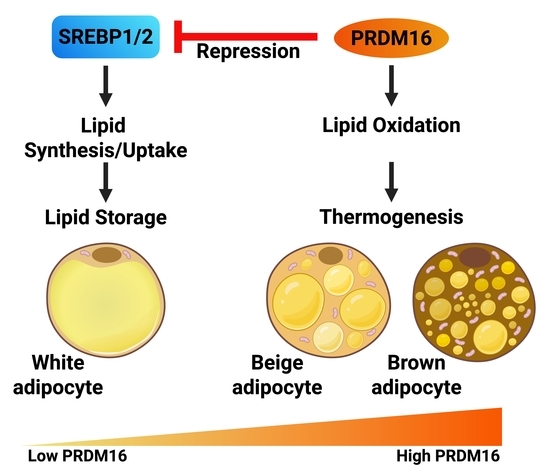
The Functional Interaction Between PRDM16 and the SREBP Pathway Controls Lipid Metabolism
Abstract
1. Introduction
2. Results
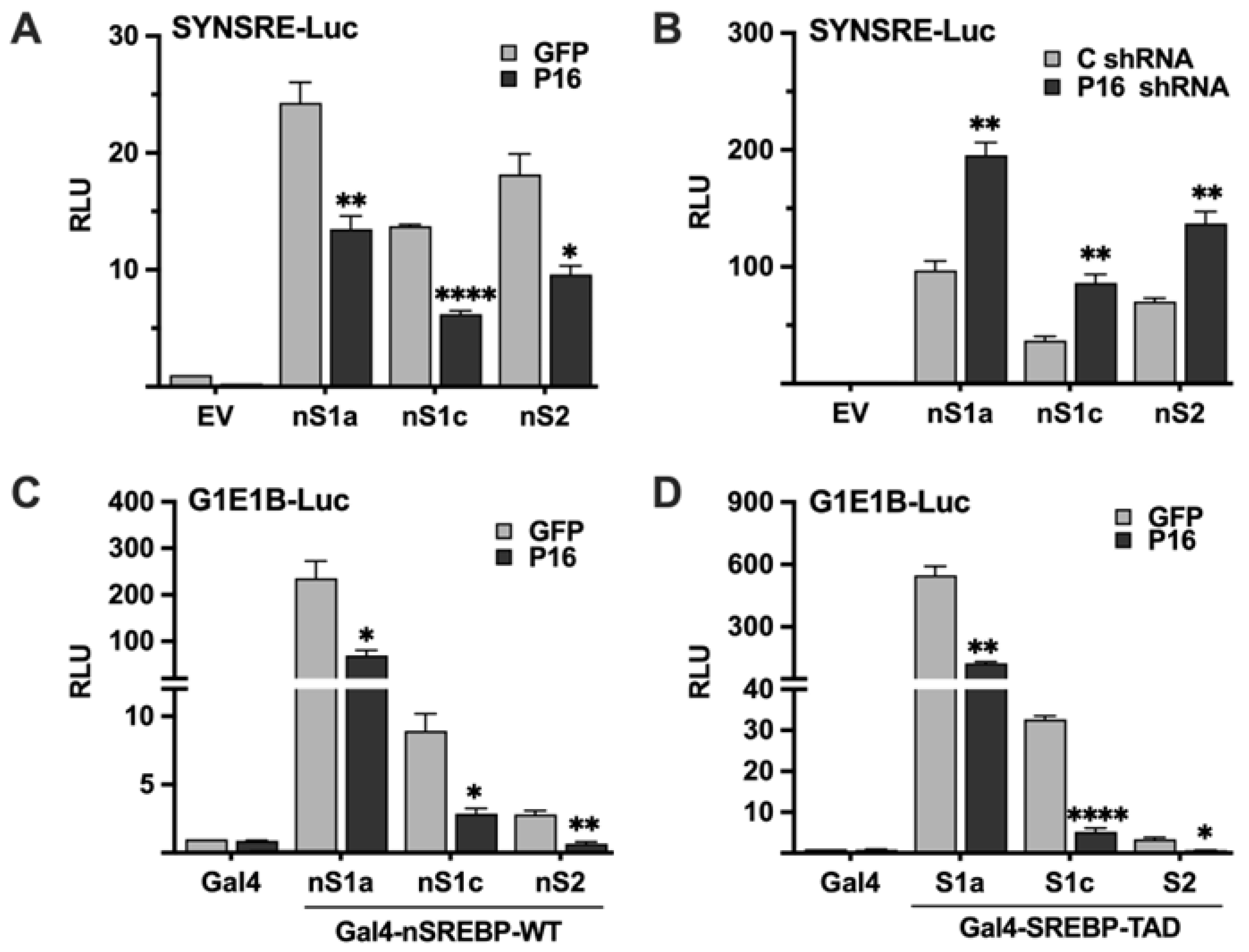
2.1. PRDM16 Represses SREBP Target Promoters in an SREBP-Dependent Manner
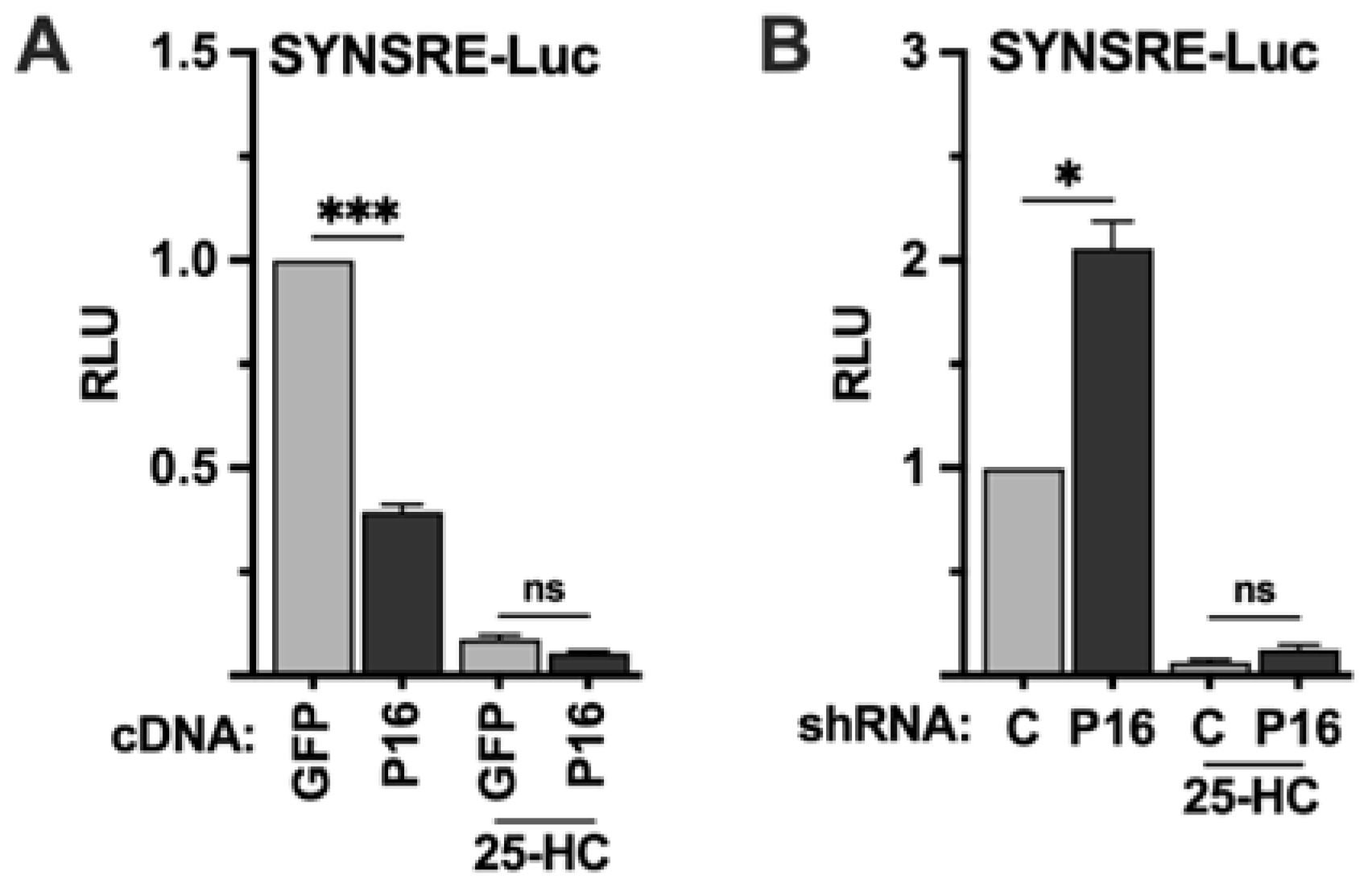
2.2. PRDM16 Is Unable to Regulate the Transcriptional Activities of SREBP1/2 Under Repressive Conditions
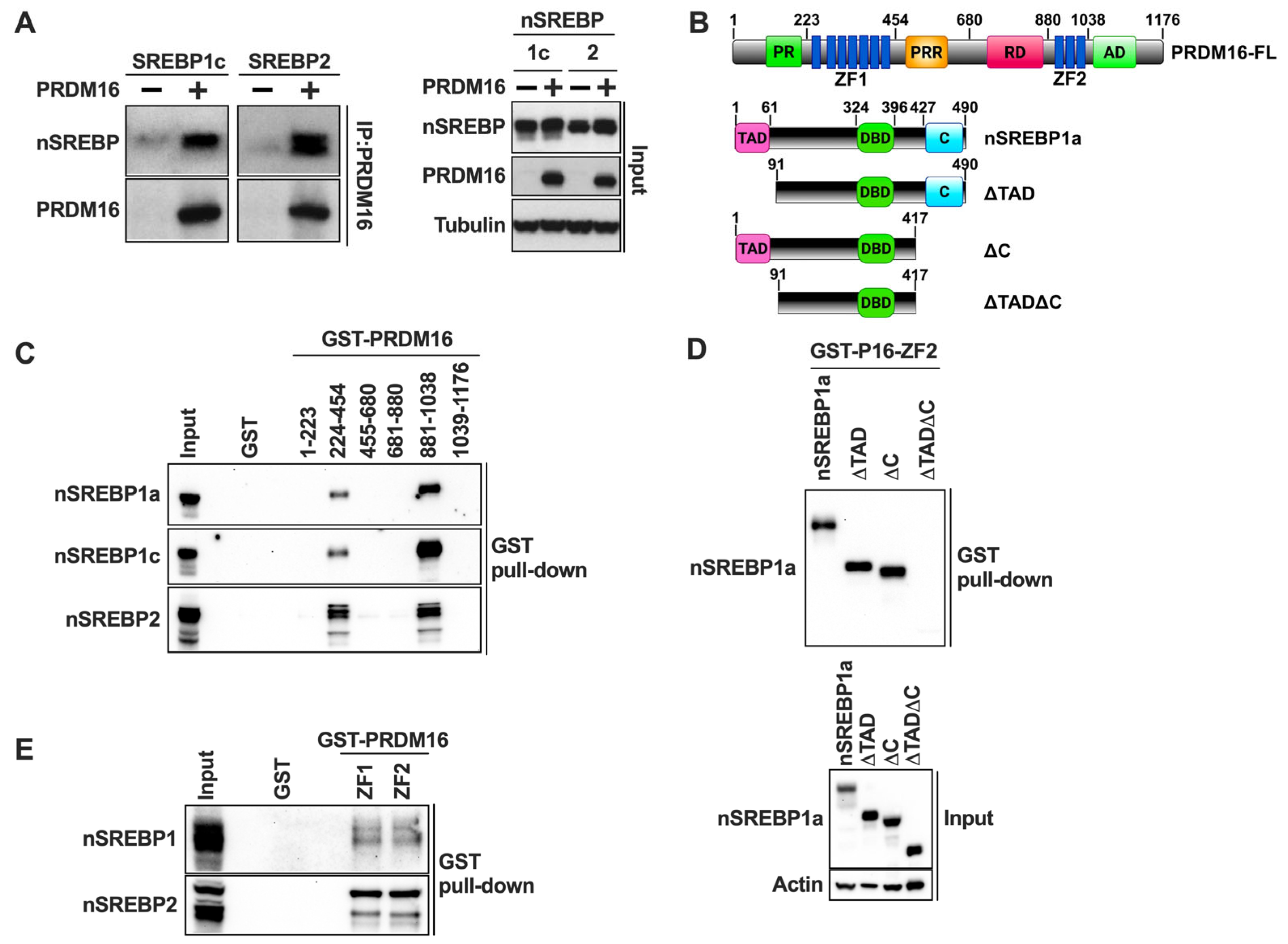
2.3. PRDM16 Targets the Nuclear Forms of SREBP1/2
2.4. PRDM16 Interacts with Nuclear SREBP1/2
2.5. PRDM16 Interacts with Nuclear SREBP1/2 Through Its Zinc Finger Domains
2.6. PRDM16 Interacts with SREBP1 Bound to Target Promoters In Vitro
2.7. Loss of PRDM16 Results in the Induction of SREBP Target Genes in an SREBP-Dependent Manner
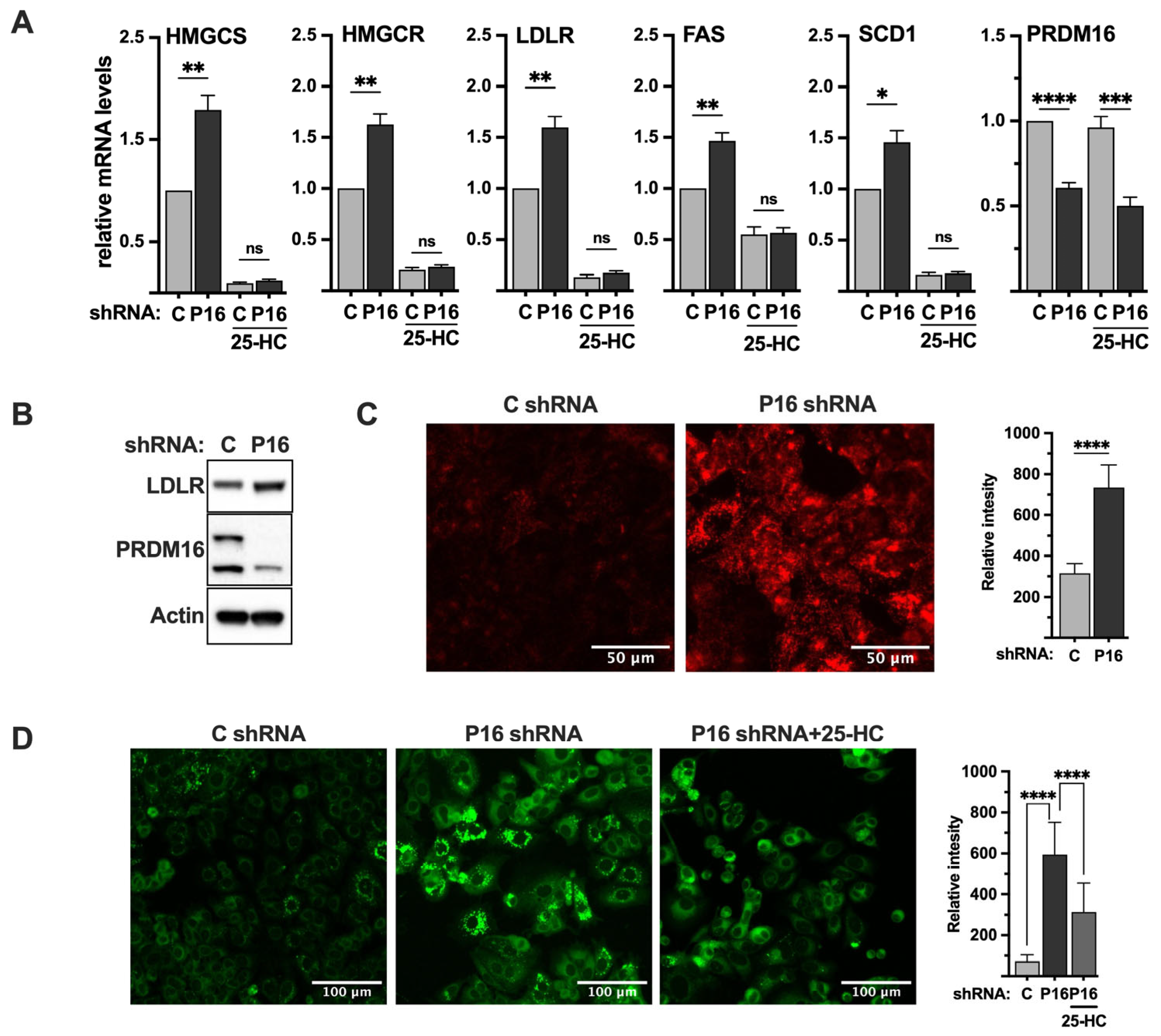
2.8. Loss of PRDM16 Results in the Induction of LDL Receptor Protein, LDL Uptake and Intracellular Lipid Accumulation
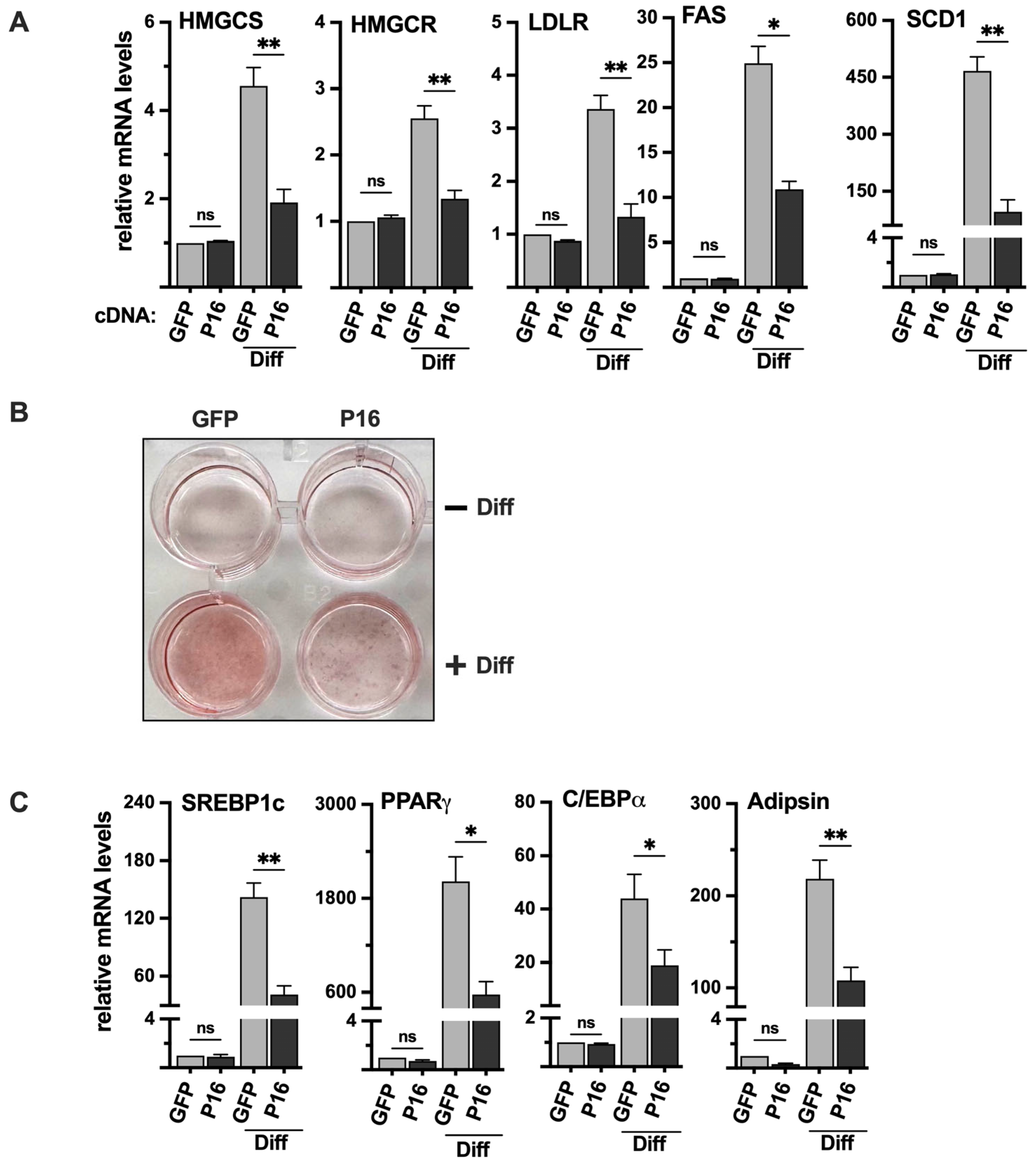
2.9. Ectopic Expression of PRDM16 Blocks the Differentiation of 3T3-L1 Cells
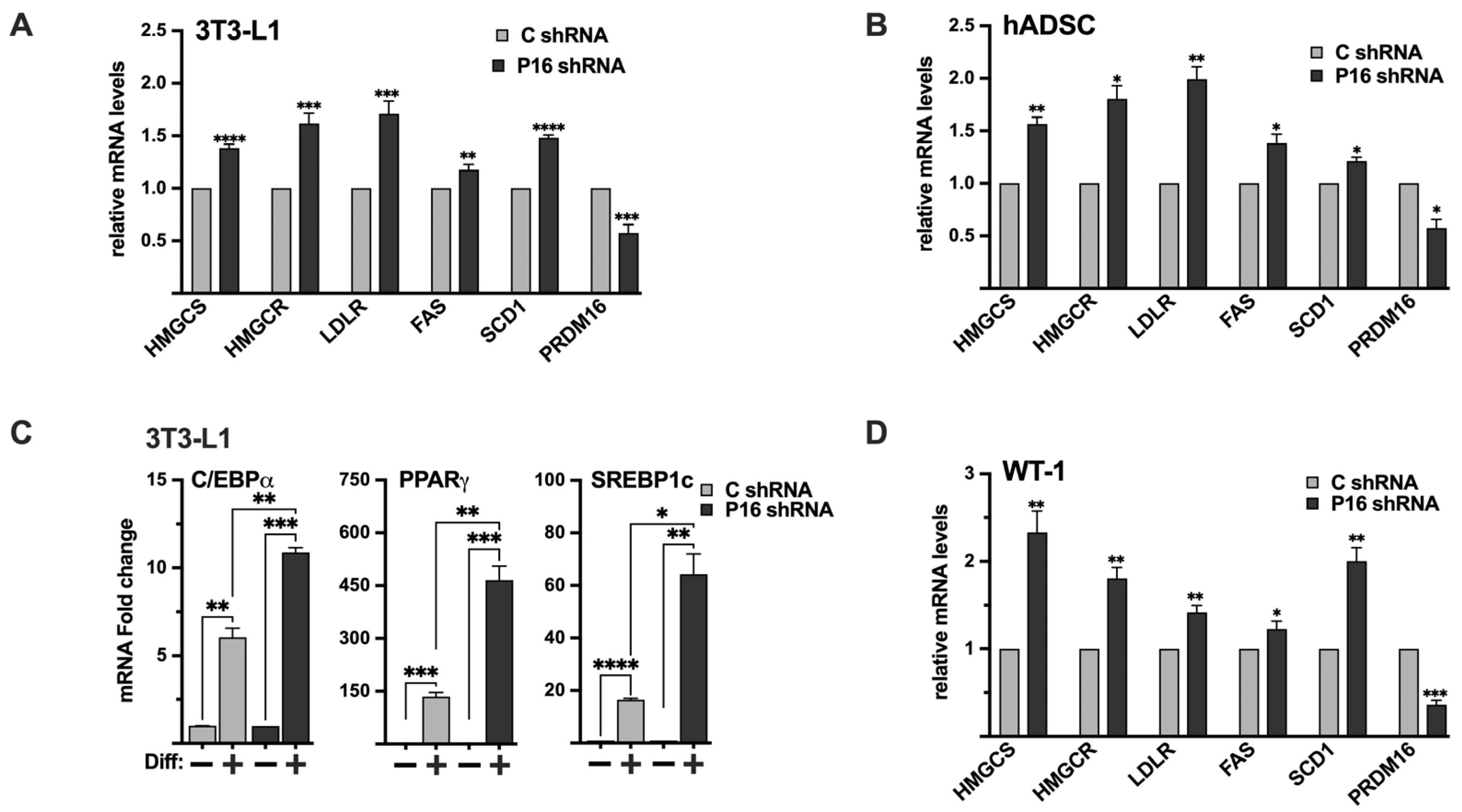
2.10. Endogenous PRDM16 Controls the Expression of SREBP Target Genes in 3T3-L1 Preadipocytes and hADSCs
2.11. Inactivation of PRDM16 in Brown Preadipocytes Enhances the Expression of SREBP Target Genes
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatments
4.2. Adipocyte Differentiation
4.3. Plasmid DNA
4.4. Lentivirus Production and Transduction
4.5. Antibodies and Reagents
4.6. Cell Lysis and Immunoblotting
4.7. Protein Purification
4.8. GST Pulldown and Co-Immunoprecipitation Assays
4.9. DNA Pulldown Assay
4.10. Electromobility Shift Assays
4.11. Luciferase and β-Galactosidase Assays
4.12. RNA Extraction and qPCR
4.13. Oil Red O Staining
4.14. LipidTox Staining of Neutral Lipids
4.15. LDL Uptake Assays
4.16. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SREBP | Sterol Regulatory Element-Binding Protein |
| PRDM | PRDI-BF1 and RIZ Homology Domain Containing |
| SCAP | SREBP Cleavage Activating Protein |
| Insig | Insulin-induced Gene |
| PPAR | Peroxisome Proliferator–Activated Receptor |
| PGC1 | Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 |
| C/EBP | CCAAT-Enhancer-Binding Protein |
| FAS | Fatty acid synthase |
| SCD1 | Stearoyl-CoA 9-desaturase |
| LDL | Low-density Lipoprotein |
| LDLR | Low-density Lipoprotein Receptor |
| HMGCS | HMG-CoA Synthase |
| HMGCR | HMG-CoA Reductase |
| 25-HC | 25-Hydroxycholesterol |
| ER | Endoplasmic Reticulum |
| GFP | Green Fluorescence Protein |
| GST | Glutathione S-Transferase |
| SRE | Sterol Regulatory Element |
| shRNA | Short Hairpin RNA |
| qPCR | Quantitative Polymerase Chain Reaction |
| LDM | Lipoprotein-Deficient Media |
| ADSC | Adipose-Derived Stem Cells |
| ZF | Zinc Finger |
| TAD | Transactivation Domain |
| EMSA | Electromobility Shift Assay |
| mRNA | Messenger RNA |
| PCR | Polymerase Chain Reaction |
| UCP1 | Uncoupling Protein 1 |
| EHMT1 | Euchromatic Histone Methyltransferase 1 |
| MED | Mediator |
| BAT | Brown Adipose Tissue |
| WAT | White Adipose Tissue |
| CtBP1/2 | C-Terminal-Binding Protein 1/2 |
| LSD1 | Lysine-Specific Histone Demethylase 1 |
| T2D | Type-2 Diabetes |
References
- Carli, F.; Della Pepa, G.; Sabatini, S.; Vidal Puig, A.; Gastaldelli, A. Lipid metabolism in MASLD and MASH: From mechanism to the clinic. JHEP Rep. 2024, 6, 101185. [Google Scholar] [CrossRef]
- Luciani, L.; Pedrelli, M.; Parini, P. Modification of lipoprotein metabolism and function driving atherogenesis in diabetes. Atherosclerosis 2024, 394, 117545. [Google Scholar] [CrossRef] [PubMed]
- Samuel, V.T.; Petersen, K.F.; Shulman, G.I. Lipid-induced insulin resistance: Unravelling the mechanism. Lancet 2010, 375, 2267–2277. [Google Scholar] [CrossRef]
- Chew, N.W.S.; Ng, C.H.; Tan, D.J.H.; Kong, G.; Lin, C.; Chin, Y.H.; Lim, W.H.; Huang, D.Q.; Quek, J.; Fu, C.E.; et al. The global burden of metabolic disease: Data from 2000 to 2019. Cell Metab. 2023, 35, 414–428.e3. [Google Scholar] [CrossRef] [PubMed]
- Srnic, N.; Westcott, F.; Caney, E.; Hodson, L. Dietary fat quantity and composition influence hepatic lipid metabolism and metabolic disease risk in humans. Dis. Models Mech. 2025, 18, dmm050878. [Google Scholar] [CrossRef]
- Choe, S.S.; Huh, J.Y.; Hwang, I.J.; Kim, J.I.; Kim, J.B. Adipose Tissue Remodeling: Its Role in Energy Metabolism and Metabolic Disorders. Front. Endocrinol. 2016, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Liu, M. Adipose tissue in control of metabolism. J. Endocrinol. 2016, 231, R77–R99. [Google Scholar] [CrossRef]
- Burchfield, J.G.; Diaz-Vegas, A.; James, D.E. The insulin signalling network. Nat. Metab. 2025, 7, 1745–1764. [Google Scholar] [CrossRef]
- Czech, M.P. Insulin action and resistance in obesity and type 2 diabetes. Nat. Med. 2017, 23, 804–814. [Google Scholar] [CrossRef]
- White, M.F.; Kahn, C.R. Insulin action at a molecular level–100 years of progress. Mol. Metab. 2021, 52, 101304. [Google Scholar] [CrossRef]
- Brown, M.S.; Goldstein, J.L. Cholesterol feedback: From Schoenheimer’s bottle to Scap’s MELADL. J. Lipid Res. 2009, 50, S15–S27. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Goldstein, J.L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA 1999, 96, 11041–11048. [Google Scholar] [CrossRef] [PubMed]
- Osborne, T.F.; Espenshade, P.J. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: What a long, strange tRIP it’s been. Genes Dev. 2009, 23, 2578–2591. [Google Scholar] [CrossRef]
- Shimano, H.; Sato, R. SREBP-regulated lipid metabolism: Convergent physiology—Divergent pathophysiology. Nat. Rev. Endocrinol. 2017, 13, 710–730. [Google Scholar] [CrossRef]
- Brown, M.S.; Radhakrishnan, A.; Goldstein, J.L. Retrospective on Cholesterol Homeostasis: The Central Role of Scap. Annu. Rev. Biochem. 2018, 87, 783–807. [Google Scholar] [CrossRef]
- Bengoechea-Alonso, M.T.; Ericsson, J. SREBP in signal transduction: Cholesterol metabolism and beyond. Curr. Opin. Cell Biol. 2007, 19, 215–222. [Google Scholar] [CrossRef]
- Espenshade, P.J. SREBPs: Sterol-regulated transcription factors. J. Cell Sci. 2006, 119, 973–976. [Google Scholar] [CrossRef]
- Goldstein, J.L.; Brown, M.S. The LDL receptor. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 431–438. [Google Scholar] [CrossRef]
- Goldstein, J.L.; Brown, M.S. A century of cholesterol and coronaries: From plaques to genes to statins. Cell 2015, 161, 161–172. [Google Scholar] [CrossRef]
- Owen, J.L.; Zhang, Y.; Bae, S.H.; Farooqi, M.S.; Liang, G.; Hammer, R.E.; Goldstein, J.L.; Brown, M.S. Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc. Natl. Acad. Sci. USA 2012, 109, 16184–16189. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Yang, J.; Horton, J.D.; Hammer, R.E.; Goldstein, J.L.; Brown, M.S. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 2002, 277, 9520–9528. [Google Scholar] [CrossRef]
- Hirano, Y.; Yoshida, M.; Shimizu, M.; Sato, R. Direct demonstration of rapid degradation of nuclear sterol regulatory element-binding proteins by the ubiquitin-proteasome pathway. J. Biol. Chem. 2001, 276, 36431–36437. [Google Scholar] [CrossRef]
- Sundqvist, A.; Bengoechea-Alonso, M.T.; Ye, X.; Lukiyanchuk, V.; Jin, J.; Harper, J.W.; Ericsson, J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab. 2005, 1, 379–391. [Google Scholar] [CrossRef]
- Dong, Q.; Majumdar, G.; O’Meally, R.N.; Cole, R.N.; Elam, M.B.; Raghow, R. Insulin-induced de novo lipid synthesis occurs mainly via mTOR-dependent regulation of proteostasis of SREBP-1c. Mol. Cell. Biochem. 2020, 463, 13–31. [Google Scholar] [CrossRef] [PubMed]
- Bengoechea-Alonso, M.T.; Ericsson, J. A phosphorylation cascade controls the degradation of active SREBP1. J. Biol. Chem. 2009, 284, 5885–5895. [Google Scholar] [CrossRef] [PubMed]
- Bengoechea-Alonso, M.T.; Aldaalis, A.; Ericsson, J. Loss of the Fbw7 tumor suppressor rewires cholesterol metabolism in cancer cells leading to activation of the PI3K-AKT signalling axis. Front. Oncol. 2022, 12, 990672. [Google Scholar] [CrossRef]
- Jang, Y.; Park, Y.K.; Lee, J.E.; Wan, D.; Tran, N.; Gavrilova, O.; Ge, K. MED1 is a lipogenesis coactivator required for postnatal adipose expansion. Genes Dev. 2021, 35, 713–728. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.B.; Spiegelman, B.M. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996, 10, 1096–1107. [Google Scholar] [CrossRef]
- Ayala-Sumuano, J.T.; Velez-Delvalle, C.; Beltrán-Langarica, A.; Marsch-Moreno, M.; Cerbón-Solorzano, J.; Kuri-Harcuch, W. Srebf1a is a key regulator of transcriptional control for adipogenesis. Sci. Rep. 2011, 1, 178. [Google Scholar] [CrossRef]
- Kim, J.B.; Wright, H.M.; Wright, M.; Spiegelman, B.M. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc. Natl. Acad. Sci. USA 1998, 95, 4333–4337. [Google Scholar] [CrossRef]
- Shimano, H.; Shimomura, I.; Hammer, R.E.; Herz, J.; Goldstein, J.L.; Brown, M.S.; Horton, J.D. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Investig. 1997, 100, 2115–2124. [Google Scholar] [CrossRef]
- Carobbio, S.; Hagen, R.M.; Lelliott, C.J.; Slawik, M.; Medina-Gomez, G.; Tan, C.Y.; Sicard, A.; Atherton, H.J.; Barbarroja, N.; Bjursell, M.; et al. Adaptive changes of the Insig1/SREBP1/SCD1 set point help adipose tissue to cope with increased storage demands of obesity. Diabetes 2013, 62, 3697–3708. [Google Scholar] [CrossRef] [PubMed]
- Crewe, C.; Zhu, Y.; Paschoal, V.A.; Joffin, N.; Ghaben, A.L.; Gordillo, R.; Oh, D.Y.; Liang, G.; Horton, J.D.; Scherer, P.E. SREBP-regulated adipocyte lipogenesis is dependent on substrate availability and redox modulation of mTORC1. JCI Insight 2019, 4, e129397. [Google Scholar] [CrossRef]
- Im, S.S.; Kwon, S.K.; Kang, S.Y.; Kim, T.H.; Kim, H.I.; Hur, M.W.; Kim, K.S.; Ahn, Y.H. Regulation of GLUT4 gene expression by SREBP-1c in adipocytes. Biochem. J. 2006, 399, 131–139. [Google Scholar] [CrossRef]
- Kuriyama, H.; Liang, G.; Engelking, L.J.; Horton, J.D.; Goldstein, J.L.; Brown, M.S. Compensatory increase in fatty acid synthesis in adipose tissue of mice with conditional deficiency of SCAP in liver. Cell Metab. 2005, 1, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Schoonjans, K.; Gelman, L.; Haby, C.; Briggs, M.; Auwerx, J. Induction of LPL gene expression by sterols is mediated by a sterol regulatory element and is independent of the presence of multiple E boxes. J. Mol. Biol. 2000, 304, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Adlanmerini, M.; Carpenter, B.J.; Remsberg, J.R.; Aubert, Y.; Peed, L.C.; Richter, H.J.; Lazar, M.A. Circadian lipid synthesis in brown fat maintains murine body temperature during chronic cold. Proc. Natl. Acad. Sci. USA 2019, 116, 18691–18699. [Google Scholar] [CrossRef]
- Kajimura, S. Promoting brown and beige adipocyte biogenesis through the PRDM16 pathway. Int. J. Obes. Suppl. 2015, 5, S11–S14. [Google Scholar] [CrossRef]
- Seale, P. Transcriptional Regulatory Circuits Controlling Brown Fat Development and Activation. Diabetes 2015, 64, 2369–2375. [Google Scholar] [CrossRef]
- Cohen, P.; Levy, J.D.; Zhang, Y.; Frontini, A.; Kolodin, D.P.; Svensson, K.J.; Lo, J.C.; Zeng, X.; Ye, L.; Khandekar, M.J.; et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 2014, 156, 304–316. [Google Scholar] [CrossRef]
- Inagaki, T.; Sakai, J.; Kajimura, S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 480–495, Erratum in Nat. Rev. Mol. Cell Biol. 2017, 18, 527. [Google Scholar] [CrossRef]
- Brun, R.P.; Kim, J.B.; Hu, E.; Spiegelman, B.M. Peroxisome proliferator-activated receptor gamma and the control of adipogenesis. Curr. Opin. Lipidol. 1997, 8, 212–218. [Google Scholar] [CrossRef]
- Rosen, E.D.; Sarraf, P.; Troy, A.E.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef]
- Guilherme, A.; Rowland, L.A.; Wang, H.; Czech, M.P. The adipocyte supersystem of insulin and cAMP signaling. Trends Cell Biol. 2023, 33, 340–354. [Google Scholar] [CrossRef]
- Santoro, A.; McGraw, T.E.; Kahn, B.B. Insulin action in adipocytes, adipose remodeling, and systemic effects. Cell Metab. 2021, 33, 748–757. [Google Scholar] [CrossRef]
- Seale, P.; Bjork, B.; Yang, W.; Kajimura, S.; Chin, S.; Kuang, S.; Scimè, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008, 454, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Seale, P.; Kajimura, S.; Yang, W.; Chin, S.; Rohas, L.M.; Uldry, M.; Tavernier, G.; Langin, D.; Spiegelman, B.M. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007, 6, 38–54. [Google Scholar] [CrossRef] [PubMed]
- Kajimura, S.; Seale, P.; Kubota, K.; Lunsford, E.; Frangioni, J.V.; Gygi, S.P.; Spiegelman, B.M. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature 2009, 460, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Ohno, H.; Shinoda, K.; Ohyama, K.; Sharp, L.Z.; Kajimura, S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 2013, 504, 163–167. [Google Scholar] [CrossRef]
- Kajimura, S.; Seale, P.; Tomaru, T.; Erdjument-Bromage, H.; Cooper, M.P.; Ruas, J.L.; Chin, S.; Tempst, P.; Lazar, M.A.; Spiegelman, B.M. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008, 22, 1397–1409. [Google Scholar] [CrossRef]
- Seale, P.; Conroe, H.M.; Estall, J.; Kajimura, S.; Frontini, A.; Ishibashi, J.; Cohen, P.; Cinti, S.; Spiegelman, B.M. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Investig. 2011, 121, 96–105. [Google Scholar] [CrossRef]
- Harms, M.J.; Ishibashi, J.; Wang, W.; Lim, H.W.; Goyama, S.; Sato, T.; Kurokawa, M.; Won, K.J.; Seale, P. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab. 2014, 19, 593–604. [Google Scholar] [CrossRef]
- Harms, M.J.; Lim, H.W.; Ho, Y.; Shapira, S.N.; Ishibashi, J.; Rajakumari, S.; Steger, D.J.; Lazar, M.A.; Won, K.J.; Seale, P. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. 2015, 29, 298–307. [Google Scholar] [CrossRef]
- Iida, S.; Chen, W.; Nakadai, T.; Ohkuma, Y.; Roeder, R.G. PRDM16 enhances nuclear receptor-dependent transcription of the brown fat-specific Ucp1 gene through interactions with Mediator subunit MED1. Genes Dev. 2015, 29, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Bard-Chapeau, E.A.; Gunaratne, J.; Kumar, P.; Chua, B.Q.; Muller, J.; Bard, F.A.; Blackstock, W.; Copeland, N.G.; Jenkins, N.A. EVI1 oncoprotein interacts with a large and complex network of proteins and integrates signals through protein phosphorylation. Proc. Natl. Acad. Sci. USA 2013, 110, E2885–E2894. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, I.; Margueron, R.; Shukeir, N.; Eisold, M.; Fritzsch, C.; Richter, F.M.; Mittler, G.; Genoud, C.; Goyama, S.; Kurokawa, M.; et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 2012, 150, 948–960. [Google Scholar] [CrossRef]
- Ferrarese, R.; Izzo, A.; Andrieux, G.; Lagies, S.; Bartmuss, J.P.; Masilamani, A.P.; Wasilenko, A.; Osti, D.; Faletti, S.; Schulzki, R.; et al. ZBTB18 inhibits SREBP-dependent lipid synthesis by halting CTBPs and LSD1 activity in glioblastoma. Life Sci. Alliance 2023, 6, e202201400. [Google Scholar] [CrossRef]
- Sekiya, M.; Kainoh, K.; Sugasawa, T.; Yoshino, R.; Hirokawa, T.; Tokiwa, H.; Nakano, S.; Nagatoishi, S.; Tsumoto, K.; Takeuchi, Y.; et al. The transcriptional corepressor CtBP2 serves as a metabolite sensor orchestrating hepatic glucose and lipid homeostasis. Nat. Commun. 2021, 12, 6315. [Google Scholar] [CrossRef]
- Toth, J.I.; Datta, S.; Athanikar, J.N.; Freedman, L.P.; Osborne, T.F. Selective coactivator interactions in gene activation by SREBP-1a and -1c. Mol. Cell Biol. 2004, 24, 8288–8300. [Google Scholar] [CrossRef]
- Yang, F.; Vought, B.W.; Satterlee, J.S.; Walker, A.K.; Jim Sun, Z.Y.; Watts, J.L.; DeBeaumont, R.; Saito, R.M.; Hyberts, S.G.; Yang, S.; et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 2006, 442, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Goldstein, J.L.; Brown, M.S. Insulin induction of SREBP-1c in rodent liver requires LXRalpha-C/EBPbeta complex. Proc. Natl. Acad. Sci. USA 2016, 113, 8182–8187. [Google Scholar] [CrossRef]
- Payne, V.A.; Au, W.S.; Lowe, C.E.; Rahman, S.M.; Friedman, J.E.; O’Rahilly, S.; Rochford, J.J. C/EBP transcription factors regulate SREBP1c gene expression during adipogenesis. Biochem. J. 2009, 425, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Biferali, B.; Bianconi, V.; Perez, D.F.; Kronawitter, S.P.; Marullo, F.; Maggio, R.; Santini, T.; Polverino, F.; Biagioni, S.; Summa, V.; et al. Prdm16-mediated H3K9 methylation controls fibro-adipogenic progenitors identity during skeletal muscle repair. Sci. Adv. 2021, 7, eabd9371. [Google Scholar] [CrossRef]
- Zeng, X.; Jedrychowski, M.P.; Chen, Y.; Serag, S.; Lavery, G.G.; Gygi, S.P.; Spiegelman, B.M. Lysine-specific demethylase 1 promotes brown adipose tissue thermogenesis via repressing glucocorticoid activation. Genes Dev. 2016, 30, 1822–1836. [Google Scholar] [CrossRef]
- Shimomura, I.; Hammer, R.E.; Richardson, J.A.; Ikemoto, S.; Bashmakov, Y.; Goldstein, J.L.; Brown, M.S. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: Model for congenital generalized lipodystrophy. Genes Dev. 1998, 12, 3182–3194. [Google Scholar] [CrossRef]
- Horton, J.D.; Shimomura, I.; Ikemoto, S.; Bashmakov, Y.; Hammer, R.E. Overexpression of sterol regulatory element-binding protein-1a in mouse adipose tissue produces adipocyte hypertrophy, increased fatty acid secretion, and fatty liver. J. Biol. Chem. 2003, 278, 36652–36660. [Google Scholar] [CrossRef]
- Holman, C.D.; Sakers, A.P.; Calhoun, R.P.; Cheng, L.; Fein, E.C.; Jacobs, C.; Tsai, L.; Rosen, E.D.; Seale, P. Aging impairs cold-induced beige adipogenesis and adipocyte metabolic reprogramming. eLife 2024, 12, RP87756. [Google Scholar] [CrossRef] [PubMed]
- Dooley, K.A.; Millinder, S.; Osborne, T.F. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J. Biol. Chem. 1998, 273, 1349–1356. [Google Scholar] [CrossRef]
- Bengoechea-Alonso, M.T.; Ericsson, J. The phosphorylation-dependent regulation of nuclear SREBP1 during mitosis links lipid metabolism and cell growth. Cell Cycle 2016, 15, 2753–2765. [Google Scholar] [CrossRef] [PubMed]
- Bengoechea-Alonso, M.T.; Ericsson, J. Cdk1/cyclin B-mediated phosphorylation stabilizes SREBP1 during mitosis. Cell Cycle 2006, 5, 1708–1718. [Google Scholar] [CrossRef]
- Bengoechea-Alonso, M.T.; Punga, T.; Ericsson, J. Hyperphosphorylation regulates the activity of SREBP1 during mitosis. Proc. Natl. Acad. Sci. USA 2005, 102, 11681–11686. [Google Scholar] [CrossRef]
- Punga, T.; Bengoechea-Alonso, M.T.; Ericsson, J. Phosphorylation and ubiquitination of the transcription factor sterol regulatory element-binding protein-1 in response to DNA binding. J. Biol. Chem. 2006, 281, 25278–25286. [Google Scholar] [CrossRef] [PubMed]
- Sundqvist, A.; Ericsson, J. Transcription-dependent degradation controls the stability of the SREBP family of transcription factors. Proc. Natl. Acad. Sci. USA 2003, 100, 13833–13838. [Google Scholar] [CrossRef] [PubMed]
- Simonsson, M.; Kanduri, M.; Gronroos, E.; Heldin, C.H.; Ericsson, J. The DNA binding activities of Smad2 and Smad3 are regulated by coactivator-mediated acetylation. J. Biol. Chem. 2006, 281, 39870–39880. [Google Scholar] [CrossRef] [PubMed]
- Ericsson, J.; Edwards, P.A. CBP is required for sterol-regulated and sterol regulatory element-binding protein-regulated transcription. J. Biol. Chem. 1998, 273, 17865–17870. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmood, H.M.; Bengoechea-Alonso, M.T.; Al-Ansari, D.E.; Machaca, K.; Ericsson, J. The Functional Interaction Between PRDM16 and the SREBP Pathway Controls Lipid Metabolism. Int. J. Mol. Sci. 2025, 26, 10246. https://doi.org/10.3390/ijms262110246
Mahmood HM, Bengoechea-Alonso MT, Al-Ansari DE, Machaca K, Ericsson J. The Functional Interaction Between PRDM16 and the SREBP Pathway Controls Lipid Metabolism. International Journal of Molecular Sciences. 2025; 26(21):10246. https://doi.org/10.3390/ijms262110246
Chicago/Turabian StyleMahmood, Hafiz Majid, Maria Teresa Bengoechea-Alonso, Dana E. Al-Ansari, Khaled Machaca, and Johan Ericsson. 2025. "The Functional Interaction Between PRDM16 and the SREBP Pathway Controls Lipid Metabolism" International Journal of Molecular Sciences 26, no. 21: 10246. https://doi.org/10.3390/ijms262110246
APA StyleMahmood, H. M., Bengoechea-Alonso, M. T., Al-Ansari, D. E., Machaca, K., & Ericsson, J. (2025). The Functional Interaction Between PRDM16 and the SREBP Pathway Controls Lipid Metabolism. International Journal of Molecular Sciences, 26(21), 10246. https://doi.org/10.3390/ijms262110246