef]
- Timmons, L.; Fire, A. Specific interference by ingested dsRNA. Nature 1998, 395, 854. [Google Scholar] [CrossRef]
- Timmons, L.; Court, D.L.; Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 2001, 263, 103–112. [Google Scholar] [CrossRef]
- Ongvarrasopone, C.; Roshorm, Y.; Panyim, S. A simple and cost effective method to generate dsRNA for RNAi studies in invertebrates. Sci. Asia 2007, 33, 35–39. [Google Scholar] [CrossRef]
- Posiri, P.; Ongvarrasopone, C.; Panyim, S. A simple one-step method for producing dsRNA from E. coli to inhibit shrimp virus replication. J. Virol. Methods 2013, 188, 64–69. [Google Scholar] [CrossRef]
- Kim, E.; Park, Y.; Kim, Y. A Transformed bacterium expressing double-stranded RNA specific to integrin β1 enhances Bt toxin efficacy against a polyphagous insect pest, Spodoptera exigua. PLoS ONE 2015, 10, e0132631. [Google Scholar] [CrossRef] [PubMed]
- Vatanparast, M.; Kim, Y. Optimization of recombinant bacteria expressing dsRNA to enhance insecticidal activity against a lepidopteran insect, Spodoptera exigua. PLoS ONE 2017, 12, e0183054. [Google Scholar] [CrossRef]
- Ahn, S.J.; Donahue, K.; Koh, Y.; Martin, R.R.; Choi, M.Y. Microbial-based double-stranded RNA production to develop cost-effective RNA interference application for insect pest management. Int. J. Insect Sci. 2019, 11, 1179543319840323. [Google Scholar] [CrossRef] [PubMed]
- Prates, L.H.F.; Merlau, M.; Rühl-Teichner, J.; Schetelig, M.F.; Häcker, I. An optimized/scale up-ready protocol for extraction of bacterially produced dsRNA at good yield and low costs. Int. J. Mol. Sci. 2023, 24, 9266. [Google Scholar] [CrossRef]
- Haskins, K.A.; Russell, J.F.; Gaddis, N.; Dressman, H.K.; Aballay, A. Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Dev. Cell 2008, 15, 87–97. [Google Scholar] [CrossRef]
- Kawashima, T.; Ikari, N.; Watanabe, Y.; Kubota, Y.; Yoshio, S.; Kanto, T.; Motohashi, S.; Shimojo, N.; Tsuji, N.M. Double-stranded RNA derived from lactic acid bacteria augments Th1 immunity via interferon-β from human dendritic cells. Front. Immunol. 2018, 9, 27. [Google Scholar] [CrossRef]
- Mijatovic-Rustempasic, S.; Tam, K.I.; Kerin, T.K.; Lewis, J.M.; Gautam, R.; Quaye, O.; Gentsch, J.R.; Bowen, M.D. Sensitive and specific quantitative detection of rotavirus A by one-step real-time reverse transcription-PCR assay without antecedent double-stranded-RNA denaturation. J. Clin. Microbiol. 2013, 51, 3047–3054. [Google Scholar] [CrossRef]
- Becskei, A.; Rahaman, S. The life and death of RNA across temperatures. Comput. Struct. Biotechnol. J. 2022, 20, 4325–4336. [Google Scholar] [CrossRef]
- Li, Y.; Breaker, R.R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2’-hydroxyl group. J. Am. Chem. Soc. 1999, 121, 5364–5372. [Google Scholar] [CrossRef]
- Videvall, E. Plasmodium parasites of birds have the most AT-rich genes of eukaryotes. Microb. Genom. 2018, 4, e000150. [Google Scholar] [CrossRef]
- Zhang, L.; Kasif, S.; Cantor, C.R.; Broude, N.E. GC/AT-content spikes as genomic punctuation marks. Proc. Natl. Acad. Sci. USA 2004, 101, 16855–16860. [Google Scholar] [CrossRef]
- Singh, R.; Ming, R.; Yu, Q. Comparative analysis of GC content variations in plant genomes. Trop. Plant Biol. 2016, 9, 136–149. [Google Scholar] [CrossRef]
- Verdonckt, T.W.; Broeck, J.V. Methods for the cost-effective production of bacteria-derived double-stranded RNA for in vitro knockdown studies. Front. Physiol. 2022, 13, 836106. [Google Scholar] [CrossRef] [PubMed]
- Fadeev, R.R.; Kudryavtseva, Y.S.; Bayazyt, K.-D.K.; Shuhalova, A.G.; Dolgikh, V.V. The optimized method to isolate heterologous dsRNA expressed in Escherichia coli HT115 (DE3). Agricult. Biol. 2024, 59, 460–472. [Google Scholar] [CrossRef]
- Draper, D.E. A guide to ions and RNA structure. RNA 2004, 10, 335–343. [Google Scholar] [CrossRef]
- Lambert, D.; Leipply, D.; Shiman, R.; Draper, D.E. The influence of monovalent cation size on the stability of RNA tertiary structures. J. Mol. Biol. 2009, 390, 791–804. [Google Scholar] [CrossRef]
- Fischer, N.M.; Polêto, M.D.; Steuer, J.; Spoel, D. Influence of Na+ and Mg2+ ions on RNA structures studied with molecular dynamics simulations. Nucleic Acids Res. 2018, 46, 4872–4882. [Google Scholar] [CrossRef]
- Kyriacou, R.G.; Mulhair, P.O.; Holland, P.W.H. GC Content Across Insect Genomes: Phylogenetic Patterns, Causes and Consequences. J. Mol. Evol. 2024, 92, 138–152. [Google Scholar] [CrossRef]
- Ruden, D.M. GC content in nuclear-encoded genes and effective number of codons (ENC) are positively correlated in AT-rich species and negatively correlated in GC-rich species. Genes 2025, 16, 432. [Google Scholar] [CrossRef]
- Baum, J.; Papenfuss, A.T.; Mair, G.R.; Janse, C.J.; Vlachou, D.; Waters, A.P.; Cowman, A.F.; Crabb, B.S.; de Koning-Ward, T.F. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 2009, 37, 3788–3798. [Google Scholar] [CrossRef]
- Cissé, O.H.; Pagni, M.; Hauser, P.M. Comparative genomics suggests that the human pathogenic fungus Pneumocystis jirovecii acquired obligate biotrophy through gene loss. Genome Biol. Evol. 2014, 6, 1938–1948. [Google Scholar] [CrossRef]
- Ma, L.; Chen, Z.; Huang, D.W.; Kutty, G.; Ishihara, M.; Wang, H.; Abouelleil, A.; Bishop, L.; Davey, E.; Deng, R.; et al. Genome analysis of three Pneumocystis species reveals adaptation mechanisms to life exclusively in mammalian hosts. Nat. Commun. 2016, 7, 10740. [Google Scholar] [CrossRef]
- Paldi, N.; Glick, E.; Oliva, M.; Zilberberg, Y.; Aubin, L.; Pettis, J.; Chen, Y.; Evans, J.D. Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) colony declines. Appl. Environ. Microbiol. 2010, 76, 5960–5964. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-García, C.; Evans, J.D.; Li, W.; Branchiccela, B.; Li, J.H.; Heerman, M.C.; Banmeke, O.; Zhao, Y.; Hamilton, M.; Higes, M.; et al. Nosemosis control in European honey bees, Apis mellifera, by silencing the gene encoding Nosema ceranae polar tube protein 3. J. Exp. Biol. 2018, 221, jeb184606. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Zhang, Y.; Duan, X.L.; Li, J.H.; Huang, W.F.; Evans, J.D.; DeGrandi-Hoffman, G.; Chen, Y.P.; Huang, S.K. RNA interference-mediated knockdown of genes encoding spore wall proteins confers protection against Nosema ceranae infection in the European honey bee, Apis mellifera. Microorganisms 2021, 9, 505. [Google Scholar] [CrossRef] [PubMed]
- Lang, H.Y.; Wang, H.; Wang, H.Q.; Zhong, Z.P.; Xie, X.; Zhang, W.; Guo, J.; Meng, L.; Hu, X.; Zhang, X.; et al. Engineered symbiotic bacteria interfering redox system inhibit microsporidia parasitism in honeybees. Nat. Commun. 2023, 14, 2778. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Wang, C.; Lang, H.; Wang, Y.; Wang, X.; Zheng, H.; Lu, Y. Liposome-based RNAi delivery in honeybee for inhibiting parasite Nosema ceranae. Synth. Syst. Biotechnol. 2024, 9, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Pompey, J.M.; Singh, U. RNA interference in Entamoeba histolytica: Implications for parasite biology and gene silencing. Future Microbiol. 2011, 6, 103–117. [Google Scholar] [CrossRef]
- Zhang, H.; Veira, J.; Bauer, S.T.; Yip, C.; Singh, U. RISC in Entamoeba histolytica: Identification of a protein-protein interaction network for the RNA interference pathway in a deep-branching Eukaryote. mBio 2021, 12, e0154021. [Google Scholar] [CrossRef]
- Wilson, I.W.; Weedall, G.D.; Lorenzi, H.; Howcroft, T.; Hon, C.C.; Deloger, M.; Guillén, N.; Paterson, S.; Clark, C.G.; Hall, N. Genetic diversity and gene family expansions in members of the genus Entamoeba. Genome Biol. Evol. 2019, 11, 688–705. [Google Scholar] [CrossRef]
- Wang, P.H.; Schulenberg, G.; Whitlock, S.; Worden, A.; Zhou, N.; Novak, S.; Chen, W. RNase If-treated quantitative PCR for dsRNA quantitation of RNAi trait in genetically modified crops. BMC Biotechnol. 2018, 18, 3. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar] [CrossRef]
- Dolgikh, V.V.; Ignatieva, A.N.; Rumiantseva, A.S.; Timofeev, S.A.; Fadeev, R.R.; Bayazyt, K.D.K. First evaluation of the Vairimorpha (Nosema) ceranae genome’s DNA replication genes as targets for RNA interference-mediated suppression of the honey bee Apis mellifera nosemosis. Protistology 2025, 19, 3–14. [Google Scholar] [CrossRef]

| Bacteria | Temperature (°C) | Heating Time (min) | 0.1% SDS | Ref. |
|---|---|---|---|---|
| E. coli HT115 (DE3) | 95 | 2 | + | [19] |
| – * | 100 | 2 | + | [14,15] |
| – | 95 | 10 | – | [17,18] |
| – | 100 | 10 | – | [16] |
| – | 100 | 60 | – | [20] |
| lactic acid bacteria | 95 | 10 | – | [21] |
| Coding Sequences | GC Content in CDS (%) | Ref. |
|---|---|---|
| Human and animal genomes (6 species) | 50.7 | [26] |
| Plant genomes (20 species) | 50.8 | [27] |
| V. ceranae | ||
| delta subunit of DNA pol. (XP_024330101.1) | 31.1 | this study |
| epsilon subunit of DNA pol. (EEQ83015.1) | 22.8 | – * |
| DNA helicase (XP_024331020.1) | 29.6 | – |
| DNA topoisomerase II (XP_024331425.1) | 28.9 | – |
| DNA ligase (EEQ82740.1) | 28.9 | – |
| N. bombycis | ||
| delta subunit of DNA pol. (EOB12369.1) | 34.6 | – |
| epsilon subunit of DNA pol. (XP_013171816.1) | 28.8 | – |
| DNA helicase (XP_013161385.1) | 28.9 | – |
| DNA topoisomerase II (XP_013181718.1) | 28.5 | – |
| DNA ligase (EOB13379.1) | 28.6 | – |
| Gene | Size (bp) | GC (%) | % bp w/o Adjacent G/C * |
|---|---|---|---|
| N. bombycis | |||
| delta subunit of DNA pol. | 561 | 37.6 | 36.4 |
| epsilon subunit of DNA pol. | 610 | 30.7 | 65.4 |
| DNA helicase | 475 | 29.9 | 62.3 |
| DNA topoisomerase II | 564 | 29.3 | 64 |
| DNA ligase | 532 | 30.8 | 62.2 |
| V. ceranae | |||
| delta subunit of DNA pol. | 590 | 31 | 55.3 |
| epsilon subunit of DNA pol. | 496 | 26 | 78.2 |
| DNA helicase | 594 | 33.8 | 56.2 |
| DNA topoisomerase II | 641 | 32.5 | 46.7 |
| DNA ligase | 549 | 29.3 | 62.7 |
| siRNAs of DNA helicase (chimeric) | 509 | 41.3 | 22.4 |
| M. persicae | |||
| nuclease | 679 | 53.6 | 9.6 |
| L. decemlineata | |||
| v-ATPaseA-mov34-actin | 869 | 47.6 | 7.8 |
| b’-COP | 401 | 43.4 | 29.4 |
| Primers | Sequence | PCR-Product Size (bp) |
|---|---|---|
| Vc ligase BamHI for | atcggaTCCGGAATAAAATCTAGAATTTAC 1,2 | 526 |
| Vc ligase HindIII rev | cgtaagCTTCTCCGTCTATTACAAAATC | |
| Nb delta BamHI for | TTTGTTATGGATCCTAAGAGAG | 539 |
| Nb delta HindIII rev | gataagCTTTTTTCATGTCTGTCTCATG | |
| Nb epsilon BamHI for | tgcggATCCTGAAAATGCACAAGATAAAG | 588 |
| Nb epsilon HindIII rev | CAATAAAATAGCTTAATAAGCTT | |
| Nb helicase HindIII for | atcaagCTTAAAGATTTTAGGAGAAATC | 453 |
| Nb helicase BamHI rev | TTAAGGATCCTAAATCTTTATAATC | |
| Nb topoII BamHI for | tcaggaTCCTATTGAGATGCACAAGGAAG | 542 |
| Nb topoII HindIII rev | cagaagCTTGTTGAAAATGTTCATCACTAAC | |
| Nb ligase HindIII for | actaagCTTTTAGTTAAATTTTTACAAGAG | 510 |
| Nb ligase BamHI rev | atcggatCCATCAAAATAAAGGCAATCAA | |
| T7 for | taatacgactcactataggg | variable |
| T7 rev | ctagttattgctcagcggtgg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fadeev, R.R.; Timofeev, S.A.; Senderskiy, I.V.; Dolgikh, V.V. GC Content and Thermal Stability of Double-Stranded RNA: Fragments of Microsporidia Vairimorpha ceranae and Nosema bombycis AT-Rich Genes Are Sensitive to Standard Heat Treatment. Int. J. Mol. Sci. 2025, 26, 10270. https://doi.org/10.3390/ijms262110270
Fadeev RR, Timofeev SA, Senderskiy IV, Dolgikh VV. GC Content and Thermal Stability of Double-Stranded RNA: Fragments of Microsporidia Vairimorpha ceranae and Nosema bombycis AT-Rich Genes Are Sensitive to Standard Heat Treatment. International Journal of Molecular Sciences. 2025; 26(21):10270. https://doi.org/10.3390/ijms262110270
Chicago/Turabian StyleFadeev, Ruslan R., Sergey A. Timofeev, Igor V. Senderskiy, and Viacheslav V. Dolgikh. 2025. "GC Content and Thermal Stability of Double-Stranded RNA: Fragments of Microsporidia Vairimorpha ceranae and Nosema bombycis AT-Rich Genes Are Sensitive to Standard Heat Treatment" International Journal of Molecular Sciences 26, no. 21: 10270. https://doi.org/10.3390/ijms262110270
APA StyleFadeev, R. R., Timofeev, S. A., Senderskiy, I. V., & Dolgikh, V. V. (2025). GC Content and Thermal Stability of Double-Stranded RNA: Fragments of Microsporidia Vairimorpha ceranae and Nosema bombycis AT-Rich Genes Are Sensitive to Standard Heat Treatment. International Journal of Molecular Sciences, 26(21), 10270. https://doi.org/10.3390/ijms262110270





