Comprehensive Epigenome-Wide Profiling Reveals Distinctive DNA Methylation Signatures and Potential Prognostic Biomarkers in Mexican Pediatric B-ALL
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics of the Analyzed Population
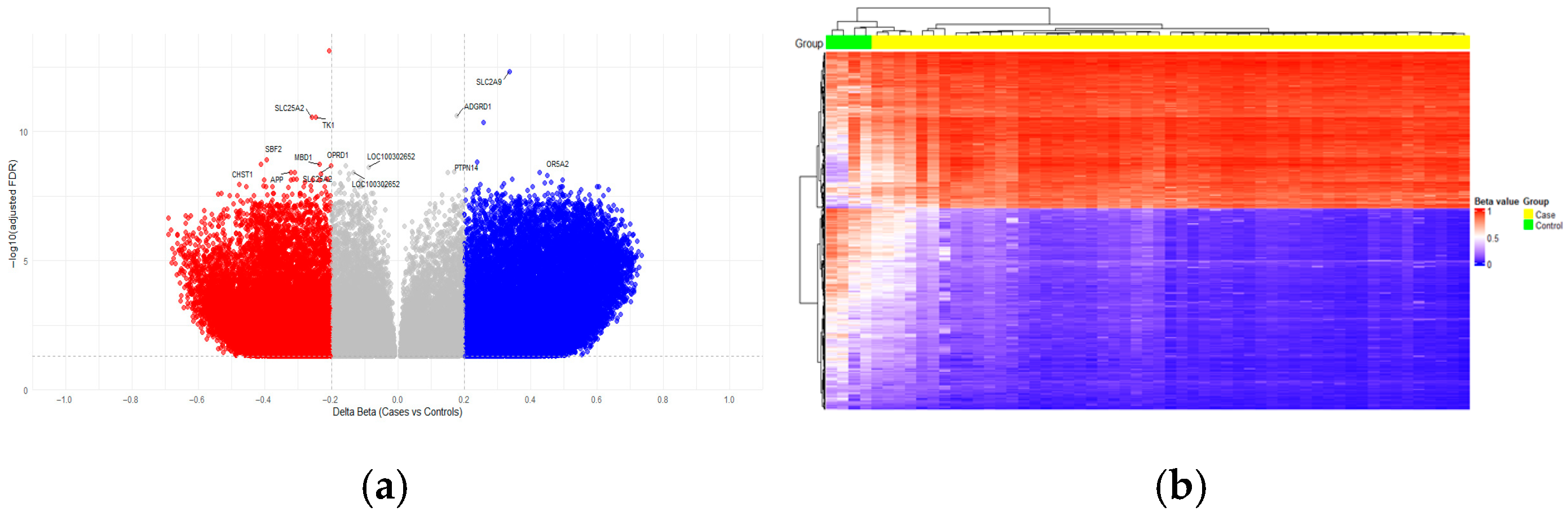
2.2. Identification of Differential Methylated CpGs in Acute Lymphoblastic Leukemia
2.2.1. Distribution of Differentially Methylated Regions in the Functional Context
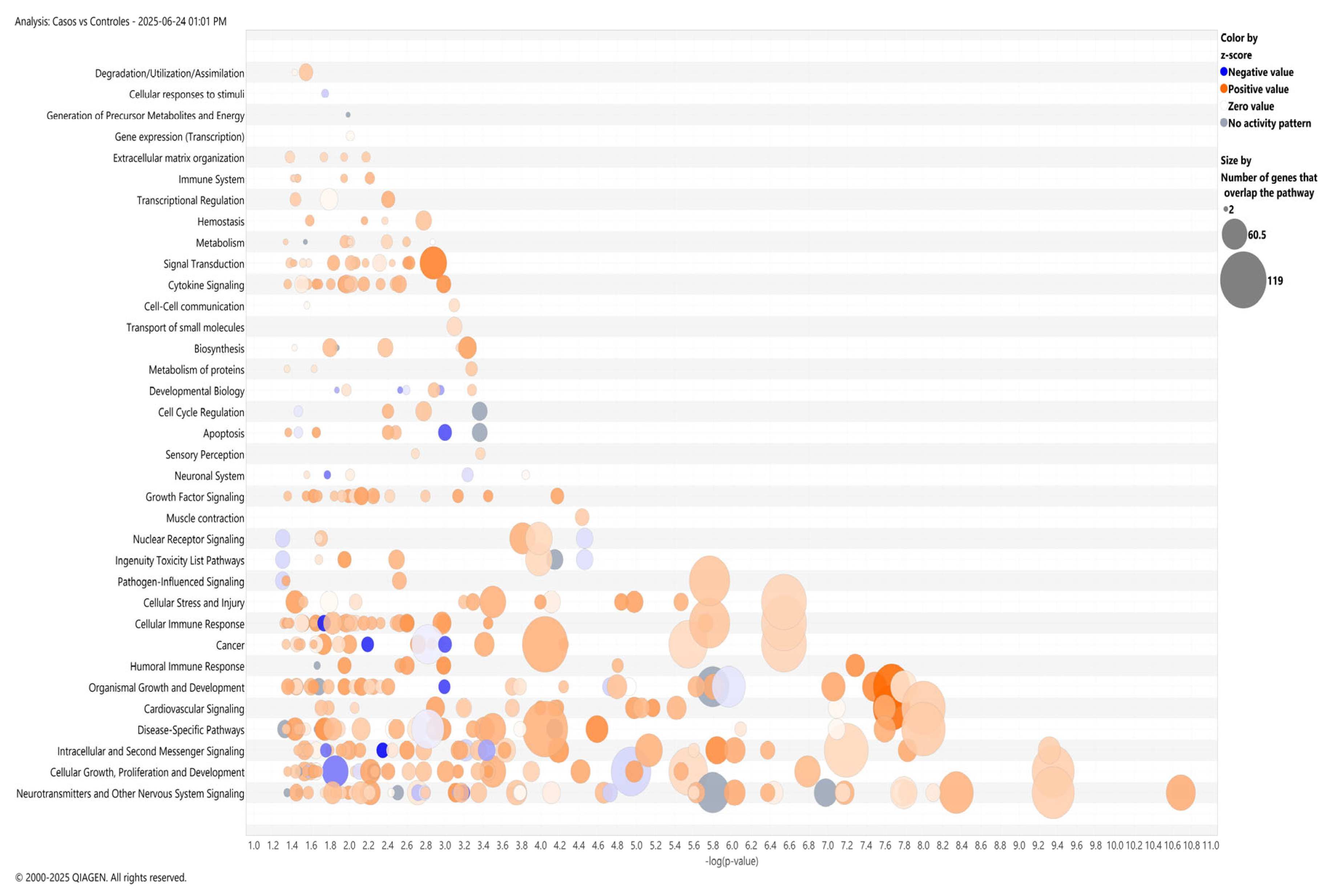
2.2.2. Pathway Enrichment and Protein–Protein Interaction Network Analysis
2.3. Methylation Analysis Based on Clinical Variables
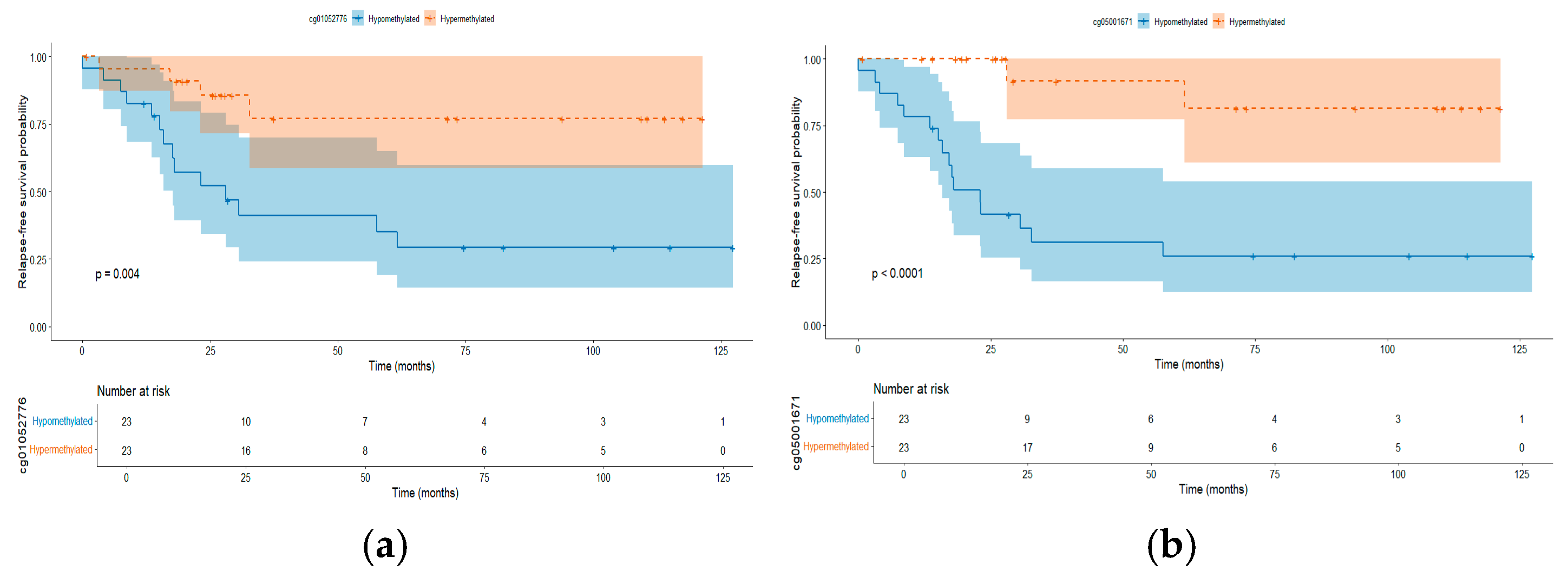
2.4. Differentially Methylated CpGs Associated with Relapse
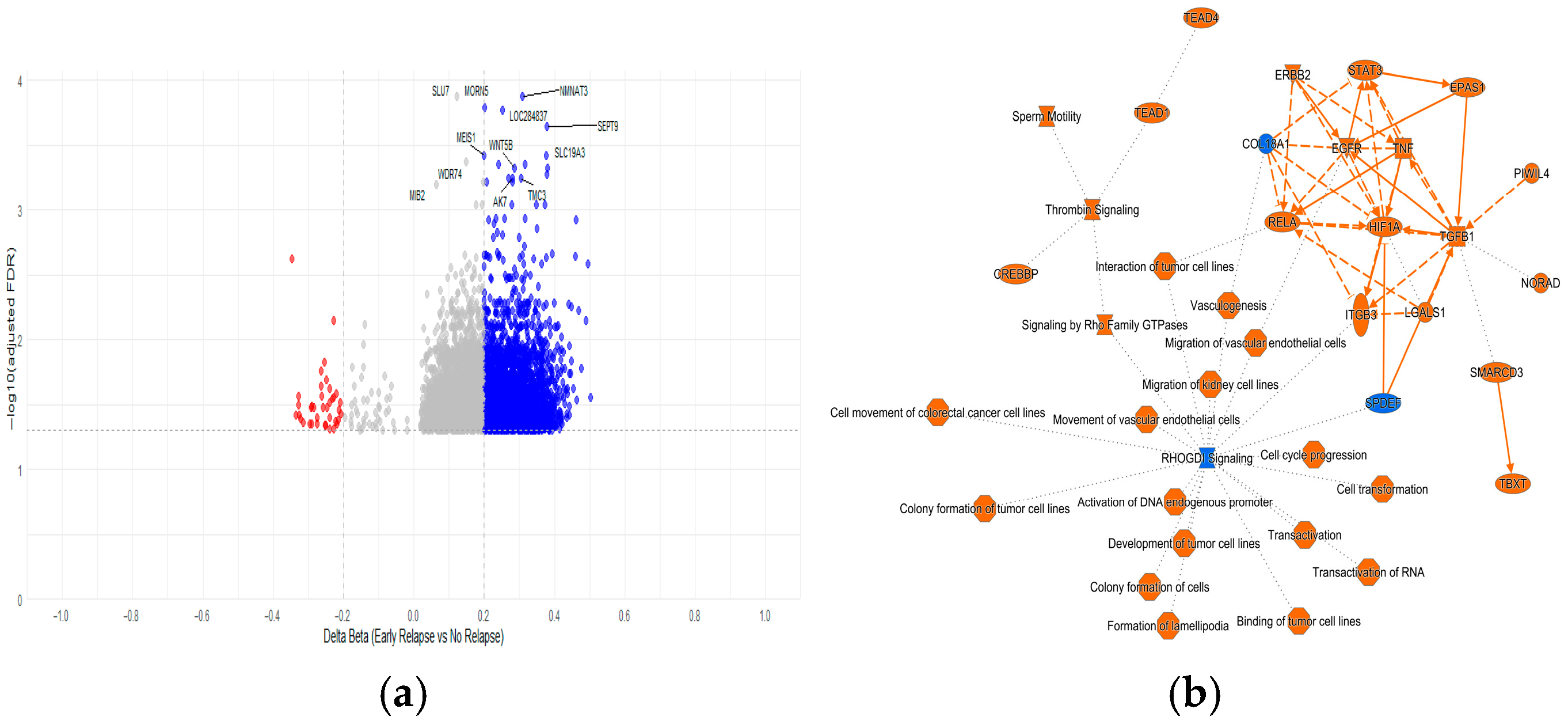
2.4.1. Methylated CpGs Profile in Early Relapse
2.4.2. Random Forest Model on Risk of Relapse
3. Discussion
4. Materials and Methods
4.1. Patient and Sample Collection
4.2. Nucleic Acid Extraction and Fusion Gene Analysis
4.3. Genome-Wide DNA Methylation Analysis
4.4. Assessment of Differentially Methylated Regions
4.5. Epigenome-Wide Association Study (EWAS)
4.6. Relapse Prediction Model in ALL Patients, Grounded in a Random Forest-Based Machine Learning Approach
4.7. Pathway Enrichment and Protein–Protein Interaction Analyses
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALL | Acute lymphoblastic leukemia |
| DNAme | DNA methylation |
| EWAS | epigenome-wide association study |
| B-ALL | B-cell ALL |
| Z-BMI | Body Mass Index Z score |
| NCI | National Cancer Institute |
| VER | Very early relapse |
| ER | Early relapse |
| DMC | Differentially methylated CpG sites |
| 5’UTR | 5′ untranslated regions |
| TSS200 | Region 200bp previous of transcription start site |
| TSS1500 | Region 1500bp previous of transcription start site |
| DMR | Differentially methylated regions |
| GO | Gene Ontology |
| IPA | Ingenuity pathway analysis |
| PPI | Protein–protein interaction |
| OOB | Out-of-bag |
| RFS | Relapse-free survival |
| MIGICCL | Mexican Inter-Institutional Group for the Identification of the Causes of Childhood Leukemia |
| BM | Bone marrow |
| PB | Peripheral blood |
| CNS | Central nervous system |
| BMIQ | Beta Mixture Quantile dilation |
| HR | Hazard ratios |
References
- Jiménez-Morales, S.; Hidalgo-Miranda, A.; Ramírez-Bello, J. Leucemia linfoblástica aguda infantil: Una aproximación genómica. Bol. Med. Hosp. Infant. Mex. 2017, 74, 13–26. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef]
- Flores-Lujano, J.; Duarte-Rodríguez, D.A.; Jiménez-Hernández, E.; Martín-Trejo, J.A.; Allende-López, A.; Peñaloza-González, J.G.; Pérez-Saldivar, M.L.; Medina-Sanson, A.; Torres-Nava, J.R.; Solís-Labastida, K.A.; et al. Persistently high incidence rates of childhood acute leukemias from 2010 to 2017 in Mexico City: A population study from the MIGICCL. Front. Public Health 2022, 10, 918921. [Google Scholar] [CrossRef]
- National Cancer Institute. Childhood Leukemia—Cancer Stat Facts. Surveillance, Epidemiology, and End Results Program. 2020. Available online: https://seer.cancer.gov/statfacts/html/childleuk.html (accessed on 26 September 2025).
- de Smith, A.J.; Jiménez-Morales, S.; Mejía-Aranguré, J.M. The genetic risk of acute lymphoblastic leukemia and its implications for children of Latin American origin. Front. Oncol. 2023, 13, 1299355. [Google Scholar] [CrossRef] [PubMed]
- Moreira, D.C.; González-Ramella, O.; Valenzuela, M.E.; Carrillo, A.K.; Faughnan, L.; Job, G.; Chen, Y.; Villegas, C.; Irigoyen, A.E.; Urbays, R.B.; et al. Evaluation of factors leading to poor outcomes for pediatric acute lymphoblastic leukemia in Mexico: A multi-institutional report of 2,116 patients. Front. Oncol. 2023, 13, 1255555. [Google Scholar] [CrossRef]
- Quiroz, E.; Aldoss, I.; Pullarkat, V.; Rego, E.; Marcucci, G.; Douer, D. The emerging story of acute lymphoblastic leukemia among the Latin American population—Biological and clinical implications. Blood Rev. 2019, 33, 98–105. [Google Scholar] [CrossRef]
- Montes-Rodríguez, I.M.; Soto-Salgado, M.; Torres-Cintrón, C.R.; Tomassini-Fernandini, J.C.; Suárez, E.; Clavell, L.A.; Cadilla, C.L. Incidence and Mortality Rates for Childhood Acute Lymphoblastic Leukemia in Puerto Rican Hispanics, 2012–2016. Cancer Epidemiol. Biomark. Prev. 2023, 32, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Hunger, S.P.; Raetz, E.A. How I treat relapsed acute lymphoblastic leukemia in the pediatric population. Blood 2020, 136, 1803–1812. [Google Scholar] [CrossRef] [PubMed]
- Rheingold, S.R.; Bhojwani, D.; Ji, L.; Xu, X.; Devidas, M.; Kairalla, J.A.; Shago, M.; Heerema, N.A.; Carroll, A.J.; Breidenbach, H.; et al. Determinants of survival after first relapse of acute lymphoblastic leukemia: A Children’s Oncology Group study. Leukemia 2024, 38, 2382–2394. [Google Scholar] [CrossRef]
- Burns, M.A.; Place, A.E.; Stevenson, K.E.; Gutiérrez, A.; Forrest, S.; Pikman, Y.; Vrooman, L.M.; Harris, M.H.; Weinberg, O.K.; Hunt, S.K.; et al. Identification of prognostic factors in childhood T-cell acute lymphoblastic leukemia: Results from DFCI ALL Consortium Protocols 05-001 and 11-001. Pediatr. Blood Cancer 2020, 68, e28719. [Google Scholar] [CrossRef]
- Toft, N.; Birgens, H.; Abrahamsson, J.; Griškevičius, L.; Hallböök, H.; Heyman, M.; Klausen, T.W.; Jónsson, Ó.; Palk, K.; Pruunsild, K.; et al. Results of NOPHO ALL2008 treatment for patients aged 1–45 years with acute lymphoblastic leukemia. Leukemia 2017, 32, 606–615. [Google Scholar] [CrossRef]
- Roberts, K.G.; Li, Y.; Payne-Turner, D.; Harvey, R.C.; Yang, Y.-L.; Pei, D.; McCastlain, K.; Ding, L.; Lu, C.; Song, G.; et al. Targetable Kinase-Activating Lesions in Ph-like Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2014, 371, 1005–1015. [Google Scholar] [CrossRef]
- Østergaard, A.; Fiocco, M.; de Groot-Kruseman, H.; Moorman, A.V.; Vora, A.; Zimmermann, M.; Schrappe, M.; Biondi, A.; Escherich, G.; Stary, J.; et al. ETV6::RUNX1 Acute Lymphoblastic Leukemia: How much therapy is needed for cure? Leukemia 2024, 38, 1477–1487. [Google Scholar] [CrossRef]
- Schmiegelow, K.; Forestier, E.; Hellebostad, M.; Heyman, M.; Kristinsson, J.; Söderhäll, S.; Taskinen, M.; on behalf of the Nordic Society of Paediatric Haematology and Oncology (NOPHO). Long-term results of NOPHO ALL-92 and ALL-2000 studies of childhood acute lymphoblastic leukemia. Leukemia 2009, 24, 345–354, Erratum in Leukemia 2010, 24, 670.. [Google Scholar] [CrossRef]
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Niksic, M. Global surveillance of trends in cancer survival: Analysis of individual records for 37,513,025 patients diagnosed with one of 18 cancers during 2000–2014 from 322 population-based registries in 71 countries (CONCORD-3). Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef]
- Lin, A.; Cheng, F.W.; Chiang, A.K.; Luk, C.; Li, R.C.; Ling, A.S.; Cheuk, D.K.; Chang, K.; Ku, D.; Lee, V.; et al. Excellent outcome of acute lymphoblastic leukaemia with TCF3-PBX1 rearrangement in Hong Kong. Pediatr. Blood Cancer 2018, 65, e27346. [Google Scholar] [CrossRef]
- Jiménez-Hernández, E.; Jaimes-Reyes, E.Z.; Arellano-Galindo, J.; García-Jiménez, X.; Tiznado-García, H.M.; Dueñas-González, M.T.; Villegas, O.M.; Sánchez-Jara, B.; Bekker-Méndez, V.C.; Ortíz-Torres, M.G.; et al. Survival of Mexican children with acute lymphoblastic leukaemia under treatment with the protocol from the Dana-Farber cancer institute 00-01. BioMed Res. Int. 2015, 2015, 576950. [Google Scholar] [CrossRef]
- Chang, T.-C.; Chen, W.; Qu, C.; Cheng, Z.; Hedges, D.; Elsayed, A.; Pounds, S.B.; Shago, M.; Rabin, K.R.; Raetz, E.A.; et al. Genomic Determinants of Outcome in Acute Lymphoblastic Leukemia. J. Clin. Oncol. 2024, 42, 3491–3503. [Google Scholar] [CrossRef]
- Jiménez-Morales, S.; Miranda-Peralta, E.; Saldaña-Alvarez, Y.; Perez-Vera, P.; Paredes-Aguilera, R.; Rivera-Luna, R.; Velázquez-Cruz, R.; Ramírez-Bello, J.; Carnevale, A.; Orozco, L. BCR-ABL, ETV6-RUNX1 and E2A-PBX1: Prevalence of the most common acute lymphoblastic leukemia fusion genes in Mexican patients. Leuk. Res. 2008, 32, 1518–1522. [Google Scholar] [CrossRef]
- Mata-Rocha, M.; Rangel-López, A.; Jimenez-Hernandez, E.; Nuñez-Enríquez, J.C.; Morales-Castillo, B.A.; Sánchez-Escobar, N.; Sepúlveda-Robles, O.A.; Bravata-Alcántara, J.C.; Nájera-Cortés, A.S.; Pérez-Saldivar, M.L.; et al. Low Prevalence of ETV6::RUNX1 Fusion Gene in a Hispanic Population. Front. Pediatr. 2022, 10, 837656. [Google Scholar] [CrossRef]
- Malhotra, P.; Jain, S.; Agarwal, A.; Sharma, A.; Agarwal, N.; Kapoor, G. Incidence and Prognostic Impact of TCF3-PBX1 Fusion in Childhood Acute Lymphoblastic Leukemia: A Single Centre Experience. Indian J. Hematol. Blood Transfus. 2021, 38, 164–168. [Google Scholar] [CrossRef]
- Lee, S.H.R.; Antillon-Klussmann, F.; Pei, D.; Yang, W.; Roberts, K.G.; Li, Z.; Devidas, M.; Yang, W.; Najera, C.; Lin, H.P.; et al. Association of Genetic Ancestry with the Molecular Subtypes and Prognosis of Childhood Acute Lymphoblastic Leukemia. JAMA Oncol. 2022, 8, 354–363, Erratum in JAMA Oncol. 2022, 8, 484. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Lim, J.Y.; Bhatia, S.; Robison, L.L.; Yang, J.J. Genomics of Racial and Ethnic Disparities in Childhood Acute Lymphoblastic Leukemia. Cancer 2013, 120, 955–962. [Google Scholar] [CrossRef] [PubMed]
- de Smith, A.J.; Wahlster, L.; Jeon, S.; Kachuri, L.; Black, S.; Langie, J.; Cato, L.D.; Nakatsuka, N.; Chan, T.-F.; Xia, G.; et al. A noncoding regulatory variant in IKZF1 increases acute lymphoblastic leukemia risk in Hispanic/Latino children. Cell Genom. 2024, 4, 100526. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Cheng, C.; Devidas, M.; Pei, D.; Fan, Y.; Yang, W.; Neale, G.; Scheet, P.; Burchard, E.G.; Torgerson, D.G.; et al. ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. J. Clin. Oncol. 2012, 30, 751–757. [Google Scholar] [CrossRef]
- Gu, Z.; Churchman, M.L.; Roberts, K.G.; Moore, I.; Zhou, X.; Nakitandwe, J.; Hagiwara, K.; Pelletier, S.; Gingras, S.; Berns, H.; et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 2019, 51, 296–307. [Google Scholar] [CrossRef]
- Shah, B.; Mattison, R.J.; Abboud, R.; Abdelmessieh, P.; Aldoss, I.; Burke, P.W.; DeAngelo, D.J.; Dinner, S.; Fathi, A.T.; Gauthier, J.; et al. Acute Lymphoblastic Leukemia, Version 2.2024, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2024, 22, 563–576. [Google Scholar] [CrossRef]
- Pölönen, P.; Di Giacomo, D.; Seffernick, A.E.; Elsayed, A.; Kimura, S.; Benini, F.; Montefiori, L.E.; Wood, B.L.; Xu, J.; Chen, C.; et al. The genomic basis of childhood T-lineage acute lymphoblastic leukaemia. Nature 2024, 632, 1082–1091. [Google Scholar] [CrossRef]
- Almamun, M.; Levinson, B.T.; van Swaay, A.C.; Johnson, N.T.; McKay, S.D.; Arthur, G.L.; Davis, J.W.; Taylor, K.H. Integrated methylome and transcriptome analysis reveals novel regulatory elements in pediatric acute lymphoblastic leukemia. Epigenetics 2015, 10, 882–890. [Google Scholar] [CrossRef]
- Bergeron, B.P.; Diedrich, J.D.; Zhang, Y.; Barnett, K.R.; Dong, Q.; Ferguson, D.C.; Autry, R.J.; Yang, W.; Hansen, B.S.; Smith, C.; et al. Epigenomic profiling of glucocorticoid responses identifies cis-regulatory disruptions impacting steroid resistance in childhood acute lymphoblastic leukemia. Leukemia 2022, 36, 2374–2383. [Google Scholar] [CrossRef]
- Tejedor, J.R.; Bueno, C.; Vinyoles, M.; Petazzi, P.; Agraz-Doblas, A.; Cobo, I.; Torres-Ruiz, R.; Bayón, G.F.; Pérez, R.F.; López-Tamargo, S.; et al. Integrative methylome-transcriptome analysis unravels cancer cell vulnerabilities in infant MLL-rearranged B cell acute lymphoblastic leukemia. J. Clin. Investig. 2021, 131, e138833. [Google Scholar] [CrossRef]
- Ghantous, A.; Nusslé, S.G.; Nassar, F.J.; Spitz, N.; Novoloaca, A.; Krali, O.; Nickels, E.; Cahais, V.; Cuenin, C.; Roy, R.; et al. Epigenome-wide analysis across the development span of pediatric acute lymphoblastic leukemia: Backtracking to birth. Mol. Cancer 2024, 23, 238. [Google Scholar] [CrossRef] [PubMed]
- Borssén, M.; Nordlund, J.; Haider, Z.; Landfors, M.; Larsson, P.; Kanerva, J.; Schmiegelow, K.; Flaegstad, T.; Jónsson, Ó.G.; Frost, B.-M.; et al. DNA methylation holds prognostic information in relapsed precursor B-cell acute lymphoblastic leukemia. Clin. Epigenet. 2018, 10, 1–7. [Google Scholar] [CrossRef]
- Jain, N.; Roberts, K.G.; Jabbour, E.; Patel, K.; Eterovic, A.K.; Chen, K.; Zweidler-McKay, P.; Lu, X.; Fawcett, G.; Wang, S.A.; et al. Ph-like acute lymphoblastic leukemia: A high-risk subtype in adults. Blood 2017, 129, 572–581. [Google Scholar] [CrossRef]
- Shoag, J.M.; Barredo, J.C.; Lossos, I.S.; Pinheiro, P.S. Acute lymphoblastic leukemia mortality in Hispanic Americans. Leuk. Lymphoma 2020, 61, 2674–2681. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Li, S.; Sok, P.; Xu, K.; Muskens, I.S.; Elliott, N.; Myint, S.S.; Pandey, P.; Hansen, H.M.; Morimoto, L.M.; Kang, A.Y.; et al. Epigenome-wide association study of acute lymphoblastic leukemia in children with down syndrome. Blood Adv. 2022, 6, 4132–4136. [Google Scholar] [CrossRef] [PubMed]
- Nordlund, J.; Bäcklin, C.L.; Wahlberg, P.; Busche, S.; Berglund, E.C.; Eloranta, M.-L.; Flaegstad, T.; Forestier, E.; Frost, B.-M.; Harila-Saari, A.; et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013, 14, r105. [Google Scholar] [CrossRef]
- Rahmani, M.; Talebi, M.; Hagh, M.F.; Feizi, A.A.H.; Solali, S. Aberrant DNA methylation of key genes and Acute Lymphoblastic Leukemia. Biomed. Pharmacother. 2018, 97, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Hogan, L.E.; Meyer, J.A.; Yang, J.; Wang, J.; Wong, N.; Yang, W.; Condos, G.; Hunger, S.P.; Raetz, E.; Saffery, R.; et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood 2011, 118, 5218–5226. [Google Scholar] [CrossRef]
- Milani, L.; Lundmark, A.; Kiialainen, A.; Nordlund, J.; Flaegstad, T.; Forestier, E.; Heyman, M.; Jonmundsson, G.; Kanerva, J.; Schmiegelow, K.; et al. DNA methylation for subtype classification and prediction of treatment outcome in patients with childhood acute lymphoblastic leukemia. Blood 2010, 115, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Bhatla, T.; Wang, J.; Morrison, D.J.; Raetz, E.A.; Burke, M.J.; Brown, P.; Carroll, W.L. Epigenetic reprogramming reverses the relapse-specific gene expression signature and restores chemosensitivity in childhood B-lymphoblastic leukemia. Blood 2012, 119, 5201–5210. [Google Scholar] [CrossRef]
- Simonin, M.; Touzart, A. Epigenetics-based stratification in pediatric T-ALL. Blood 2025, 145, 2108–2110. [Google Scholar] [CrossRef]
- Tian, Y.; Morris, T.J.; Webster, A.P.; Yang, Z.; Beck, S.; Feber, A.; Teschendorff, A.E. ChAMP: Updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 2017, 33, 3982–3984. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, M.E.; Chen, S.-C.; Andersson, A.K.; Phillips, L.A.; Li, Y.; Sotzen, J.; Kundu, M.; Downing, J.R.; Melnick, A.; Mullighan, C.G. Integrated genetic and epigenetic analysis of childhood acute lymphoblastic leukemia. J. Clin. Investig. 2013, 123, 3099–3111. [Google Scholar] [CrossRef]
- Chatterton, Z.; Morenos, L.; Mechinaud, F.; Ashley, D.M.; Craig, J.M.; Sexton-Oates, A.; Halemba, M.S.; Parkinson-Bates, M.; Ng, J.; Morrison, D.; et al. Epigenetic deregulation in pediatric acute lymphoblastic leukemia. Epigenetics 2014, 9, 459–467. [Google Scholar] [CrossRef]
- Caliskan, G.; Pawitan, Y.; Vu, T.N. Similarities and differences of bone marrow and peripheral blood samples from acute myeloid leukemia patients in terms of cellular heterogeneity and ex-vivo drug sensitivity. eJHaem 2024, 5, 721–727. [Google Scholar] [CrossRef]
- Takeuchi, S.; Matsushita, M.; Zimmermann, M.; Ikezoe, T.; Komatsu, N.; Seriu, T.; Schrappe, M.; Bartram, C.R.; Koeffler, H.P. Clinical significance of aberrant DNA methylation in childhood acute lymphoblastic leukemia. Leuk. Res. 2011, 35, 1345–1349. [Google Scholar] [CrossRef][Green Version]
- Hetzel, S.; Mattei, A.L.; Kretzmer, H.; Qu, C.; Chen, X.; Fan, Y.; Wu, G.; Roberts, K.G.; Luger, S.; Litzow, M.; et al. Acute lymphoblastic leukemia displays a distinct highly methylated genome. Nat. Cancer 2022, 3, 768–782. [Google Scholar] [CrossRef]
- Schäfer, V.; Ernst, J.; Rinke, J.; Winkelmann, N.; Beck, J.F.; Hochhaus, A.; Gruhn, B.; Ernst, T. EZH2 mutations and promoter hypermethylation in childhood acute lymphoblastic leukemia. J. Cancer Res. Clin. Oncol. 2016, 142, 1641–1650. [Google Scholar] [CrossRef]
- Roman-Gomez, J.; Jimenez-Velasco, A.; Barrios, M.; Prosper, F.; Heiniger, A.; Torres, A.; Agirre, X. Poor prognosis in acute lymphoblastic leukemia may relate to promoter hypermethylation of cancer-related genes. Leuk. Lymphoma 2007, 48, 1269–1282. [Google Scholar] [CrossRef]
- Ng, Ö.H.; Fırtına, S.; Can, I.; Karakaş, Z.; Ağaoğlu, L.; Doğru, Ö.; Celkan, T.; Akçay, A.; Yıldırmak, Y.; Timur, Ç.; et al. WNT5A hipermetilasyonun Çocukluk Çağı akut lenfoblastik lösemideki olası rolü. Turk. J. Hematol. 2015, 32, 127–135. [Google Scholar] [CrossRef]
- Roman-Gomez, J.; Castillejo, J.A.; Jimenez, A.; Barrios, M.; Heiniger, A.; Torres, A. The role of DNA hypermethylation in the pathogenesis and prognosis of acute lymphoblastic leukemia. Leuk. Lymphoma 2003, 44, 1855–1864. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, S.E.; Reyes, A.; Qi, Y.; Adriaens, C.; Hegazi, E.; Pelka, K.; Chen, J.H.; Zou, L.S.; Drier, Y.; Hecht, V.; et al. Large-Scale Topological Changes Restrain Malignant Progression in Colorectal Cancer. Cell 2020, 182, 1474–1489.e23. [Google Scholar] [CrossRef]
- May-Hau, D.I.; Bárcenas-López, D.A.; Núñez-Enríquez, J.C.; Bekker-Méndez, V.C.; Beltrán-Anaya, F.O.; Jiménez-Hernández, E.; Ortíz-Maganda, M.P.; Guerra-Castillo, F.X.; Medina-Sanson, A.; Flores-Lujano, J.; et al. Underexpression of LINC00173 in TCF3/PBX1-Positive Cases Is Associated With Poor Prognosis in Children With B-Cell Precursor Acute Lymphoblastic Leukemia. Front. Oncol. 2022, 12, 887766. [Google Scholar] [CrossRef]
- Bárcenas-López, D.A.; Núñez-Enríquez, J.C.; Hidalgo-Miranda, A.; Beltrán-Anaya, F.O.; May-Hau, D.I.; Jiménez-Hernández, E.; Bekker-Méndez, V.C.; Flores-Lujano, J.; Medina-Sansón, A.; Tamez-Gómez, E.L.; et al. Transcriptome Analysis Identifies LINC00152 as a Biomarker of Early Relapse and Mortality in Acute Lymphoblastic Leukemia. Genes 2020, 11, 302. [Google Scholar] [CrossRef]
- Georgiadis, P.; Liampa, I.; Hebels, D.G.; Krauskopf, J.; Chatziioannou, A.; Valavanis, I.; de Kok, T.M.; Kleinjans, J.C.; Bergdahl, I.A.; Melin, B.; et al. Evolving DNA methylation and gene expression markers of B-cell chronic lymphocytic leukemia are present in pre-diagnostic blood samples more than 10 years prior to diagnosis. BMC Genom. 2017, 18, 728. [Google Scholar] [CrossRef]
- Yang, L.; Rodriguez, B.; Mayle, A.; Park, H.J.; Lin, X.; Luo, M.; Jeong, M.; Curry, C.V.; Kim, S.-B.; Ruau, D.; et al. DNMT3A Loss Drives Enhancer Hypomethylation in FLT3-ITD-Associated Leukemias. Cancer Cell 2016, 29, 922–934, Erratum in Cancer Cell 2016, 30, 363–365.. [Google Scholar] [CrossRef]
- Bastian, L.; Schroeder, M.P.; Eckert, C.; Schlee, C.; Tanchez, J.O.; Kämpf, S.; Wagner, D.L.; Schulze, V.; Isaakidis, K.; Lázaro-Navarro, J.; et al. PAX5 biallelic genomic alterations define a novel subgroup of B-cell precursor acute lymphoblastic leukemia. Leukemia 2019, 33, 1895–1909. [Google Scholar] [CrossRef]
- Guilhamon, P.; Eskandarpour, M.; Halai, D.; Wilson, G.A.; Feber, A.; Teschendorff, A.E.; Gomez, V.; Hergovich, A.; Tirabosco, R.; Amary, M.F.; et al. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat. Commun. 2013, 4, 2166. [Google Scholar] [CrossRef]
- Navarrete-Meneses, M.d.P.; Pérez-Vera, P. Epigenetic alterations in acute lymphoblastic leukemia. Bol. Med. Hosp. Infant. Mex. 2017, 74, 243–264. [Google Scholar] [CrossRef]
- Núñez-Enríquez, J.C.; Bárcenas-López, D.A.; Hidalgo-Miranda, A.; Jiménez-Hernández, E.; Bekker-Méndez, V.C.; Flores-Lujano, J.; Solis-Labastida, K.A.; Martínez-Morales, G.B.; Sánchez-Muñoz, F.; Espinoza-Hernández, L.E.; et al. Gene Expression Profiling of Acute Lymphoblastic Leukemia in Children with Very Early Relapse. Arch. Med. Res. 2016, 47, 644–655. [Google Scholar] [CrossRef]
- Brown, P.; Levis, M.; Shurtleff, S.; Campana, D.; Downing, J.; Small, D. FLT3 inhibition selectively kills childhood acute lymphoblastic leukemia cells with high levels of FLT3 expression. Blood 2005, 105, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.; Evans, K.; Earley, E.J.; Smith, C.M.; Erickson, S.W.; Stearns, T.; Philip, V.M.; Neuhauser, S.B.; Chuang, J.H.; Jocoy, E.L.; et al. In vivo activity of the dual SYK/FLT3 inhibitor TAK-659 against pediatric acute lymphoblastic leukemia xenografts. Pediatr. Blood Cancer 2023, 70, e30503. [Google Scholar] [CrossRef]
- Au, A.C.; Hernandez, P.A.; Lieber, E.; Nadroo, A.M.; Shen, Y.-M.; Kelley, K.A.; Gelb, B.D.; Diaz, G.A. Protein tyrosine phosphatase PTPN14 Is a regulator of lymphatic function and choanal development in humans. Am. J. Hum. Genet. 2010, 87, 436–444. [Google Scholar] [CrossRef]
- Olafsdottir, T.; Stacey, S.N.; Sveinbjornsson, G.; Thorleifsson, G.; Norland, K.; Sigurgeirsson, B.; Thorisdottir, K.; Kristjansson, A.K.; Tryggvadottir, L.; Sarin, K.Y.; et al. Loss-of-function variants in the tumor-suppressor gene PTPN14 confer increased cancer risk. Cancer Res. 2021, 81, 1954–1964. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Du, Z.; Huang, J.; Cheng, Y.; Yi, W.; Li, T.; Yang, J.; Chen, C. Comprehensive analysis of PTPN family expression and prognosis in acute myeloid leukemia. Front. Genet. 2023, 13, 1087938. [Google Scholar] [CrossRef]
- Kleppe, M.; Lahortiga, I.; El Chaar, T.; De Keersmaecker, K.; Mentens, N.; Graux, C.; Van Roosbroeck, K.; Ferrando, A.A.; Langerak, A.W.; Meijerink, J.P.P.; et al. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat. Genet. 2010, 42, 530–535. [Google Scholar] [CrossRef]
- Laczmanska, I.; Karpinski, P.; Bebenek, M.; Sedziak, T.; Ramsey, D.; Szmida, E.; Sasiadek, M.M. Protein tyrosine phosphatase receptor-like genes are frequently hypermethylated in sporadic colorectal cancer. J. Hum. Genet. 2012, 58, 11–15. [Google Scholar] [CrossRef]
- Tsai, Y.-H.; Mitra, P.; Taniar, D.; Pai, T.-W. DNA methylation biomarker analysis from low-survival-rate cancers based on genetic functional approaches. Front. Bioinform. 2025, 5, 1523524. [Google Scholar] [CrossRef]
- Izquierdo, A.G.; Boughanem, H.; Diaz-Lagares, A.; Arranz-Salas, I.; Esteller, M.; Tinahones, F.J.; Casanueva, F.F.; Macias-Gonzalez, M.; Crujeiras, A.B. DNA methylome in visceral adipose tissue can discriminate patients with and without colorectal cancer. Epigenetics 2021, 17, 665–676. [Google Scholar] [CrossRef]
- Lin, Y.; Wu, Y.; Zhang, Q.; Tu, X.; Chen, S.; Pan, J.; Xu, N.; Lin, M.; She, P.; Niu, G.; et al. RPTOR blockade suppresses brain metastases of NSCLC by interfering the ceramide metabolism via hijacking YY1 binding. J. Exp. Clin. Cancer Res. 2024, 43, 1, Erratum in J. Exp. Clin. Cancer Res. 2024, 43, 37.. [Google Scholar] [CrossRef]
- Wang, H.; Wang, W.; Wang, Z.; Li, X. Transcriptomic correlates of cell cycle checkpoints with distinct prognosis, molecular characteristics, immunological regulation, and therapeutic response in colorectal adenocarcinoma. Front. Immunol. 2023, 14, 1291859. [Google Scholar] [CrossRef]
- Copeland, S.E.; Snow, S.M.; Wan, J.; Matkowskyj, K.A.; Halberg, R.B.; Weaver, B.A. MAD1 upregulation sensitizes to inflammation-mediated tumor formation. PLoS Genet. 2024, 20, e1011437. [Google Scholar] [CrossRef]
- Liping, X.; Jia, L.; Qi, C.; Liang, Y.; Dongen, L.; Jianshuai, J. Cell Cycle Genes Are Potential Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. BioMed. Res. Int. 2020, 2020, 6206157, Erratum in Biomed. Res. Int. 2020, 2020, 8679532. [Google Scholar] [CrossRef]
- Lupo, P.J.; Brown, A.L.; Arroyo, V.M.; Kamdar, K.Y.; Belmont, J.W.; Scheurer, M.E.; Leisenring, W.M.; Gramatges, M.M.; Okcu, M.F.; Yasui, Y.; et al. DNA methylation and obesity in survivors of pediatric acute lymphoblastic leukemia: A report from the Childhood Cancer Survivor Study. Genes Chromosom. Cancer 2018, 58, 52–59. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhou, Q.; Sun, Y.; Lai, F.; Wang, Z.; Hao, Z.; Li, G. MethMarkerDB: A comprehensive cancer DNA methylation biomarker database. Nucleic Acids Res. 2023, 52, D1380–D1392. [Google Scholar] [CrossRef]
- Schraw, J.M.; Yiu, T.T.; Lupo, P.J.; Tsavachidis, S.; Rau, R.; Bondy, M.L.; Rabin, K.R.; Shen, L.; Scheurer, M.E. Maternal folate genes and aberrant DNA hypermethylation in pediatric acute lymphoblastic leukemia. PLoS ONE 2018, 13, e0197408. [Google Scholar] [CrossRef]
- Zhou, M.; Ge, X.; Xu, X.; Sheng, B.; Wang, H.; Shi, H.; Liu, S.; Tan, B.; Xu, K.; Wang, J. A hot and cold tumor-related prognostic signature for stage II colorectal cancer. Oncol. Lett. 2024, 28, 419. [Google Scholar] [CrossRef]
- Susmi, T.F.; Rahman, A.; Khan, M.R.; Yasmin, F.; Islam, S.; Nasif, O.; Alharbi, S.A.; Batiha, G.E.-S.; Hossain, M.U. Prognostic and clinicopathological insights of phosphodiesterase 9A gene as novel biomarker in human colorectal cancer. BMC Cancer 2021, 21, 577. [Google Scholar] [CrossRef]
- Ma, X.; Edmonson, M.; Yergeau, D.; Muzny, D.M.; Hampton, O.A.; Rusch, M.; Song, G.; Easton, J.; Harvey, R.C.; Wheeler, D.A.; et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat. Commun. 2015, 6, 6604. [Google Scholar] [CrossRef]
- Lombardo, B.; Esposito, D.; Iossa, S.; Vitale, A.; Verdesca, F.; Perrotta, C.; Di Leo, L.; Costa, V.; Pastore, L.; Franzé, A. Intragenic deletion in macrod2: A family with complex phenotypes including microcephaly, intellectual disability, polydactyly, renal and pancreatic malformations. Cytogenet. Genome Res. 2019, 158, 25–31. [Google Scholar] [CrossRef]
- Lu, Z.; Xie, J.; Wu, G.; Shen, J.; Collins, R.; Chen, W.; Kang, X.; Luo, M.; Zou, Y.; Huang, L.J.-S.; et al. Fasting selectively blocks development of acute lymphoblastic leukemia via leptin-receptor upregulation. Nat. Med. 2016, 23, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liao, Q.; Wen, X.; Fan, J.; Yuan, T.; Tong, X.; Jia, R.; Chai, P.; Fan, X. Hijacking of the nervous system in cancer: Mechanism and therapeutic targets. Mol. Cancer 2025, 24, 44. [Google Scholar] [CrossRef] [PubMed]
- Gossai, N.P.; Gordon, P.M. The role of the central nervous system microenvironment in pediatric acute lymphoblastic leukemia. Front. Pediatr. 2017, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Ranta, S.; Nilsson, F.; Harila-Saari, A.; Saft, L.; Tani, E.; Söderhäll, S.; Heyman, M. Detection of Central Nervous System Involvement in Childhood Acute Lymphoblastic Leukemia by Cytomorphology and Flow Cytometry of the Cerebrospinal Fluid. Pediatric Blood Cancer 2015, 62, 951–956. [Google Scholar] [CrossRef]
- Wang, H.-M.; Feng, M.; Liu, Y.-L.; Wei, C.-F.; Qian, M.-J.; Li, T.; Yang, X.-P.; Cui, S.; Liu, C.-Y.; Yi, F.; et al. Up-Regulation of GABAergic Signal Events in Bone Marrow Lymphocytes in Childhood Acute Lymphoblastic Leukemia. Chin. J. Physiol. 2016, 59, 119–125. [Google Scholar] [CrossRef]
- Liu, X.; Guo, H.; Kang, M.; Fu, W.; Li, H.; Ji, H.; Zhao, J.; Fang, Y.; Du, M.; Xue, Y. Multi-step gene set analysis identified HTR3 family genes involving childhood acute lymphoblastic leukemia susceptibility. Arch. Toxicol. 2024, 99, 299–307. [Google Scholar] [CrossRef]
- Stanulović, V.S.; Al Omair, S.; Reed, M.A.; Roberts, J.; Potluri, S.; Fulton-Ward, T.; Gudgeon, N.; Bishop, E.L.; Roels, J.; Perry, T.A.; et al. The glutamate/aspartate transporter EAAT1 is crucial for T-cell acute lymphoblastic leukemia proliferation and survival. Haematologica 2024, 109, 3505–3519. [Google Scholar] [CrossRef]
- Tang, J.; Peng, W.; Ji, J.; Peng, C.; Wang, T.; Yang, P.; Gu, J.; Feng, Y.; Jin, K.; Wang, X.; et al. GPR176 Promotes Cancer Progression by Interacting with G Protein GNAS to Restrain Cell Mitophagy in Colorectal Cancer. Adv. Sci. 2023, 10, 2205627. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Carpizo, V.; Ruiz-Llorente, S.; Sarmentero, J.; González-Corpas, A.; Barrero, M.J. CREBBP/EP300 bromodomain inhibition affects the proliferation of AR-positive breast cancer cell lines. Mol. Cancer Res. 2019, 17, 720–730. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, Z.; Tian, Y.; Cong, L.; Zheng, Y.; Wu, Z.; Shan, G.; Xia, Y.; Zhu, Y.; Li, X.; et al. MAPK1 promotes the metastasis and invasion of gastric cancer as a bidirectional transcription factor. BMC Cancer 2023, 23, 959. [Google Scholar] [CrossRef]
- Lin, H.-P.; Lin, C.-Y.; Huo, C.; Jan, Y.-J.; Tseng, J.-C.; Jiang, S.S.; Kuo, Y.-Y.; Chen, S.-C.; Wang, C.-T.; Chan, T.-M.; et al. AKT3 promotes prostate cancer proliferation cells through regulation of Akt, B-Raf & TSC1/TSC2. Oncotarget 2015, 6, 27097–27112. [Google Scholar] [CrossRef]
- Chen, S.; Ran, J.; Fan, Z.; Liu, M.; Wu, L.; Li, Q.; Peng, J.; Hu, Z. Functional status analysis of RNH1 in bladder cancer for predicting immunotherapy response. Sci. Rep. 2023, 13, 12625. [Google Scholar] [CrossRef]
- Lin, X.; Dinglin, X.; Cao, S.; Zheng, S.; Wu, C.; Chen, W.; Li, Q.; Hu, Q.; Zheng, F.; Wu, Z.; et al. Enhancer-Driven lncRNA BDNF-AS Induces Endocrine Resistance and Malignant Progression of Breast Cancer through the RNH1/TRIM21/mTOR Cascade. Cell Rep. 2020, 31, 107753. [Google Scholar] [CrossRef]
- Haddad, S.A.; Ruiz-Narváez, E.A.; Haiman, C.A.; Sucheston-Campbell, L.E.; Bensen, J.T.; Zhu, Q.; Liu, S.; Yao, S.; Bandera, E.V.; Rosenberg, L.; et al. An exome-wide analysis of low frequency and rare variants in relation to risk of breast cancer in African American Women: The AMBER Consortium. Carcinogenesis 2016, 37, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Nordlund, J.; Bäcklin, C.L.; Zachariadis, V.; Cavelier, L.; Dahlberg, J.; Öfverholm, I.; Barbany, G.; Nordgren, A.; Övernäs, E.; Abrahamsson, J.; et al. DNA methylation-based subtype prediction for pediatric acute lymphoblastic leukemia. Clin. Epigenet. 2015, 7, 11. [Google Scholar] [CrossRef]
- Kunz, J.B.; Rausch, T.; Bandapalli, O.R.; Eilers, J.; Pechanska, P.; Schuessele, S.; Assenov, Y.; Stütz, A.M.; Kirschner-Schwabe, R.; Hof, J.; et al. Pediatric T-cell lymphoblastic leukemia evolves into relapse by clonal selection, acquisition of mutations and promoter hypomethylation. Haematologica 2015, 100, 1442–1450. [Google Scholar] [CrossRef]
- Liapodimitri, A.; Camacho, O.; Lunsford, K.; Tetens, A.R.; Craig-Schwartz, J.; Skalitzky, K.; Rabin, K.R.; Feinberg, A.; Koldobskiy, M. Mapping the Dynamic DNA Methylation Landscape in Pre-B ALL from Diagnosis to Relapse Reveals a Role for Aberrant PRC2 Regulation. Blood 2024, 144, 4127. [Google Scholar] [CrossRef]
- Orgueira, A.M.; Krali, O.; Míguez, C.P.; Raíndo, A.P.; Arias, J.Á.D.; Pérez, M.S.G.; Encinas, M.M.P.; Sanmartín, M.F.; Sinnet, D.; Heyman, M.; et al. Refining risk prediction in pediatric acute lymphoblastic leukemia through DNA methylation profiling. Clin. Epigenet. 2024, 16, 49. [Google Scholar] [CrossRef]
- Han, S.; Williamson, B.D.; Fong, Y. Improving random forest predictions in small datasets from two-phase sampling designs. BMC Med. Inform. Decis. Mak. 2021, 21, 322. [Google Scholar] [CrossRef]
- Barreñada, L.; Dhiman, P.; Timmerman, D.; Boulesteix, A.-L.; Van Calster, B. Understanding overfitting in random forest for probability estimation: A visualization and simulation study. Diagn. Progn. Res. 2024, 8, 14, Erratum in Diagn. Progn. Res. 2025, 9, 9.. [Google Scholar] [CrossRef]
- Kim, A.; Benavente, C.A. Oncogenic Roles of UHRF1 in Cancer. Epigenomes 2024, 8, 26. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, Z.; Meng, L.; Wang, Y.; Ashaq, M.S.; Li, Y.; Zhao, B. Identification of ubiquitination-related hub genes in chronic myeloid leukemia cell by bioinformatics analysis. J. Cancer 2024, 15, 3750–3759. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Luo, Y.; Zeng, C.; He, H.; Zhang, X. UHRF1 regulates the transcriptional repressor HBP1 through MIF in T acute lymphoblastic leukemia. Oncol. Rep. 2021, 46, 131. [Google Scholar] [CrossRef] [PubMed]
- PDQ Pediatric Treatment Editorial Board. Childhood Acute Lymphoblastic Leukemia Treatment (PDQ®): Health Professional Version. In PDQ Cancer Information Summaries; National Cancer Institute: Bethesda, MD, USA, 2002. [Google Scholar]
- Nordlund, J.; Christofer, B.; Raine, A. Chapter 10 Computational and Statistical Analysis of Array-Based DNA Methylation Data. Methods Mol. Biol. 2019, 1878, 173–191. [Google Scholar]
- Wu, M.C.; Kuan, P. Chapter 16 A Guide to Illumina BeadChip Data Analysis. Methods Mol. Biol. 2018, 1708, 303–330. [Google Scholar] [PubMed]
- Jaffe, A.E.; Murakami, P.; Lee, H.; Leek, J.T.; Fallin, M.D.; Feinberg, A.P.; Irizarry, R.A. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Leuk. Res. 2012, 41, 200–209. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed]
- Ingenuity Pathway Analysis|QIAGEN Digital Insights. Available online: https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ (accessed on 26 September 2025).
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Mansell, G.; Gorrie-Stone, T.J.; Bao, Y.; Kumari, M.; Schalkwyk, L.S.; Mill, J.; Hannon, E. Guidance for DNA methylation studies: Statistical insights from the Illumina EPIC array. BMC Genom. 2019, 20, 27. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. Statistical Powe Analysis for the Behavioral Sciences, 2nd ed.; Lawrence Erlbaum Associates: Mahwah, NJ, USA, 1988; Volume 3. [Google Scholar]


| Variable | n (%) |
|---|---|
| Sex | |
| Male | 24 (45) |
| Female | 29 (55) |
| Age | |
| 1–9 years | 46 (87) |
| >10 years | 7 (13) |
| WBC count at diagnosis (1 × 103/µL) | |
| <50 | 32 (60) |
| >50 | 21 (40) |
| WHO ALL classification | |
| ETV6::RUNX1 | 11 (20) |
| TCF3::PBX1 | 6 (12) |
| BCR::ABL1 | 2 (4) |
| KMT2A::AFF1 | 1 (2) |
| Undetermined * | 33 (62) |
| NCI risk classification at diagnosis | |
| Standard risk | 28 (53) |
| High risk | 25 (47) |
| WHO Z-BMI | |
| Underweight | 7 (13) |
| Normal weight | 25 (48) |
| Overweight | 2 (4) |
| Obesity | 11 (20) |
| No data | 8 (15) |
| Death | |
| Yes | 8 (18) |
| No | 38 (82) |
| Relapse status | |
| Relapse ** | 18(34) |
| VER | 11 (61%) |
| ER | 7 (39%) |
| Non-relapse | 28 (53) |
| Unknown | 7 (13) |
| CpG ID | Chromosome | Gene | Genomic Region | CpG Island Region | Δβ | FDR |
|---|---|---|---|---|---|---|
| cg10030658 | 6 | IGR | island | −0.21 | 7.30 × 10−14 | |
| cg26342454 | 4 | SLC2A9 | 5′UTR | opensea | 0.34 | 4.59 × 10−13 |
| cg00030420 | 5 | SLC25A2 | TSS200 | island | −0.26 | 2.81 × 10−11 |
| cg06098276 | 17 | TK1 | Body | opensea | −0.25 | 2.81 × 10−11 |
| cg07007382 | 6 | IGR | opensea | 0.26 | 4.24 × 10−11 | |
| cg03990261 | 11 | SBF2 | Body | opensea | −0.39 | 1.20 × 10−9 |
| cg09974432 | 1 | PTPN14 | Body | opensea | 0.24 | 1.52 × 10−9 |
| cg09039751 | 11 | CHST1 | Body | island | −0.41 | 1.88 × 10−9 |
| cg11093980 | 18 | MBD1 | Body | shore | −0.23 | 1.88 × 10−9 |
| cg21636683 | 5 | SLC25A2 | TSS200 | island | −0.20 | 2.09 × 10−9 |
| cg23082883 | 7 | IGR | opensea | −0.31 | 3.70 × 10−9 | |
| cg19423170 | 21 | APP | Body | opensea | −0.32 | 3.70 × 10−9 |
| cg00706994 | 11 | OR5A2 | 1stExon | opensea | 0.43 | 3.70 × 10−9 |
| cg16284437 | 18 | SALL3 | Body | shore | −0.23 | 4.42 × 10−9 |
| cg12919873 | 21 | IGR | opensea | 0.45 | 4.98 × 10−9 | |
| cg02918872 | 5 | SOX30 | TSS200 | island | −0.21 | 6.97 × 10−9 |
| cg26767916 | 7 | AOAH | Body | opensea | −0.30 | 6.97 × 10−9 |
| cg09764435 | 1 | GALNT2 | Body | opensea | 0.34 | 6.97 × 10−9 |
| cg25432232 | 19 | AURKC | 5′UTR | island | −0.31 | 7.01 × 10−9 |
| cg16732663 | 1 | SLC26A9 | Body | opensea | 0.50 | 7.36 × 10−9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fong-López, A.A.; Núñez-Enríquez, J.C.; Bekker-Méndez, V.C.; Flores-Lujano, J.; Mata-Rocha, M.; Jiménez-Hernández, E.; Ortíz-Maganda, M.P.; Guerra-Castillo, F.X.; Medina-Sanson, A.; Martín-Trejo, J.A.; et al. Comprehensive Epigenome-Wide Profiling Reveals Distinctive DNA Methylation Signatures and Potential Prognostic Biomarkers in Mexican Pediatric B-ALL. Int. J. Mol. Sci. 2025, 26, 10261. https://doi.org/10.3390/ijms262110261
Fong-López AA, Núñez-Enríquez JC, Bekker-Méndez VC, Flores-Lujano J, Mata-Rocha M, Jiménez-Hernández E, Ortíz-Maganda MP, Guerra-Castillo FX, Medina-Sanson A, Martín-Trejo JA, et al. Comprehensive Epigenome-Wide Profiling Reveals Distinctive DNA Methylation Signatures and Potential Prognostic Biomarkers in Mexican Pediatric B-ALL. International Journal of Molecular Sciences. 2025; 26(21):10261. https://doi.org/10.3390/ijms262110261
Chicago/Turabian StyleFong-López, Alan Alberto, Juan Carlos Núñez-Enríquez, Vilma Carolina Bekker-Méndez, Janet Flores-Lujano, Minerva Mata-Rocha, Elva Jiménez-Hernández, Mónica Patricia Ortíz-Maganda, Francisco Xavier Guerra-Castillo, Aurora Medina-Sanson, Jorge Alfonso Martín-Trejo, and et al. 2025. "Comprehensive Epigenome-Wide Profiling Reveals Distinctive DNA Methylation Signatures and Potential Prognostic Biomarkers in Mexican Pediatric B-ALL" International Journal of Molecular Sciences 26, no. 21: 10261. https://doi.org/10.3390/ijms262110261
APA StyleFong-López, A. A., Núñez-Enríquez, J. C., Bekker-Méndez, V. C., Flores-Lujano, J., Mata-Rocha, M., Jiménez-Hernández, E., Ortíz-Maganda, M. P., Guerra-Castillo, F. X., Medina-Sanson, A., Martín-Trejo, J. A., Peñaloza-González, J. G., Velázquez-Aviña, M. M., Torres-Nava, J. R., Espinosa-Elizondo, R. M., Pérez-Saldívar, M. L., Flores-Villegas, L. V., Merino-Pasaye, L. E., Duarte-Rodríguez, D. A., Sepúlveda-Robles, O. A., ... Jiménez-Morales, S. (2025). Comprehensive Epigenome-Wide Profiling Reveals Distinctive DNA Methylation Signatures and Potential Prognostic Biomarkers in Mexican Pediatric B-ALL. International Journal of Molecular Sciences, 26(21), 10261. https://doi.org/10.3390/ijms262110261










