Immunometabolic Dysregulation in B-Cell Acute Lymphoblastic Leukemia Revealed by Single-Cell RNA Sequencing: Perspectives on Subtypes and Potential Therapeutic Targets
Abstract
1. Introduction
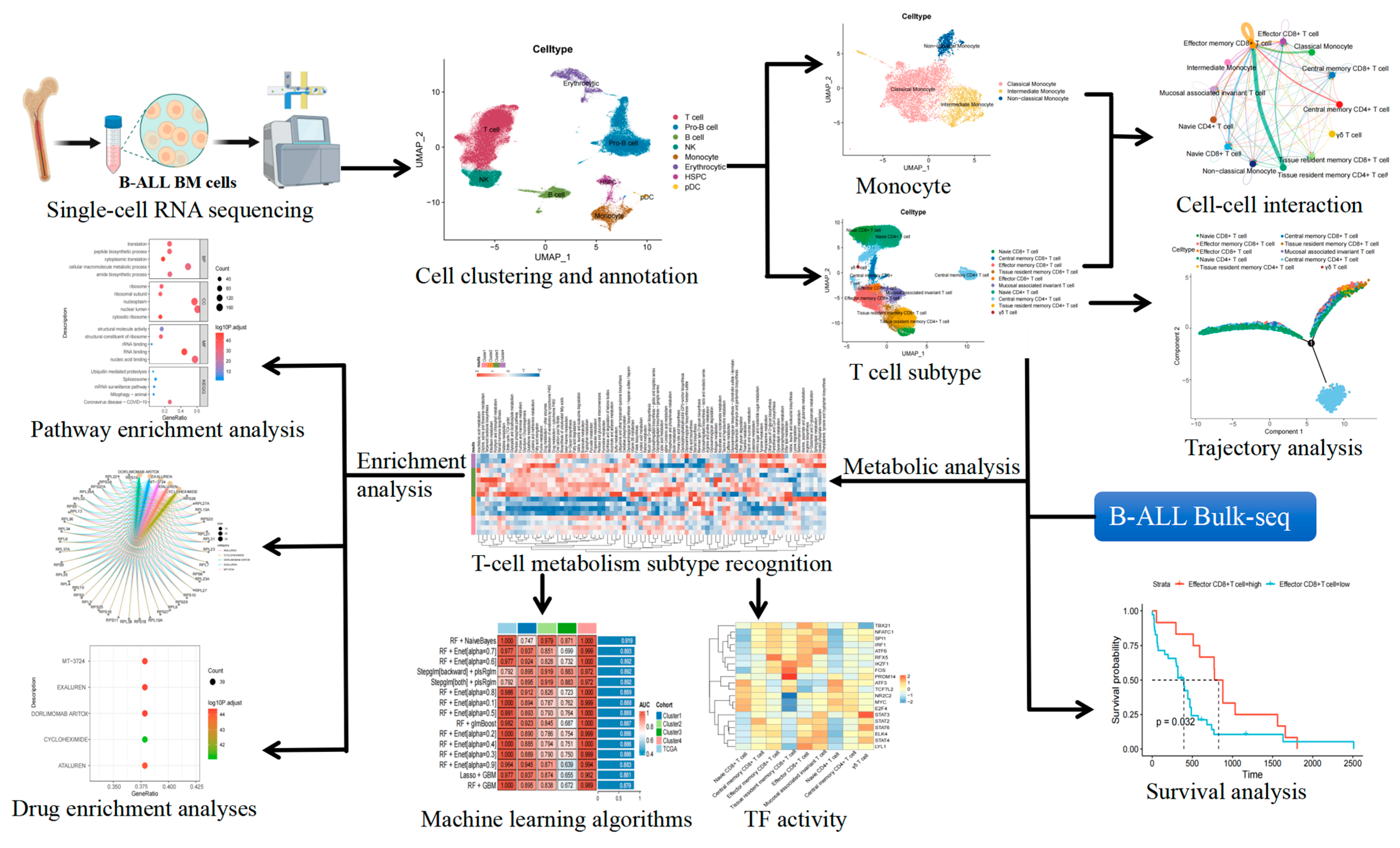
2. Results
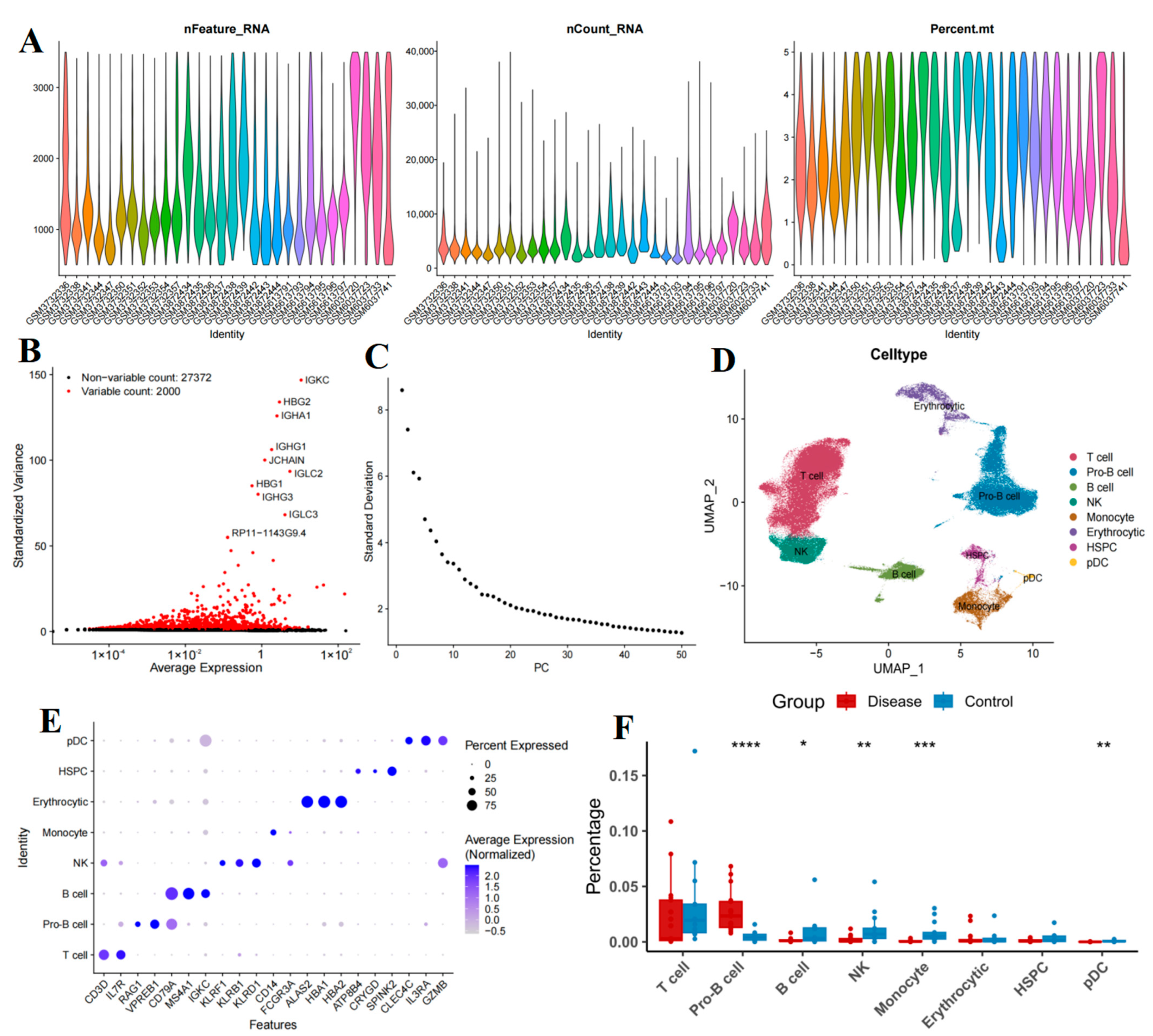
2.1. Cellular Composition of Bone Marrow Samples
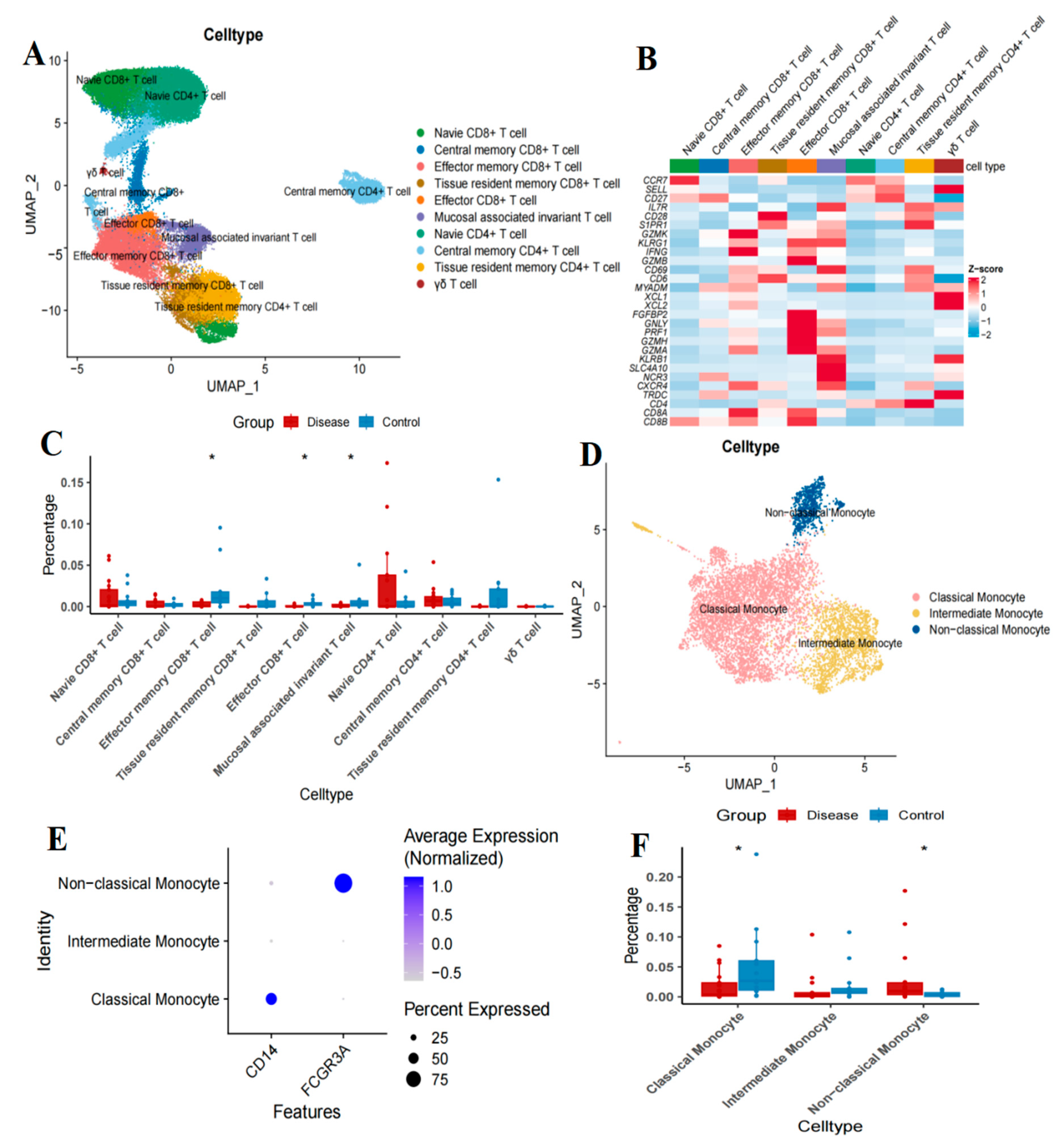
2.2. Altered Proportions of T Cell Subtypes in Patients with B-Cell Acute Lymphoblastic Leukemia
2.3. Trajectory of T-Cell Maturation in B-ALL Bone Marrow
2.4. Prognostic Value of T Cell Subsets for B-Cell Acute Lymphoblastic Leukemia
2.5. Changes in Monocyte Subsets in Patients with B-Cell Acute Lymphoblastic Leukemia
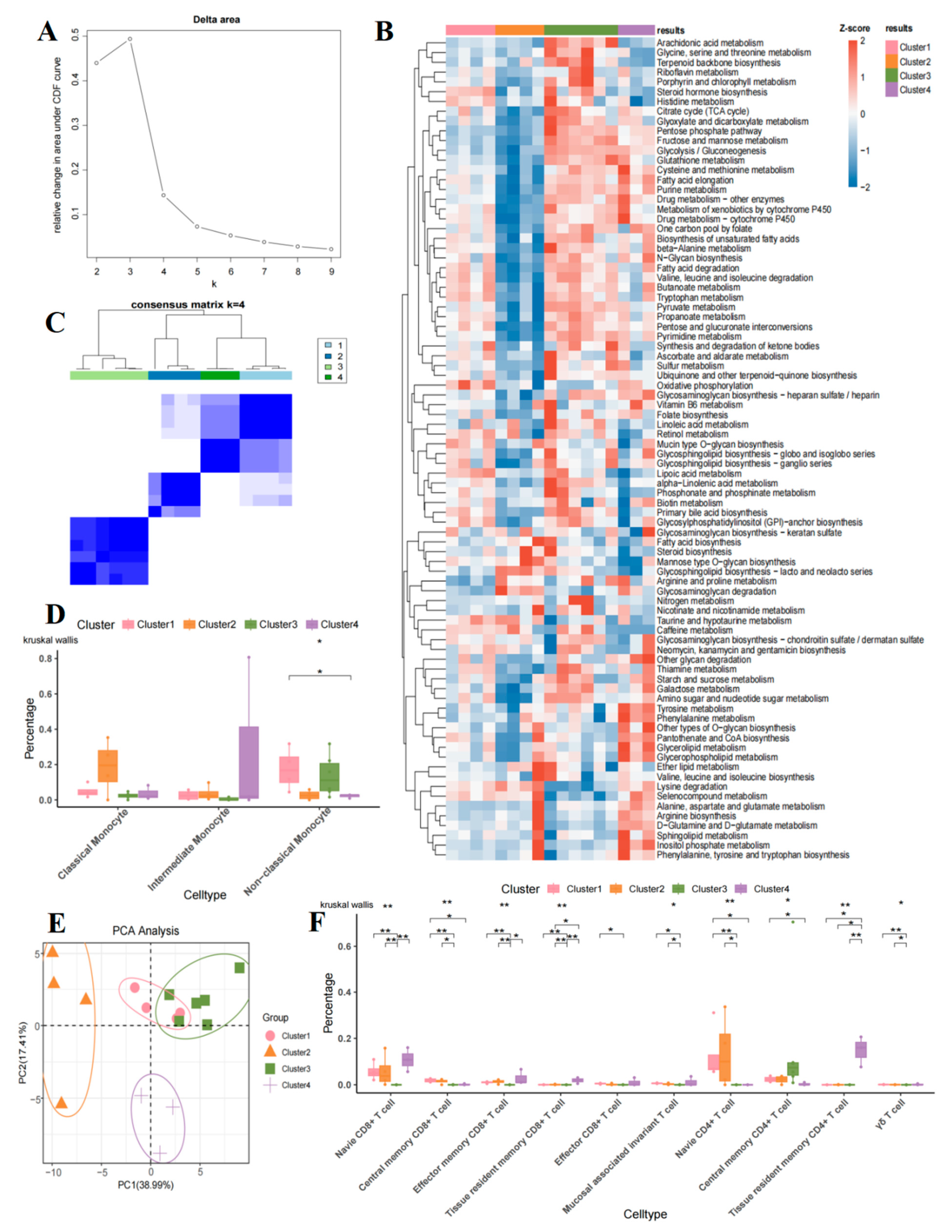
2.6. Metabolic Heterogeneity of T Cell Subsets in B-Cell Acute Lymphoblastic Leukemia
2.7. Comparison of T Cell and Monocyte Subsets in Four Metabolic Groups
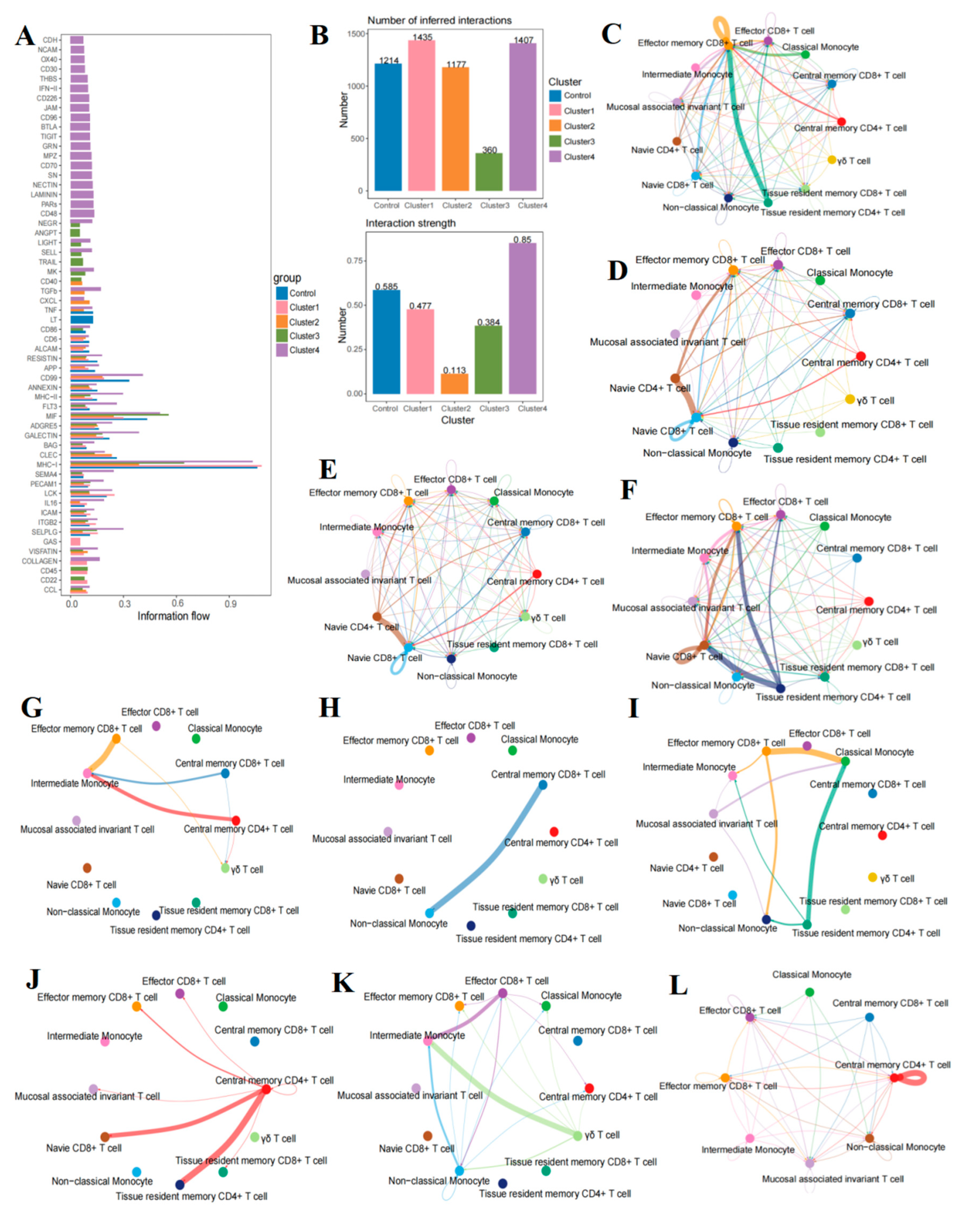
2.8. The Communication Between T Cells and Monocytes in B-Cell Acute Lymphoblastic Leukemia
2.9. Intensive Pathway Mediation in B-Cell Acute Lymphoblastic Leukemia Group D
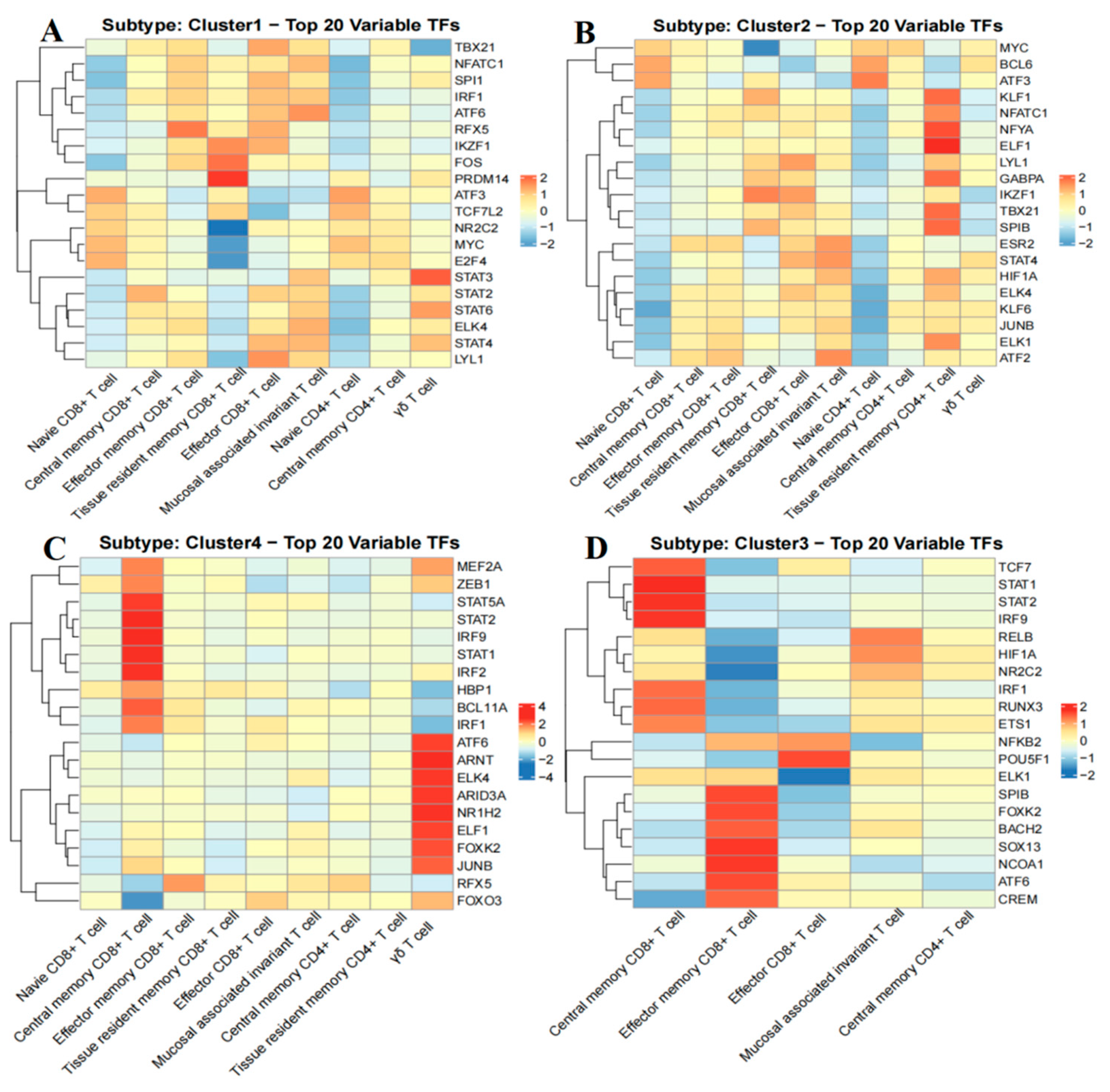
2.10. Analysis of Transcription Factor Activity in B-Cell Acute Lymphoblastic Leukemia Groups
2.11. Differentially Expressed Genes Among Various Groups of B-Cell Acute Lymphoblastic Leukemia
2.12. Advanced Machine Learning Models for Subtype Classification
2.13. Drug Enrichment Analysis for Personalized Treatment of B-Cell Acute Lymphoblastic Leukemia Subtypes
3. Discussion
4. Materials and Methods
4.1. Data Collection
4.2. Single-Cell RNA Sequencing Quality Control and Cell-Type Annotation
4.3. T Cell and Monocyte Clustering and Annotation
4.4. Pseudo-Time Trajectories Analysis
4.5. Survival Analysis
4.6. Metabolic Heterogeneity Analysis
4.7. Consensus Clustering Analysis
4.8. Cell–Cell Communication Analysis
4.9. Calculation of Transcription Factor Activity
4.10. Machine Learning Algorithms
4.11. Differential Gene Expression Analysis
4.12. Enrichment Analysis
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yasuda, T.; Sanada, M.; Tsuzuki, S.; Hayakawa, F. Oncogenic lesions and molecular subtypes in adults with B-cell acute lymphoblastic leukemia. Cancer Sci. 2023, 114, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Passet, M.; Kim, R.; Clappier, E. Genetic subtypes of B-cell acute lymphoblastic leukemia in adults. Blood 2025, 145, 1451–1463. [Google Scholar] [CrossRef]
- Lv, M.; Zhu, S.; Peng, H.; Cheng, Z.; Zhang, G.; Wang, Z. B-cell acute lymphoblastic leukemia-related microRNAs: Uncovering their diverse and special roles. Am. J. Cancer Res. 2021, 11, 1104–1120. [Google Scholar]
- Isidro-Hernandez, M.; Casado-Garcia, A.; Oak, N.; Aleman-Arteaga, S.; Ruiz-Corzo, B.; Martinez-Cano, J.; Mayado, A.; Sanchez, E.G.; Blanco, O.; Gaspar, M.L.; et al. Immune stress suppresses innate immune signaling in preleukemic precursor B-cells to provoke leukemia in predisposed mice. Nat. Commun. 2023, 14, 5159. [Google Scholar] [CrossRef]
- Witkowski, M.T.; Dolgalev, I.; Evensen, N.A.; Ma, C.; Chambers, T.; Roberts, K.G.; Sreeram, S.; Dai, Y.; Tikhonova, A.N.; Lasry, A.; et al. Extensive Remodeling of the Immune Microenvironment in B Cell Acute Lymphoblastic Leukemia. Cancer Cell 2020, 37, 867–882.E12. [Google Scholar] [CrossRef] [PubMed]
- Dander, E.; Fallati, A.; Gulic, T.; Pagni, F.; Gaspari, S.; Silvestri, D.; Cricri, G.; Bedini, G.; Portale, F.; Buracchi, C.; et al. Monocyte-macrophage polarization and recruitment pathways in the tumour microenvironment of B-cell acute lymphoblastic leukaemia. Br. J. Haematol. 2021, 193, 1157–1171. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; You, E.; Park, C.J.; Cho, Y.U.; Jang, S.; Im, H.J.; Seo, J.J.; Park, H.S.; Lee, J.H. Increased expression of immune checkpoint programmed cell death protein-1 (PD-1) on T cell subsets of bone marrow aspirates in patients with B-Lymphoblastic leukemia, especially in relapse and at diagnosis. Cytom. B Clin. Cytom. 2020, 98, 336–347. [Google Scholar] [CrossRef]
- Ocadlikova, D.; Lussana, F.; Fracchiolla, N.; Bonifacio, M.; Santoro, L.; Delia, M.; Chiaretti, S.; Pasciolla, C.; Cignetti, A.; Forghieri, F.; et al. Blinatumomab differentially modulates peripheral blood and bone marrow immune cell repertoire: A Campus ALL study. Br. J. Haematol. 2023, 203, 637–650. [Google Scholar] [CrossRef]
- Gerdemann, U.; Alvarez-Calderon, F. Uncovering CD4+ T-cell exhaustion in B-ALL. Blood 2022, 140, 294–296. [Google Scholar] [CrossRef]
- Atre, T.; Reid, G.S.D. T cell exhaustion in pediatric B-ALL: Current knowledge and future perspectives. Front. Immunol. 2025, 16, 1531145. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, J.; Zhao, B.; Liu, F.; Liu, T.; Duan, Y.; Chen, Y.; Chen, X.; Zou, Y.; Zhang, L.; et al. Single-cell transcriptomic analysis of the immune microenvironment in pediatric acute leukemia. Cancer Lett. 2024, 596, 217018. [Google Scholar] [CrossRef]
- Li, B.; Schmidt, N.W. Epidermal Fatty Acid Binding Protein (E-FABP) Is Not Required for the Generation or Maintenance of Effector and Memory T Cells following Infection with Listeria monocytogenes. PLoS ONE 2016, 11, e0162427. [Google Scholar] [CrossRef]
- Ecker, C.; Guo, L.; Voicu, S.; Gil-de-Gomez, L.; Medvec, A.; Cortina, L.; Pajda, J.; Andolina, M.; Torres-Castillo, M.; Donato, J.L.; et al. Differential Reliance on Lipid Metabolism as a Salvage Pathway Underlies Functional Differences of T Cell Subsets in Poor Nutrient Environments. Cell Rep. 2018, 23, 741–755. [Google Scholar] [CrossRef]
- Ricciardi, S.; Manfrini, N.; Alfieri, R.; Calamita, P.; Crosti, M.C.; Gallo, S.; Muller, R.; Pagani, M.; Abrignani, S.; Biffo, S. The Translational Machinery of Human CD4(+) T Cells Is Poised for Activation and Controls the Switch from Quiescence to Metabolic Remodeling. Cell Metab. 2018, 28, 895–906.E5. [Google Scholar] [CrossRef]
- Voss, K.; Luthers, C.R.; Pohida, K.; Snow, A.L. Fatty Acid Synthase Contributes to Restimulation-Induced Cell Death of Human CD4 T Cells. Front. Mol. Biosci. 2019, 6, 106. [Google Scholar] [CrossRef]
- Tabas, I.; Lichtman, A.H. Monocyte-Macrophages and T Cells in Atherosclerosis. Immunity 2017, 47, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Trompette, A.; Gollwitzer, E.S.; Pattaroni, C.; Lopez-Mejia, I.C.; Riva, E.; Pernot, J.; Ubags, N.; Fajas, L.; Nicod, L.P.; Marsland, B.J. Dietary Fiber Confers Protection against Flu by Shaping Ly6c(-) Patrolling Monocyte Hematopoiesis and CD8(+) T Cell Metabolism. Immunity 2018, 48, 992–1005.E8. [Google Scholar] [CrossRef]
- Burel, J.G.; Pomaznoy, M.; Lindestam Arlehamn, C.S.; Weiskopf, D.; da Silva Antunes, R.; Jung, Y.; Babor, M.; Schulten, V.; Seumois, G.; Greenbaum, J.A.; et al. Circulating T cell-monocyte complexes are markers of immune perturbations. eLife 2019, 8, e46045. [Google Scholar] [CrossRef]
- Gruss, H.J.; Herrmann, F. CD30 ligand, a member of the TNF ligand superfamily, with growth and activation control CD30+ lymphoid and lymphoma cells. Leuk. Lymphoma 1996, 20, 397–409. [Google Scholar] [CrossRef]
- Abdalhabib, E.K.; Algarni, A.; Saboor, M.; Alanazi, F.; Ibrahim, I.K.; Alfeel, A.H.; Alanazi, A.M.; Alanazi, A.M.; Alruwaili, A.M.; Alanazi, M.H.; et al. Association of TNF-alpha rs1800629 with Adult Acute B-Cell Lymphoblastic Leukemia. Genes 2022, 13, 1237. [Google Scholar] [CrossRef] [PubMed]
- Kostel Bal, S.; Giuliani, S.; Block, J.; Repiscak, P.; Hafemeister, C.; Shahin, T.; Kasap, N.; Ransmayr, B.; Miao, Y.; van de Wetering, C.; et al. Biallelic NFATC1 mutations cause an inborn error of immunity with impaired CD8+ T-cell function and perturbed glycolysis. Blood 2023, 142, 827–845. [Google Scholar] [CrossRef]
- Xu, C.; Zhao, W.; Peng, L.; Yin, T.; Guo, J.; Li, Y.; Liu, L.; Yang, J.; Xu, C.; Du, M. PRDM14 extinction enables the initiation of trophoblast stem cell formation. Cell Mol. Life Sci. 2024, 81, 208. [Google Scholar] [CrossRef]
- Fang, D.; Cui, K.; Cao, Y.; Zheng, M.; Kawabe, T.; Hu, G.; Khillan, J.S.; Li, D.; Zhong, C.; Jankovic, D.; et al. Differential regulation of transcription factor T-bet induction during NK cell development and T helper-1 cell differentiation. Immunity 2022, 55, 639–655 e637. [Google Scholar] [CrossRef]
- Zhang, J.; Lyu, T.; Cao, Y.; Feng, H. Role of TCF-1 in differentiation, exhaustion, and memory of CD8(+) T cells: A review. FASEB J. 2021, 35, e21549. [Google Scholar] [CrossRef]
- Hong, H.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Kanagawa, O.; Nakagawa, M.; Okita, K.; Yamanaka, S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 2009, 460, 1132–1135. [Google Scholar] [CrossRef]
- Sprokholt, J.K.; Kaptein, T.M.; van Hamme, J.L.; Overmars, R.J.; Gringhuis, S.I.; Geijtenbeek, T.B.H. RIG-I-like receptor activation by dengue virus drives follicular T helper cell formation and antibody production. PLoS Pathog. 2017, 13, e1006738. [Google Scholar] [CrossRef]
- Maurer, B.; Nivarthi, H.; Wingelhofer, B.; Pham, H.T.T.; Schlederer, M.; Suske, T.; Grausenburger, R.; Schiefer, A.I.; Prchal-Murphy, M.; Chen, D.; et al. High activation of STAT5A drives peripheral T-cell lymphoma and leukemia. Haematologica 2020, 105, 435–447. [Google Scholar] [CrossRef]
- Parker, M.E.; Mehta, N.U.; Liao, T.C.; Tomaszewski, W.H.; Snyder, S.A.; Busch, J.; Ciofani, M. Restriction of innate Tgammadelta17 cell plasticity by an AP-1 regulatory axis. Nat. Immunol. 2025, 26, 1299–1314. [Google Scholar] [CrossRef] [PubMed]
- Clausen, B.E.; Waldburger, J.M.; Schwenk, F.; Barras, E.; Mach, B.; Rajewsky, K.; Forster, I.; Reith, W. Residual MHC class II expression on mature dendritic cells and activated B cells in RFX5-deficient mice. Immunity 1998, 8, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Penzo, M.; Montanaro, L.; Trere, D.; Derenzini, M. The Ribosome Biogenesis-Cancer Connection. Cells 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Liu, Y.; Yu, X.Y.; Pan, X.; Zhang, Y.; Tu, J.; Song, Y.H.; Li, Y. Ribosome biogenesis in disease: New players and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 15. [Google Scholar] [CrossRef]
- Chen, C.; Hao, X.; Lai, X.; Liu, L.; Zhu, J.; Shao, H.; Huang, D.; Gu, H.; Zhang, T.; Yu, Z.; et al. Oxidative phosphorylation enhances the leukemogenic capacity and resistance to chemotherapy of B cell acute lymphoblastic leukemia. Sci. Adv. 2021, 7, eabd6280. [Google Scholar] [CrossRef]
- Ling, Y.; Xu, N.; Zhao, K.; Han, L.; Zhang, Q.; Fan, Z.; Huang, F.; Chen, Z.; Xuan, L.; Liu, H.; et al. Allogeneic hematopoietic cell transplant overcomes the poor prognostic value of CDKN2 deletion in adult B-lineage acute lymphoblastic leukemia. Cancer Lett. 2021, 510, 59–66. [Google Scholar] [CrossRef]
- Cox, C.V.; Blair, A. A primitive cell origin for B-cell precursor ALL? Stem Cell Rev. 2005, 1, 189–196. [Google Scholar] [CrossRef]
- Paietta, E.; Roberts, K.G.; Wang, V.; Gu, Z.; Buck, G.A.N.; Pei, D.; Cheng, C.; Levine, R.L.; Abdel-Wahab, O.; Cheng, Z.; et al. Molecular classification improves risk assessment in adult BCR-ABL1-negative B-ALL. Blood 2021, 138, 948–958. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ai, Y.; Abdel-Wahab, O. Molecular impact of mutations in RNA splicing factors in cancer. Mol. Cell 2024, 84, 3667–3680. [Google Scholar] [CrossRef]
- Tseng, C.C.; Obeng, E.A. RNA splicing as a therapeutic target in myelodysplastic syndromes. Semin. Hematol. 2024, 61, 431–441. [Google Scholar] [CrossRef]
- Gu, Z.; Churchman, M.L.; Roberts, K.G.; Moore, I.; Zhou, X.; Nakitandwe, J.; Hagiwara, K.; Pelletier, S.; Gingras, S.; Berns, H.; et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 2019, 51, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Besse, L.; Besse, A.; Kraus, M.; Maurits, E.; Overkleeft, H.S.; Bornhauser, B.; Bourquin, J.P.; Driessen, C. High Immunoproteasome Activity and sXBP1 in Pediatric Precursor B-ALL Predicts Sensitivity towards Proteasome Inhibitors. Cells 2021, 10, 2853. [Google Scholar] [CrossRef]
- Zhuo, Z.; Wang, J.; Zhang, Y.; Meng, G. Integrative alternative splicing analysis reveals new prognosis signature in B-cell acute lymphoblastic leukemia. Int. J. Biol. Sci. 2024, 20, 4496–4512. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.; Hao, Y.; He, C.; Li, L.; Zhu, G. Exosome-orchestrated hypoxic tumor microenvironment. Mol. Cancer 2019, 18, 57. [Google Scholar] [CrossRef]
- Li, C.; Hou, X.; Zhang, P.; Li, J.; Liu, X.; Wang, Y.; Guan, Q.; Zhou, Y. Exosome-based Tumor Therapy: Opportunities and Challenges. Curr. Drug Metab. 2020, 21, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, M.L.; Cook, R.S.; Johnson, D.B.; Balko, J.M. Biological Consequences of MHC-II Expression by Tumor Cells in Cancer. Clin. Cancer Res. 2019, 25, 2392–2402. [Google Scholar] [CrossRef]
- Dersh, D.; Phelan, J.D.; Gumina, M.E.; Wang, B.; Arbuckle, J.H.; Holly, J.; Kishton, R.J.; Markowitz, T.E.; Seedhom, M.O.; Fridlyand, N.; et al. Genome-wide Screens Identify Lineage- and Tumor-Specific Genes Modulating MHC-I- and MHC-II-Restricted Immunosurveillance of Human Lymphomas. Immunity 2021, 54, 116–131 e110. [Google Scholar] [CrossRef]
- Davis, R.E.; Ngo, V.N.; Lenz, G.; Tolar, P.; Young, R.M.; Romesser, P.B.; Kohlhammer, H.; Lamy, L.; Zhao, H.; Yang, Y.; et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2010, 463, 88–92. [Google Scholar] [CrossRef]
- Mayes, K.; Qiu, Z.; Alhazmi, A.; Landry, J.W. ATP-dependent chromatin remodeling complexes as novel targets for cancer therapy. Adv. Cancer Res. 2014, 121, 183–233. [Google Scholar] [CrossRef]
- Tian, Y.; Xu, L.; Li, X.; Li, H.; Zhao, M. SMARCA4: Current status and future perspectives in non-small-cell lung cancer. Cancer Lett. 2023, 554, 216022. [Google Scholar] [CrossRef]
- Huang, Y.; Luo, L.; Xu, Y.; Li, J.; Wu, Z.; Zhao, C.; Wen, J.; Jiang, P.; Zhu, H.; Wang, L.; et al. UHRF1-mediated epigenetic reprogramming regulates glycolysis to promote progression of B-cell acute lymphoblastic leukemia. Cell Death Dis. 2025, 16, 351. [Google Scholar] [CrossRef]
- Li, X.; Meng, W.; Wang, X.; Huang, S.; Wang, J.; Liang, H.; Si, D. Ribosomal protein L9 is a potential therapeutic target for B-ALL through the activation of the p53 signaling pathway. Front. Immunol. 2025, 16, 1560706. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Di, S.; Nie, F.; Meng, W.; Huang, S. RPS15a knockdown impedes the progression of B-ALL by inducing p53-mediated nucleolar stress. Biochem. Biophys. Res. Commun. 2025, 763, 151768. [Google Scholar] [CrossRef] [PubMed]
- Bortolozzi, R.; Mattiuzzo, E.; Trentin, L.; Accordi, B.; Basso, G.; Viola, G. Ribociclib, a Cdk4/Cdk6 kinase inhibitor, enhances glucocorticoid sensitivity in B-acute lymphoblastic leukemia (B-All). Biochem. Pharmacol. 2018, 153, 230–241. [Google Scholar] [CrossRef]
- Wilde, L.; Porazzi, P.; Trotta, R.; De Dominici, M.; Palmisiano, N.; Keiffer, G.; Rancani, K.; Yingling, K.; Calabretta, B.; Kasner, M. A phase I study of the combination of palbociclib and dexamethasone for the treatment of relapsed or refractory B-cell acute lymphoblastic leukemia. Leuk. Res. 2023, 129, 107075. [Google Scholar] [CrossRef] [PubMed]
- Raetz, E.A.; Teachey, D.T.; Minard, C.; Liu, X.; Norris, R.E.; Denic, K.Z.; Reid, J.; Evensen, N.A.; Gore, L.; Fox, E.; et al. Palbociclib in combination with chemotherapy in pediatric and young adult patients with relapsed/refractory acute lymphoblastic leukemia and lymphoma: A Children’s Oncology Group study (AINV18P1). Pediatr. Blood Cancer 2023, 70, e30609. [Google Scholar] [CrossRef]
- Ketzer, F.; Abdelrasoul, H.; Vogel, M.; Marienfeld, R.; Muschen, M.; Jumaa, H.; Wirth, T.; Ushmorov, A. CCND3 is indispensable for the maintenance of B-cell acute lymphoblastic leukemia. Oncogenesis 2022, 11, 1. [Google Scholar] [CrossRef]
- Shi, Y.F.; Wang, N.; Huang, Z.Y.; Chen, R.R.; Huang, Y.S.; Zhu, Y.Y.; Xing, C.Y.; Liang, B.; Yu, K.; Feng, J.H. Normal Absolute Monocyte Count at the Time of Relapse is Associated with Improved Survival After First Salvage Therapy in Adult Patients with Early Relapsed B-Lineage Acute Lymphoblastic Leukemia. Cancer Manag. Res. 2020, 12, 7097–7105. [Google Scholar] [CrossRef] [PubMed]
- Metayer, L.E.; Brown, R.D.; Carlebur, S.; Burke, G.A.A.; Brown, G.C. Mechanisms of cell death induced by arginase and asparaginase in precursor B-cell lymphoblasts. Apoptosis 2019, 24, 145–156. [Google Scholar] [CrossRef]
- Kantarjian, H.; Pui, C.H.; Jabbour, E. Acute lymphocytic leukaemia. Lancet 2025, 406, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Reizis, B. Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity 2019, 50, 37–50. [Google Scholar] [CrossRef]
- Olingy, C.E.; Dinh, H.Q.; Hedrick, C.C. Monocyte heterogeneity and functions in cancer. J. Leukoc. Biol. 2019, 106, 309–322. [Google Scholar] [CrossRef]
- Shimasaki, N.; Jain, A.; Campana, D. NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 2020, 19, 200–218. [Google Scholar] [CrossRef]
- Blaeschke, F.; Stenger, D.; Kaeuferle, T.; Willier, S.; Lotfi, R.; Kaiser, A.D.; Assenmacher, M.; Doring, M.; Feucht, J.; Feuchtinger, T. Induction of a central memory and stem cell memory phenotype in functionally active CD4(+) and CD8(+) CAR T cells produced in an automated good manufacturing practice system for the treatment of CD19(+) acute lymphoblastic leukemia. Cancer Immunol. Immunother. 2018, 67, 1053–1066. [Google Scholar] [CrossRef]
- Shi, S.; Xing, H.; Xu, X.; Chai, J.; Lu, Z.; Wang, J.; Wang, B. CXCR6 defines therapeutic subtypes of CD4(+) cytotoxic T cell lineage for adoptive cell transfer therapy in pediatric B cell acute lymphoblastic leukemia. Int. Immunopharmacol. 2024, 132, 111972. [Google Scholar] [CrossRef]
- Han, J.; Khatwani, N.; Searles, T.G.; Turk, M.J.; Angeles, C.V. Memory CD8(+) T cell responses to cancer. Semin. Immunol. 2020, 49, 101435. [Google Scholar] [CrossRef]
- St Paul, M.; Ohashi, P.S. The Roles of CD8(+) T Cell Subsets in Antitumor Immunity. Trends Cell Biol. 2020, 30, 695–704. [Google Scholar] [CrossRef]
- Li, Y.R.; Zhou, K.; Wilson, M.; Kramer, A.; Zhu, Y.; Dawson, N.; Yang, L. Mucosal-associated invariant T cells for cancer immunotherapy. Mol. Ther. 2023, 31, 631–646. [Google Scholar] [CrossRef]
- Nakamura, K.; Smyth, M.J. Myeloid immunosuppression and immune checkpoints in the tumor microenvironment. Cell Mol. Immunol. 2020, 17, 1–12. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Y.; Li, Z.; Huang, B.; Xu, L.; Lai, J.; Lu, Y.; Zha, X.; Liu, B.; Lan, Y.; et al. Single-Cell RNA-Seq of T Cells in B-ALL Patients Reveals an Exhausted Subset with Remarkable Heterogeneity. Adv. Sci. 2021, 8, e2101447. [Google Scholar] [CrossRef] [PubMed]
- Youngblood, B.; Hale, J.S.; Ahmed, R. T-cell memory differentiation: Insights from transcriptional signatures and epigenetics. Immunology 2013, 139, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.D.; O’Sullivan, D.; Pearce, E.L. T cell metabolism drives immunity. J. Exp. Med. 2015, 212, 1345–1360. [Google Scholar] [CrossRef] [PubMed]
- Belizario, J.E.; Brandao, W.; Rossato, C.; Peron, J.P. Thymic and Postthymic Regulation of Naive CD4(+) T-Cell Lineage Fates in Humans and Mice Models. Mediat. Inflamm. 2016, 2016, 9523628. [Google Scholar] [CrossRef]
- Koh, C.H.; Lee, S.; Kwak, M.; Kim, B.S.; Chung, Y. CD8 T-cell subsets: Heterogeneity, functions, and therapeutic potential. Exp. Mol. Med. 2023, 55, 2287–2299. [Google Scholar] [CrossRef]
- Kok, L.; Masopust, D.; Schumacher, T.N. The precursors of CD8(+) tissue resident memory T cells: From lymphoid organs to infected tissues. Nat. Rev. Immunol. 2022, 22, 283–293. [Google Scholar] [CrossRef]
- Laky, K.; Fleischacker, C.; Fowlkes, B.J. TCR and Notch signaling in CD4 and CD8 T-cell development. Immunol. Rev. 2006, 209, 274–283. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Valignat, M.P.; Zhang, L.; Gelard, L.; Zhang, F.; Le Guen, V.; Audebert, S.; Camoin, L.; Fossum, E.; Bogen, B.; et al. ARHGAP45 controls naive T- and B-cell entry into lymph nodes and T-cell progenitor thymus seeding. EMBO Rep. 2021, 22, e52196. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, Y.; Chamoto, K.; Oura, T.; Honjo, T. Critical role of the CD44(low)CD62L(low) CD8(+) T cell subset in restoring antitumor immunity in aged mice. Proc. Natl. Acad. Sci. USA 2021, 118, e2103730118. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, L.E.; Young, R.M.; Hawkins, T.A.; Stickney, H.L.; Cavodeassi, F.; Schwarz, Q.; Pullin, L.M.; Villegas, R.; Moro, E.; Argenton, F.; et al. Lef1-dependent Wnt/beta-catenin signalling drives the proliferative engine that maintains tissue homeostasis during lateral line development. Development 2011, 138, 3931–3941. [Google Scholar] [CrossRef]
- Kim, E.; Cheng, Y.; Bolton-Gillespie, E.; Cai, X.; Ma, C.; Tarangelo, A.; Le, L.; Jambhekar, M.; Raman, P.; Hayer, K.E.; et al. Rb family proteins enforce the homeostasis of quiescent hematopoietic stem cells by repressing Socs3 expression. J. Exp. Med. 2017, 214, 1901–1912. [Google Scholar] [CrossRef]
- Liu, Q.; Sun, Z.; Chen, L. Memory T cells: Strategies for optimizing tumor immunotherapy. Protein Cell 2020, 11, 549–564. [Google Scholar] [CrossRef]
- Corrado, M.; Pearce, E.L. Targeting memory T cell metabolism to improve immunity. J. Clin. Invest. 2022, 132, e148546. [Google Scholar] [CrossRef]
- Viel, S.; Vivier, E.; Walzer, T.; Marcais, A. Targeting metabolic dysfunction of CD8 T cells and natural killer cells in cancer. Nat. Rev. Drug Discov. 2025, 24, 190–208. [Google Scholar] [CrossRef]
- Peralta, R.M.; Xie, B.; Lontos, K.; Nieves-Rosado, H.; Spahr, K.; Joshi, S.; Ford, B.R.; Quann, K.; Frisch, A.T.; Dean, V.; et al. Dysfunction of exhausted T cells is enforced by MCT11-mediated lactate metabolism. Nat. Immunol. 2024, 25, 2297–2307. [Google Scholar] [CrossRef]
- Tang, Y.; Chen, Z.; Zuo, Q.; Kang, Y. Regulation of CD8+ T cells by lipid metabolism in cancer progression. Cell Mol. Immunol. 2024, 21, 1215–1230. [Google Scholar] [CrossRef]
- Scharping, N.E.; Menk, A.V.; Moreci, R.S.; Whetstone, R.D.; Dadey, R.E.; Watkins, S.C.; Ferris, R.L.; Delgoffe, G.M. The Tumor Microenvironment Represses T Cell Mitochondrial Biogenesis to Drive Intratumoral T Cell Metabolic Insufficiency and Dysfunction. Immunity 2016, 45, 374–388. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Xu, J.; Liu, S.; Lv, Y.; Zhang, R.; Zhou, X.; Zhang, Y.; Weng, S.; Xu, H.; Ba, Y.; et al. Infiltrating treg reprogramming in the tumor immune microenvironment and its optimization for immunotherapy. Biomark. Res. 2024, 12, 97. [Google Scholar] [CrossRef] [PubMed]
- Vodnala, S.K.; Eil, R.; Kishton, R.J.; Sukumar, M.; Yamamoto, T.N.; Ha, N.H.; Lee, P.H.; Shin, M.; Patel, S.J.; Yu, Z.; et al. T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 2019, 363, eaau0135. [Google Scholar] [CrossRef] [PubMed]
- Reina-Campos, M.; Scharping, N.E.; Goldrath, A.W. CD8(+) T cell metabolism in infection and cancer. Nat. Rev. Immunol. 2021, 21, 718–738. [Google Scholar] [CrossRef]
- Dander, E.; Palmi, C.; D’Amico, G.; Cazzaniga, G. The Bone Marrow Niche in B-Cell Acute Lymphoblastic Leukemia: The Role of Microenvironment from Pre-Leukemia to Overt Leukemia. Int. J. Mol. Sci. 2021, 22, 4426. [Google Scholar] [CrossRef]
- Sperling, S.; Fiedler, P.; Lechner, M.; Pollithy, A.; Ehrenberg, S.; Schiefer, A.I.; Kenner, L.; Feuchtinger, A.; Kuhn, R.; Swinerd, G.; et al. Chronic CD30 signaling in B cells results in lymphomagenesis by driving the expansion of plasmablasts and B1 cells. Blood 2019, 133, 2597–2609. [Google Scholar] [CrossRef]
- Sam, I.; Ben Hamouda, N.; Alkatrib, M.; Gonnin, C.; Siska, P.J.; Oudard, S.; Lebbe, C.; Tartour, E. The CD70-CD27 Axis in Cancer Immunotherapy: Predictive Biomarker and Therapeutic Target. Clin. Cancer Res. 2025, 31, 2872–2881. [Google Scholar] [CrossRef]
- Allahbakhshian Farsani, M.; Kamel, M.; Mehrpouri, M.; Heris, R.S.; Hamidpour, M.; Salari, S.; Mohamadi, M.H. The Expression of Interferon Gamma (IFN-gamma) and Interleukin 6 (IL6) in Patients with Acute Lymphoblastic Leukemia (ALL). Pathol. Oncol. Res. 2020, 26, 461–466. [Google Scholar] [CrossRef]
- Chen, L. PD-L1 and the dawn of modern cancer immunotherapy. Nat. Med. 2025, 31, 1378. [Google Scholar] [CrossRef]
- Guan, X.; Hu, R.; Choi, Y.; Srivats, S.; Nabet, B.Y.; Silva, J.; McGinnis, L.; Hendricks, R.; Nutsch, K.; Banta, K.L.; et al. Anti-TIGIT antibody improves PD-L1 blockade through myeloid and T(reg) cells. Nature 2024, 627, 646–655. [Google Scholar] [CrossRef]
- Jablonska, E.; Dakowicz, L.; Ratajczak-Wrona, W.; Garley, M.; Sawicka-Powierza, J.; Krawczuk-Rybak, M. TNF superfamily proteins in the serum of patients with B-ALL—Preliminary study. Clin. Lab. 2014, 60, 1757–1764. [Google Scholar] [CrossRef]
- Fischer, R.; Kontermann, R.E.; Pfizenmaier, K. Selective Targeting of TNF Receptors as a Novel Therapeutic Approach. Front. Cell Dev. Biol. 2020, 8, 401. [Google Scholar] [CrossRef]
- Wu, X.; Li, T.; Jiang, R.; Yang, X.; Guo, H.; Yang, R. Targeting MHC-I molecules for cancer: Function, mechanism, and therapeutic prospects. Mol. Cancer 2023, 22, 194. [Google Scholar] [CrossRef]
- Escherich, C.S.; Li, Z.; Barnett, K.R.; Li, Y.; Walker, M.; Yoshimura, S.; Yang, W.; Huang, X.; Yu, J.; Stock, W.; et al. Differentiation-dependent EBF1 activity determines CD22 transcription and leukemia sensitivity to inotuzumab ozogamicin. Blood 2025, 146, 471–481. [Google Scholar] [CrossRef]
- Yin, H.; Wang, J.; Tan, Y.; Jiang, M.; Zhang, H.; Meng, G. Transcription factor abnormalities in B-ALL leukemogenesis and treatment. Trends Cancer 2023, 9, 855–870. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, H.; Li, Z.; Bai, L.; Wang, Q.; Li, J.; Jiang, M.; Xue, Q.; Cheng, N.; Zhang, W.; et al. Functional, structural, and molecular characterizations of the leukemogenic driver MEF2D-HNRNPUL1 fusion. Blood 2022, 140, 1390–1407. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Zhang, Z.; Hernandez, K.; Savage, J.; Li, S.; Miller, D.; Agrawal, S.; Ortuno, F.; Staudt, L.M.; Heath, A.; Grossman, R.L. Uniform genomic data analysis in the NCI Genomic Data Commons. Nat. Commun. 2021, 12, 1226. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587 e3529. [Google Scholar] [CrossRef]
- Aran, D.; Looney, A.P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R.P.; Wolters, P.J.; Abate, A.R.; et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019, 20, 163–172. [Google Scholar] [CrossRef]
- Lasry, A.; Nadorp, B.; Fornerod, M.; Nicolet, D.; Wu, H.; Walker, C.J.; Sun, Z.; Witkowski, M.T.; Tikhonova, A.N.; Guillamot-Ruano, M.; et al. An inflammatory state remodels the immune microenvironment and improves risk stratification in acute myeloid leukemia. Nat. Cancer 2023, 4, 27–42, Erratum in: Nat. Cancer 2023, 4, 149. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, X.; Zheng, L.; Zhang, Y.; Li, Y.; Fang, Q.; Gao, R.; Kang, B.; Zhang, Q.; Huang, J.Y.; et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018, 564, 268–272. [Google Scholar] [CrossRef]
- Zheng, L.; Qin, S.; Si, W.; Wang, A.; Xing, B.; Gao, R.; Ren, X.; Wang, L.; Wu, X.; Zhang, J.; et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 2021, 374, abe6474. [Google Scholar] [CrossRef]
- Ezdoglian, A.; Tsang, A.S.M.; Khodadust, F.; Burchell, G.; Jansen, G.; de Gruijl, T.; Labots, M.; van der Laken, C.J. Monocyte-related markers as predictors of immune checkpoint inhibitor efficacy and immune-related adverse events: A systematic review and meta-analysis. Cancer Metastasis Rev. 2025, 44, 35. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; Pliner, H.A.; Trapnell, C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 2017, 14, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Sessler, T.; Quinn, G.P.; Wappett, M.; Rogan, E.; Sharkey, D.; Ahmaderaghi, B.; Lawler, M.; Longley, D.B.; McDade, S.S. surviveR: A flexible shiny application for patient survival analysis. Sci. Rep. 2023, 13, 22093. [Google Scholar] [CrossRef]
- Muhl, D.; Herold, M.; Herold, Z.; Hornyak, L.; Szasz, A.M.; Dank, M. Longitudinal Analysis of 1alpha,25-dihidroxyvitamin D(3) and Homocysteine Changes in Colorectal Cancer. Cancers 2022, 14, 658. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, S.; Ma, J.; Chen, Z.; Song, G.; Rao, D.; Cheng, Y.; Huang, S.; Liu, Y.; Jiang, S.; et al. Spatiotemporal Immune Landscape of Colorectal Cancer Liver Metastasis at Single-Cell Level. Cancer Discov. 2022, 12, 134–153. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Ni, S.; Zhao, K.; Qian, J.; Duan, Y. Bioinformatics-Led Discovery of Osteoarthritis Biomarkers and Inflammatory Infiltrates. Front. Immunol. 2022, 13, 871008. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Turei, D.; Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019, 29, 1363–1375. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Freshour, S.L.; Kiwala, S.; Cotto, K.C.; Coffman, A.C.; McMichael, J.F.; Song, J.J.; Griffith, M.; Griffith, O.L.; Wagner, A.H. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021, 49, D1144–D1151. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, D.; Hu, D.; Wang, J.; Peng, J.; Wang, S. Immunometabolic Dysregulation in B-Cell Acute Lymphoblastic Leukemia Revealed by Single-Cell RNA Sequencing: Perspectives on Subtypes and Potential Therapeutic Targets. Int. J. Mol. Sci. 2025, 26, 9996. https://doi.org/10.3390/ijms26209996
Sun D, Hu D, Wang J, Peng J, Wang S. Immunometabolic Dysregulation in B-Cell Acute Lymphoblastic Leukemia Revealed by Single-Cell RNA Sequencing: Perspectives on Subtypes and Potential Therapeutic Targets. International Journal of Molecular Sciences. 2025; 26(20):9996. https://doi.org/10.3390/ijms26209996
Chicago/Turabian StyleSun, Dingya, Dun Hu, Jialu Wang, Jun Peng, and Shan Wang. 2025. "Immunometabolic Dysregulation in B-Cell Acute Lymphoblastic Leukemia Revealed by Single-Cell RNA Sequencing: Perspectives on Subtypes and Potential Therapeutic Targets" International Journal of Molecular Sciences 26, no. 20: 9996. https://doi.org/10.3390/ijms26209996
APA StyleSun, D., Hu, D., Wang, J., Peng, J., & Wang, S. (2025). Immunometabolic Dysregulation in B-Cell Acute Lymphoblastic Leukemia Revealed by Single-Cell RNA Sequencing: Perspectives on Subtypes and Potential Therapeutic Targets. International Journal of Molecular Sciences, 26(20), 9996. https://doi.org/10.3390/ijms26209996




