Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients II: A Preliminary Investigation of the Complementary Low Molecular Mass Fraction
Abstract
1. Introduction
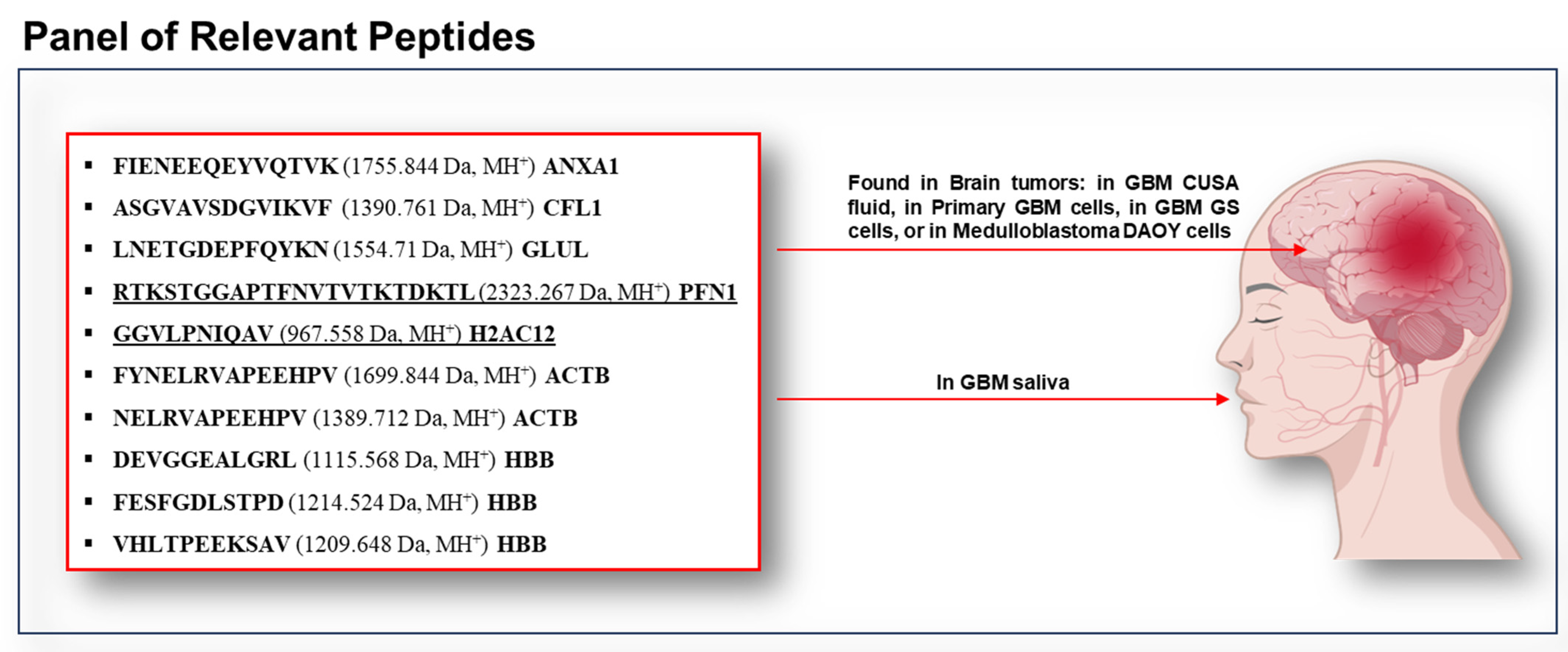
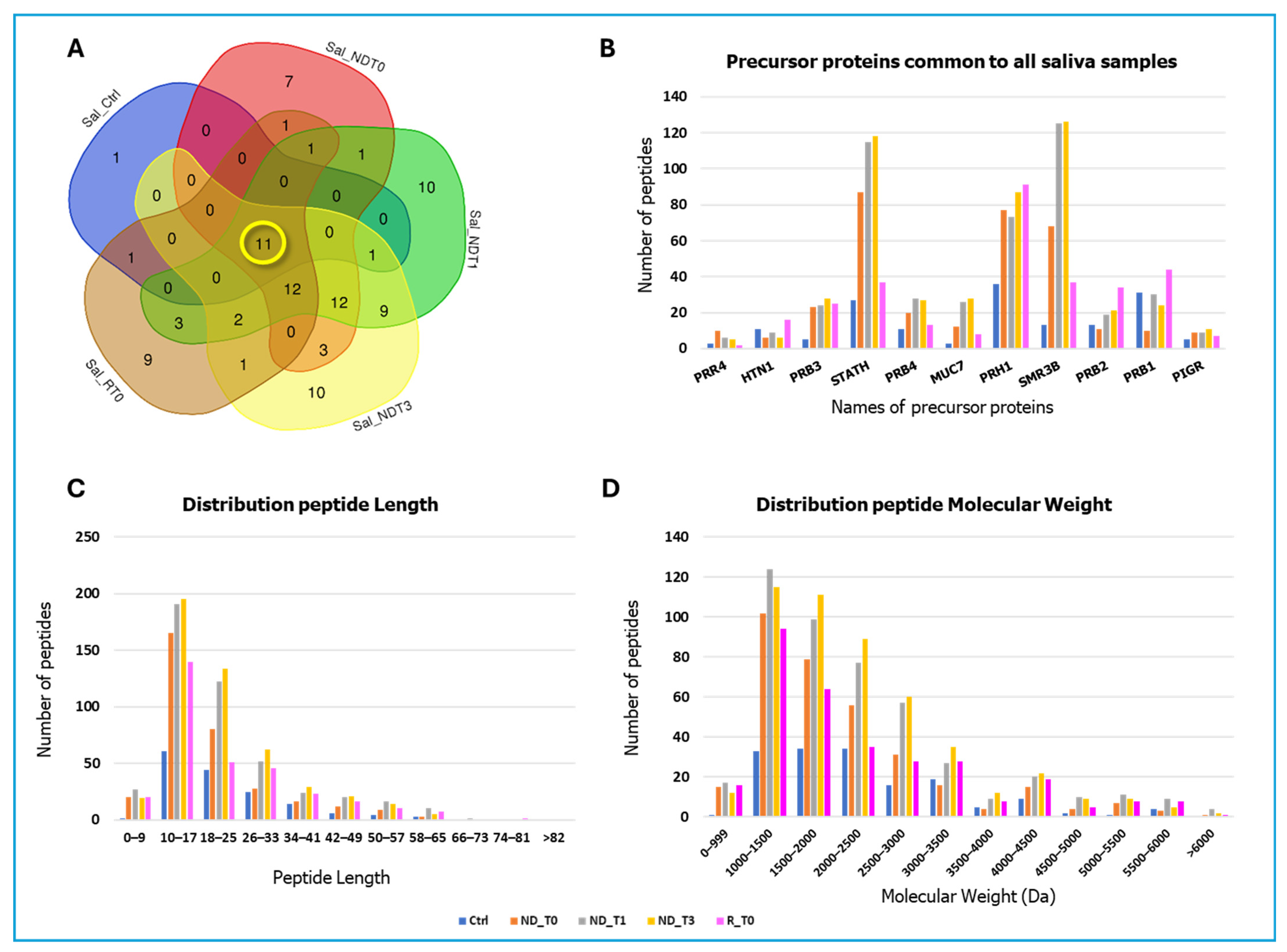
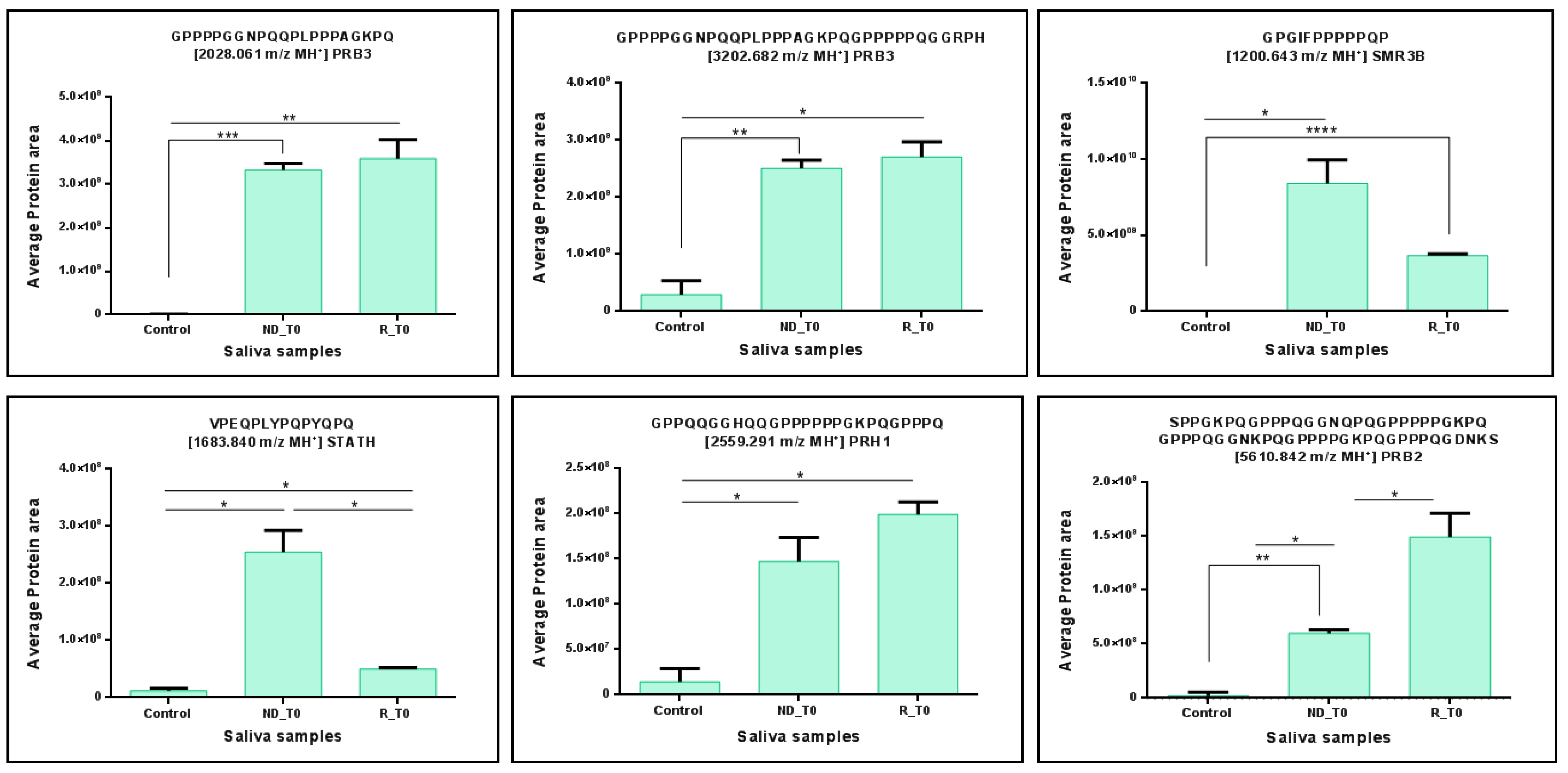
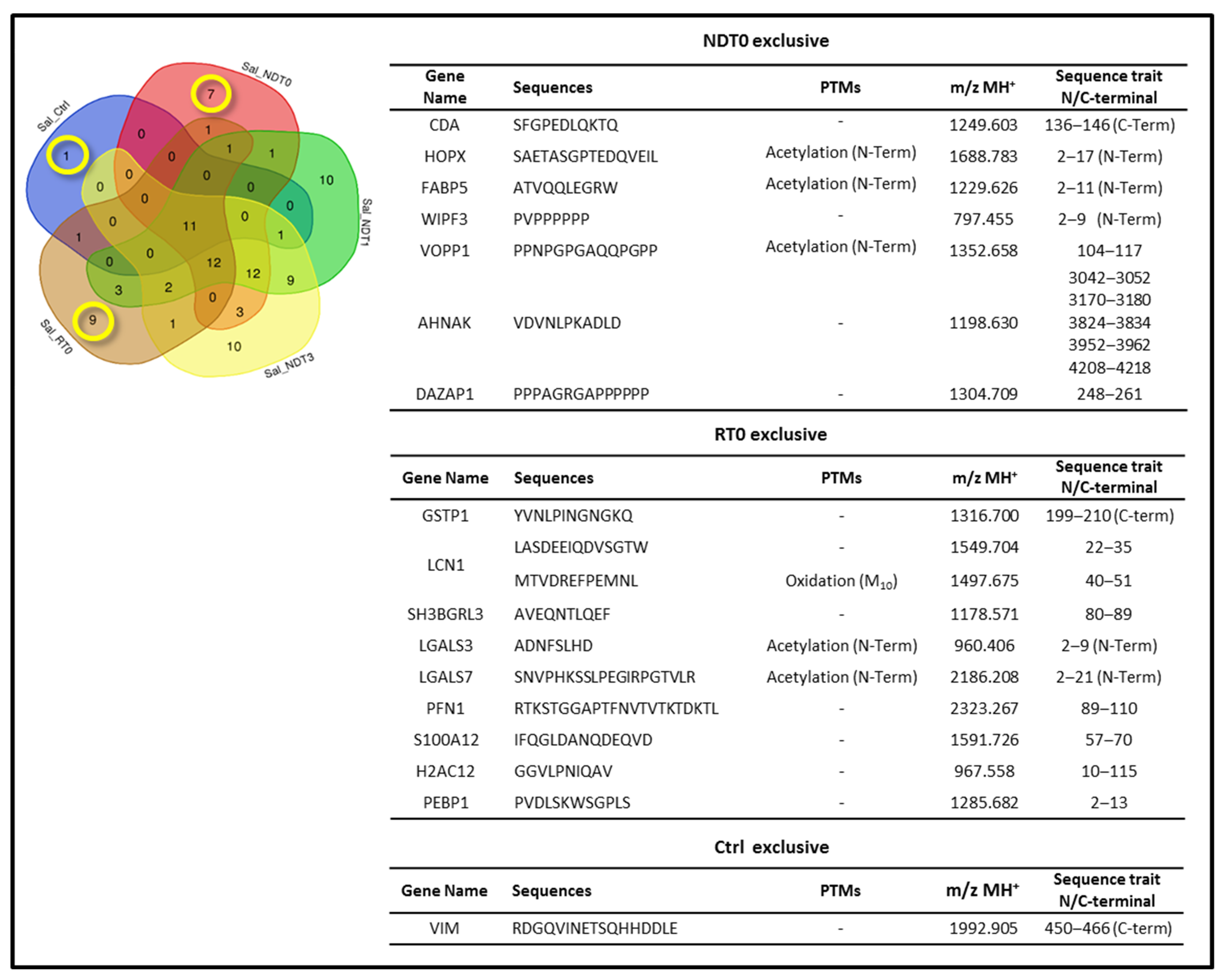
2. Results and Discussion
3. Materials and Methods
3.1. Sample Collection
3.2. Chemicals
3.3. Sample Preparation
3.4. LC-MS Analyses
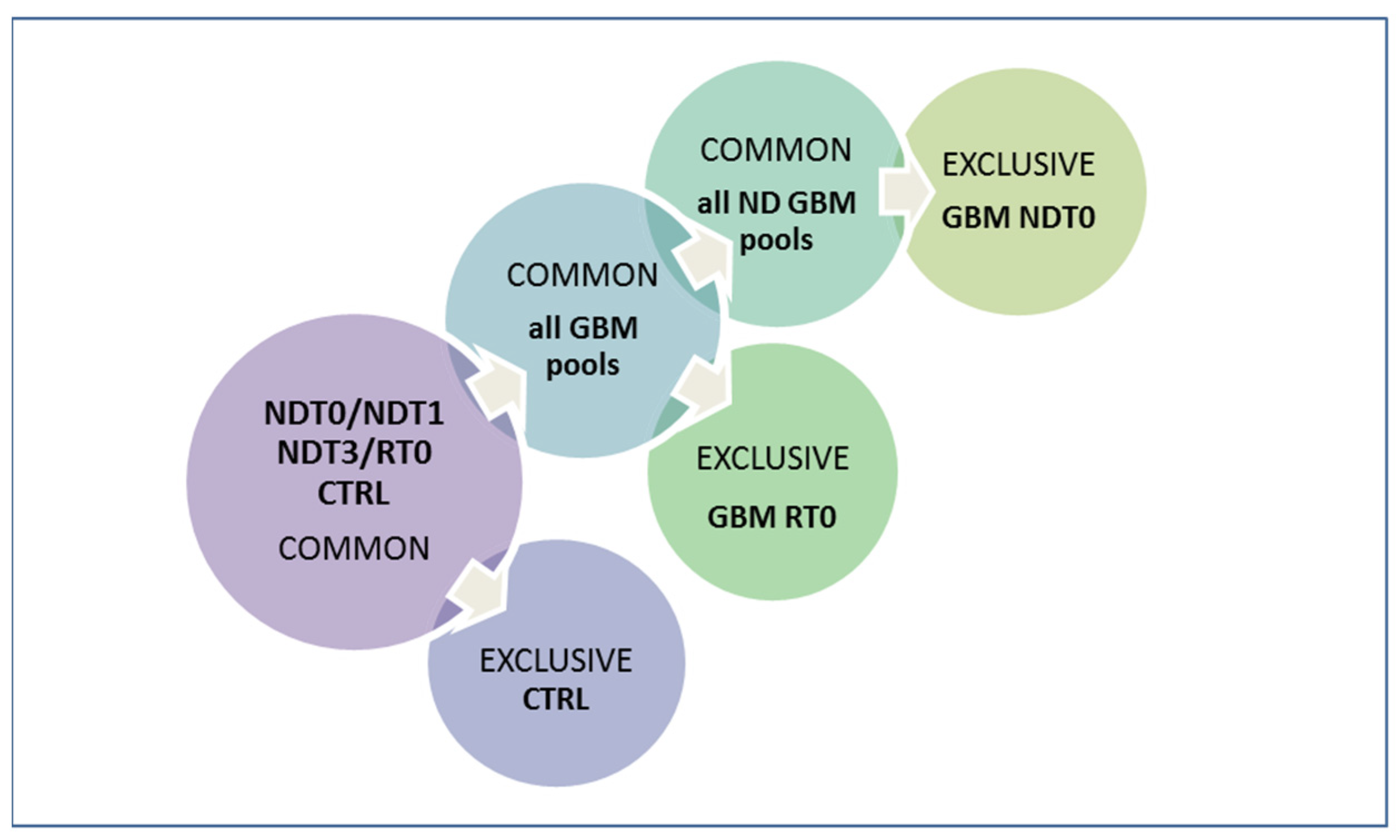
3.5. Data and Bioinformatics Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GBM | Glioblastoma IDH wild type |
| ND GBM | Newly Diagnosed GBM |
| R GBM | Recurrent GBM |
| T0 | Pre-surgery saliva |
| T1 | 1-month post-surgery saliva |
| T3 | 3-month post-surgery saliva |
| CUSA | Cavitron Ultrasonic Surgical Aspirator |
| FASP | Filter-Aided Sample Preparation |
| LC | Liquid Chromatography |
| MS | Mass Spectrometry |
| CNS | Central Nervous System |
| FA | Formic acid |
| ACN | Acetonitrile |
| PIC | Protease inhibitor cocktail |
| FDR | False Discovery Rate |
References
- Park, Y.W.; Vollmuth, P.; Foltyn-Dumitru, M.; Sahm, F.; Ahn, S.S.; Chang, J.H.; Kim, S.H. The 2021 WHO Classification for Gliomas and Implications on Imaging Diagnosis: Part 1-Key Points of the Fifth Edition and Summary of Imaging Findings on Adult-Type Diffuse Gliomas. J. Magn. Reson. Imaging 2023, 58, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Salari, N.; Ghasemi, H.; Fatahian, R.; Mansouri, K.; Dokaneheifard, S.; Shiri, M.H.; Hemmati, M.; Mohammadi, M. The global prevalence of primary central nervous system tumors: A systematic review and meta-analysis. Eur. J. Med. Res. 2023, 28, 39. [Google Scholar] [CrossRef]
- Stoyanov, G.S.; Lyutfi, E.; Georgieva, R.; Georgiev, R.; Dzhenkov, D.L.; Petkova, L.; Ivanov, B.D.; Kaprelyan, A.; Ghenev, P. Reclassification of Glioblastoma Multiforme According to the 2021 World Health Organization Classification of Central Nervous System Tumors: A Single Institution Report and Practical Significance. Cureus 2022, 14, 21822. [Google Scholar] [CrossRef]
- Bikfalvi, A.; da Costa, C.A.; Avril, T.; Barnier, J.V.; Bauchet, L.; Brisson, L.; Cartron, P.F.; Castel, H.; Chevet, E.; Chneiweiss, H.; et al. Challenges in glioblastoma research: Focus on the tumor microenvironment. Trends Cancer. 2023, 9, 9–27. [Google Scholar] [CrossRef]
- Raucher, D. Tumor targeting peptides: Novel therapeutic strategies in glioblastoma. Curr. Opin. Pharmacol. 2019, 47, 14–19. [Google Scholar] [CrossRef]
- Rodríguez-Camacho, A.; Flores-Vázquez, J.G.; Moscardini-Martelli, J.; Torres-Ríos, J.A.; Olmos-Guzmán, A.; Ortiz-Arce, C.S.; Cid-Sánchez, D.R.; Pérez, S.R.; Macías-González, M.D.S.; Hernández-Sánchez, L.C.; et al. Glioblastoma Treatment: State-of-the-Art and Future Perspectives. Int. J. Mol. Sci. 2022, 23, 7207. [Google Scholar] [CrossRef]
- Pineda, E.; Domenech, M.; Hernández, A.; Comas, S.; Balaña, C. Recurrent Glioblastoma: Ongoing Clinical Challenges and Future Prospects. Onco. Target. Ther. 2023, 16, 71–86. [Google Scholar] [CrossRef]
- Hishii, M.; Matsumoto, T.; Arai, H. Diagnosis and Treatment of Early-Stage Glioblastoma. Asian J. Neurosurg. 2019, 14, 589–592. [Google Scholar] [CrossRef]
- Hu, C.C.; Wang, S.G.; Gao, Z.; Qing, M.F.; Pan, S.; Liu, Y.Y.; Li, F. Emerging salivary biomarkers for early detection of oral squamous cell carcinoma. World J. Clin. Oncol. 2025, 16, 103803. [Google Scholar] [CrossRef] [PubMed]
- Wormwood, K.L.; Aslebagh, R.; Channaveerappa, D.; Dupree, E.J.; Borland, M.M.; Ryan, J.P.; Darie, C.C.; Woods, A.G. Salivary proteomics and biomarkers in neurology and psychiatry. Proteom. Clin. Appl. 2015, 9, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Boroumand, M.; Olianas, A.; Cabras, T.; Manconi, B.; Fanni, D.; Faa, G.; Desiderio, C.; Messana, I.; Castagnola, M. Saliva, a bodily fluid with recognized and potential diagnostic applications. J. Sep. Sci. 2021, 44, 3677–3690. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Guo, H.; Zhao, Y.; Liu, Z.; Wang, C.; Bu, J.; Sun, T.; Wei, J. Liquid biopsy in cancer current: Status, challenges and future prospects. Signal Transduct. Target. Ther. 2024, 9, 336. [Google Scholar] [CrossRef]
- Seyhan, A.A. Circulating Liquid Biopsy Biomarkers in Glioblastoma: Advances and Challenges. Int. J. Mol. Sci. 2024, 25, 7974. [Google Scholar] [CrossRef]
- Wong, D.T. Salivary Diagnostics: Amazing as it might seem, doctors can detect and monitor diseases using molecules found in a sample of spit. Am. Sci. 2008, 96, 37–43. [Google Scholar] [CrossRef]
- Ghorbani, A.; Avery, L.M.; Sohaei, D.; Soosaipillai, A.; Richer, M.; Horbinski, C.; McCortney, K.; Xu, W.; Diamandis, E.P.; Prassas, I. Discovery of novel glioma serum biomarkers by proximity extension assay. Clin. Proteom. 2023, 20, 12. [Google Scholar] [CrossRef]
- Macklin, A.; Khan, S.; Kislinger, T. Recent advances in mass spectrometry based clinical proteomics: Applications to cancer research. Clin. Proteom. 2020, 17, 17. [Google Scholar] [CrossRef]
- Nakayasu, E.S.; Gritsenko, M.; Piehowski, P.D.; Gao, Y.; Orton, D.J.; Schepmoes, A.A.; Fillmore, T.L.; Frohnert, B.I.; Rewers, M.; Krischer, J.P.; et al. Tutorial: Best practices and considerations for mass-spectrometry-based protein biomarker discovery and validation. Nat. Protoc. 2021, 16, 3737–3760. [Google Scholar] [CrossRef]
- La Rocca, G.; Simboli, G.A.; Vincenzoni, F.; Rossetti, D.V.; Urbani, A.; Ius, T.; Della Pepa, G.M.; Olivi, A.; Sabatino, G.; Desiderio, C. Glioblastoma CUSA Fluid Protein Profiling: A Comparative Investigation of the Core and Peripheral Tumor Zones. Cancers 2020, 13, 30. [Google Scholar] [CrossRef]
- Moresi, F.; Rossetti, D.V.; Vincenzoni, F.; Simboli, G.A.; La Rocca, G.; Olivi, A.; Urbani, A.; Sabatino, G.; Desiderio, C. Investigating Glioblastoma Multiforme Sub-Proteomes: A Computational Study of CUSA Fluid Proteomic Data. Int. J. Mol. Sci. 2022, 23, 2058. [Google Scholar] [CrossRef] [PubMed]
- Muntiu, A.; Vincenzoni, F.; Rossetti, D.V.; Urbani, A.; La Rocca, G.; Albanese, A.; Mazzucchi, E.; Olivi, A.; Sabatino, G.; Desiderio, C. Proteomic analysis of the low molecular mass fraction of newly diagnosed and recurrent glioblastoma CUSA fluid: A pilot investigation of the peptidomic profile. Int. J. Mol. Sci. 2025, 26, 6055. [Google Scholar] [CrossRef] [PubMed]
- Muntiu, A.; Moresi, F.; Vincenzoni, F.; Rossetti, D.V.; Iavarone, F.; Messana, I.; Castagnola, M.; La Rocca, G.; Mazzucchi, E.; Olivi, A.; et al. Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients: A Pilot Investigation. Int. J. Mol. Sci. 2024, 25, 12984. [Google Scholar] [CrossRef]
- Iavarone, F.; Desiderio, C.; Vitali, A.; Messana, I.; Martelli, C.; Castagnola, M.; Cabras, T. Cryptides: Latent peptides everywhere. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 246–263. [Google Scholar] [CrossRef] [PubMed]
- Marcu, A.; Bichmann, L.; Kuchenbecker, L.; Kowalewski, D.J.; Freudenmann, L.K.; Backert, L.; Mühlenbruch, L.; Szolek, A.; Lübke, M.; Wagner, P.; et al. HLA Ligand Atlas: A benign reference of HLA-presented peptides to improve T-cell-based cancer immunotherapy. J. Immunother. Cancer 2021, 9, 2071. [Google Scholar] [CrossRef]
- Luo, X.; Huang, Y.; Li, H.; Luo, Y.; Zuo, Z.; Ren, J.; Xie, Y. SPENCER: A comprehensive database for small peptides encoded by noncoding RNAs in cancer patients. Nucleic Acids Res. 2022, 50, D1373–D1381. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, S.; Meng, N.; He, Y.; Lu, R.; Yan, G.R. ncRNA-Encoded Peptides or Proteins and Cancer. Mol. Ther. 2019, 27, 1718–1725. [Google Scholar] [CrossRef]
- Oliveros, J.C. (2007–2015) Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 28 January 2025).
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- Saitoh, E.; Taniguchi, M.; Ochiai, A.; Kato, T.; Imai, A.; Isemura, S. Bioactive peptides hidden in human salivary proteins. J. Oral Biosci. 2017, 59, 71–79. [Google Scholar] [CrossRef]
- Oudhoff, M.J.; Kroeze, K.L.; Nazmi, K.; van den Keijbus, P.A.; van ’t Hof, W.; Fernandez-Borja, M.; Hordijk, P.L.; Gibbs, S.; Bolscher, J.G.; Veerman, E.C. Structure-activity analysis of histatin, a potent wound healing peptide from human saliva: Cyclization of histatin potentiates molar activity 1000-fold. FASEB J. 2009, 23, 3928–3935. [Google Scholar] [CrossRef]
- Arora, A.; Patil, V.; Kundu, P.; Kondaiah, P.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Pal, D.; Somasundaram, K. Serum biomarkers identification by iTRAQ and verification by MRM: S100A8/S100A9 levels predict tumor-stroma involvement and prognosis in Glioblastoma. Sci. Rep. 2019, 9, 2749. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, C.; Ma, G.; Zhang, Q. Expression of SPRR3 is associated with tumor cell proliferation and invasion in glioblastoma multiforme. Oncol. Lett. 2014, 7, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Ghezelbash, M.; Masoudian, N.; Pooladi, M. Beta actin expression profile in malignant human glioma tumors. Int. Clin. Neurosci. J. 2018, 5, 72–77. [Google Scholar] [CrossRef]
- Haubitz, M.; Good, D.M.; Woywodt, A.; Haller, H.; Rupprecht, H.; Theodorescu, D.; Dakna, M.; Coon, J.J.; Mischak, H. Identification and validation of urinary biomarkers for differential diagnosis and evaluation of therapeutic intervention in anti-neutrophil cytoplasmic antibody-associated vasculitis. Mol. Cell Proteom. 2009, 8, 2296–2307. [Google Scholar] [CrossRef]
- Siu, K.W.M.; Matta, A.; De Souza, L.V. Biomarkers for Head-and-Neck Cancers and Precancers. International Patent Application No. WO 2009097692A1, 13 August 2009. Available online: https://patents.google.com/patent/WO2009097692A1/en (accessed on 29 September 2025).
- Kumar, A.; Kumar, V.; Arora, M.; Kumar, M.; Ammalli, P.; Thakur, B.; Prasad, J.; Kumari, S.; Sharma, M.C.; Kale, S.S.; et al. Overexpression of prothymosin-α in glioma is associated with tumor aggressiveness and poor prognosis. Biosci. Rep. 2022, 42, BSR20212685. [Google Scholar] [CrossRef]
- Kim, S.A.; Lee, Y.; Jung, D.E.; Park, K.H.; Park, J.Y.; Gang, J.; Jeon, S.B.; Park, E.C.; Kim, Y.G.; Lee, B.; et al. Pancreatic adenocarcinoma up-regulated factor (PAUF), a novel up-regulated secretory protein in pancreatic ductal adenocarcioma. Cancer Sci. 2009, 100, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Escudero-Paniagua, B.; Bartolomé, R.A.; Rodríguez, S.; De Los Ríos, V.; Pintado, L.; Jaén, M.; Lafarga, M.; Fernández-Aceñero, M.J.; Casal, J.I. PAUF/ZG16B promotes colorectal cancer progression through alterations of the mitotic functions and the Wnt/β-catenin pathway. Carcinogenesis 2020, 41, 203–213. [Google Scholar] [CrossRef]
- Barderas, R.; Mendes, M.; Torres, S.; Bartolomé, R.A.; López-Lucendo, M.; Villar-Vázquez, R.; Peláez-García, A.; Fuente, E.; Bonilla, F.; Casal, J.I. In-depth characterization of the secretome of colorectal cancer metastatic cells identifies key proteins in cell adhesion, migration, and invasion. Mol. Cell Proteom. 2013, 12, 1602–1620. [Google Scholar] [CrossRef]
- Choi, C.H.; Chung, J.Y.; Park, H.S.; Jun, M.; Lee, Y.Y.; Kim, B.G.; Hewitt, S.M. Pancreatic adenocarcinoma up-regulated factor expression is associated with disease-specific survival in cervical cancer patients. Hum. Pathol. 2015, 46, 884–893. [Google Scholar] [CrossRef]
- Jin, H.J.; Jung, S.; DebRoy, A.R.; Davuluri, R.V. Identification and validation of regulatory SNPs that modulate transcription factor chromatin binding and gene expression in prostate cancer. Oncotarget 2016, 7, 54616–54626. [Google Scholar] [CrossRef]
- Sasahira, T.; Kurihara, M.; Nishiguchi, Y.; Nakashima, C.; Kirita, T.; Kuniyasu, H. Pancreatic adenocarcinoma up-regulated factor has oncogenic functions in oral squamous cell carcinoma. Histopathology 2017, 70, 539–548. [Google Scholar] [CrossRef]
- Choi, C.H.; Kang, T.H.; Song, J.S.; Kim, Y.S.; Chung, E.J.; Ylaya, K.; Kim, S.; Koh, S.S.; Chung, J.Y.; Kim, J.H.; et al. Elevated expression of pancreatic adenocarcinoma upregulated factor (PAUF) is associated with poor prognosis and chemoresistance in epithelial ovarian cancer. Sci. Rep. 2018, 8, 12161. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Shi, C.; Liu, X.; Liang, C.; Yang, C.; Wan, X.; Li, L.; Liu, Y. Identification of ZG16B as a prognostic biomarker in breast cancer. Open Med. 2020, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Baras, A.; Moskaluk, C.A. Intracellular localization of GASP/ECOP/VOPP1. J. Mol. Histol. 2010, 41, 153–164. [Google Scholar] [CrossRef]
- Fang, Z.; Wu, L.; Dai, H.; Hu, P.; Wang, B.; Han, Q.; Xu, Y.; Lv, S.; Zhu, Y.; Gan, M.; et al. The role of vesicular overexpressed in cancer pro-survival protein 1 in hepatocellular carcinoma proliferation. Cancer Biomark. 2020, 28, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Bonin, F.; Taouis, K.; Azorin, P.; Petitalot, A.; Tariq, Z.; Nola, S.; Bouteille, N.; Tury, S.; Vacher, S.; Bièche, I.; et al. VOPP1 promotes breast tumorigenesis by interacting with the tumor suppressor WWOX. BMC Biol. 2018, 16, 109. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, O.; Lefort, K.; Mariotto, A.; Huber, M.; Hohl, D. HOPX Exhibits Oncogenic Activity during Squamous Skin Carcinogenesis. J. Investig. Dermatol. 2021, 141, 2354–2368. [Google Scholar] [CrossRef]
- Yap, L.F.; Lai, S.L.; Patmanathan, S.N.; Gokulan, R.; Robinson, C.M.; White, J.B.; Chai, S.J.; Rajadurai, P.; Prepageran, N.; Liew, Y.T.; et al. HOPX functions as a tumour suppressor in head and neck cancer. Sci. Rep. 2016, 6, 38758. [Google Scholar] [CrossRef]
- De Toni, A.; Zbinden, M.; Epstein, J.A.; Ruiz i Altaba, A.; Prochiantz, A.; Caillé, I. Regulation of survival in adult hippocampal and glioblastoma stem cell lineages by the homeodomain-only protein HOP. Neural Dev. 2008, 3, 13. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, S.; Sun, J.; Wang, X.; Guan, M.; Yin, J.; Tang, B. Oncogenic role and potential regulatory mechanism of fatty acid binding protein 5 based on a pan-cancer analysis. Sci. Rep. 2023, 13, 4060. [Google Scholar] [CrossRef]
- Martelli, C.; D’Angelo, L.; Barba, M.; Baranzini, M.; Inserra, I.; Iavarone, F.; Vincenzoni, F.; Tamburrini, G.; Massimi, L.; Rocco, C.D.; et al. Top-down proteomic characterization of DAOY medulloblastoma tumor cell line. EuPA Open Proteom. 2016, 12, 13–21. [Google Scholar] [CrossRef]
- Ramos, I.; Stamatakis, K.; Oeste, C.L.; Pérez-Sala, D. Vimentin as a Multifaceted Player and Potential Therapeutic Target in Viral Infections. Int. J. Mol. Sci. 2020, 21, 4675. [Google Scholar] [CrossRef]
- Sivagnanam, A.; Shyamsundar, V.; Kesavan, P.; Krishnamurthy, A.; Thangaraj, S.V.; Venugopal, D.C.; Kasirajan, H.; Ramani, P.; Sarma, V.R.; Ramshankar, V. 2D-DIGE-Based Proteomic Profiling with Validations Identifies Vimentin as a Secretory Biomarker Useful for Early Detection and Poor Prognosis in Oral Cancers. J. Oncol. 2022, 2022, 4215097. [Google Scholar] [CrossRef] [PubMed]
- Ellis, S.; Way, R.; Nel, M.; Burleigh, A.; Doykov, I.; Kembou-Ringert, J.; Woodall, M.; Masonou, T.; Case, K.M.; Ortez, A.T.; et al. Salivary IgA and vimentin differentiate in vitro SARS-CoV-2 infection: A study of 290 convalescent COVID-19 patients. Mucosal Immunol. 2024, 17, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Byun, Y.; Chen, F.; Chang, R.; Trivedi, M.; Green, K.J.; Cryns, V.L. Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ. 2001, 8, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, J.; Yan, Y.; Xiang, L.; Zhai, X.; Cai, L.; Sun, Z.; Pi, M.; Xiong, Q.; Zhou, H.; et al. Vimentin Fragmentation and Its Role in Amyloid-Beta Plaque Deposition in Alzheimer’s Disease. Int. J. Mol. Sci. 2025, 26, 2857. [Google Scholar] [CrossRef]
- Girol, A.P.; Mimura, K.K.; Drewes, C.C.; Bolonheis, S.M.; Solito, E.; Farsky, S.H.; Gil, C.D.; Oliani, S.M. Anti-inflammatory mechanisms of the annexin A1 protein and its mimetic peptide Ac2-26 in models of ocular inflammation in vivo and in vitro. J. Immunol. 2013, 190, 5689–5701. [Google Scholar] [CrossRef]
- Huang, J.J.; Xia, C.J.; Wei, Y.; Yao, Y.; Dong, M.W.; Lin, K.Z.; Yu, L.S.; Gao, Y.; Fan, Y.Y. Annexin A1-derived peptide Ac2-26 facilitates wound healing in diabetic mice. Wound Repair Regen. 2020, 28, 772–779. [Google Scholar] [CrossRef]
- Gimenes, A.D.; Andrade, B.F.D.; Pinotti, J.V.P.; Oliani, S.M.; Galvis-Alonso, O.Y.; Gil, C.D. Annexin A1-derived peptide Ac2-26 in a pilocarpine-induced status epilepticus model: Anti-inflammatory and neuroprotective effects. J. Neuroinflamm. 2019, 16, 32. [Google Scholar] [CrossRef]
- Qin, C.X.; Rosli, S.; Deo, M.; Cao, N.; Walsh, J.; Tate, M.; Alexander, A.E.; Donner, D.; Horlock, D.; Li, R.; et al. Cardioprotective Actions of the Annexin-A1 N-Terminal Peptide, Ac2-26, Against Myocardial Infarction. Front. Pharmacol. 2019, 10, 269. [Google Scholar] [CrossRef]
- Liu, L.; An, D.; Xu, J.; Shao, B.; Li, X.; Shi, J. Ac2-26 Induces IKKβ Degradation Through Chaperone-Mediated Autophagy Via HSPB1 in NCM-Treated Microglia. Front. Mol. Neurosci. 2018, 11, 76. [Google Scholar] [CrossRef]
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016, 19, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Moraes, L.A.; Kar, S.; Foo, S.L.; Gu, T.; Toh, Y.Q.; Ampomah, P.B.; Sachaphibulkij, K.; Yap, G.; Zharkova, O.; Lukman, H.M.; et al. Annexin-A1 enhances breast cancer growth and migration by promoting alternative macrophage polarization in the tumour microenvironment. Sci. Rep. 2017, 7, 17925. [Google Scholar] [CrossRef]
- Neidert, M.C.; Kowalewski, D.J.; Silginer, M.; Kapolou, K.; Backert, L.; Freudenmann, L.K.; Peper, J.K.; Marcu, A.; Wang, S.S.; Walz, J.S.; et al. The natural HLA ligandome of glioblastoma stem-like cells: Antigen discovery for T cell-based immunotherapy. Acta Neuropathol. 2018, 135, 923–938. [Google Scholar] [CrossRef]
- Jiang, S.; Li, H.; Zhang, L.; Mu, W.; Zhang, Y.; Chen, T.; Wu, J.; Tang, H.; Zheng, S.; Liu, Y.; et al. Generic Diagramming Platform (GDP): A comprehensive database of high-quality biomedical graphics. Nucleic Acids Res. 2025, 53, D1670–D1676. [Google Scholar] [CrossRef]
- Ding, X.; Zhang, C.; Chen, H.; Ren, M.; Liu, X. Cathepsins Trigger Cell Death and Regulate Radioresistance in Glioblastoma. Cells 2022, 11, 4108. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.E.; Iwadate, Y.; Machida, T.; Hiwasa, T.; Nimura, Y.; Nagai, Y.; Takiguchi, M.; Tanzawa, H.; Yamaura, A.; Seki, N. Cathepsin D is a potential serum marker for poor prognosis in glioma patients. Cancer Res. 2005, 65, 5190–5194. [Google Scholar] [CrossRef]
- Mohamed, M.M.; Sloane, B.F. Cysteine cathepsins: Multifunctional enzymes in cancer. Nat. Rev. Cancer 2006, 6, 764–775. [Google Scholar] [CrossRef]
- Muntiu, A.; Papait, A.; Vincenzoni, F.; Rossetti, D.V.; Romele, P.; Cargnoni, A.; Silini, A.; Parolini, O.; Desiderio, C. Proteomic analysis of the human amniotic mesenchymal stromal cell secretome by integrated approaches via filter-aided sample preparation. J. Proteom. 2025, 310, 105339. [Google Scholar] [CrossRef]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef]
- Bauça, J.M.; Martínez-Morillo, E.; Diamandis, E.P. Peptidomics of urine and other biofluids for cancer diagnostics. Clin. Chem. 2014, 60, 1052–1061. [Google Scholar] [CrossRef] [PubMed]
- Umapathy, V.R.; Natarajan, P.M.; Swamikannu, B. Review Insights on Salivary Proteomics Biomarkers in Oral Cancer Detection and Diagnosis. Molecules 2023, 28, 5283. [Google Scholar] [CrossRef] [PubMed]
- Rao, J. Molecular mechanisms of glioma invasiveness: The role of proteases. Nat. Rev. Cancer 2003, 3, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Mentlein, R.; Hattermann, K.; Held-Feindt, J. Lost in disruption: Role of proteases in glioma invasion and progression. Biochim. Biophys. Acta 2012, 1825, 178–185. [Google Scholar] [CrossRef] [PubMed]
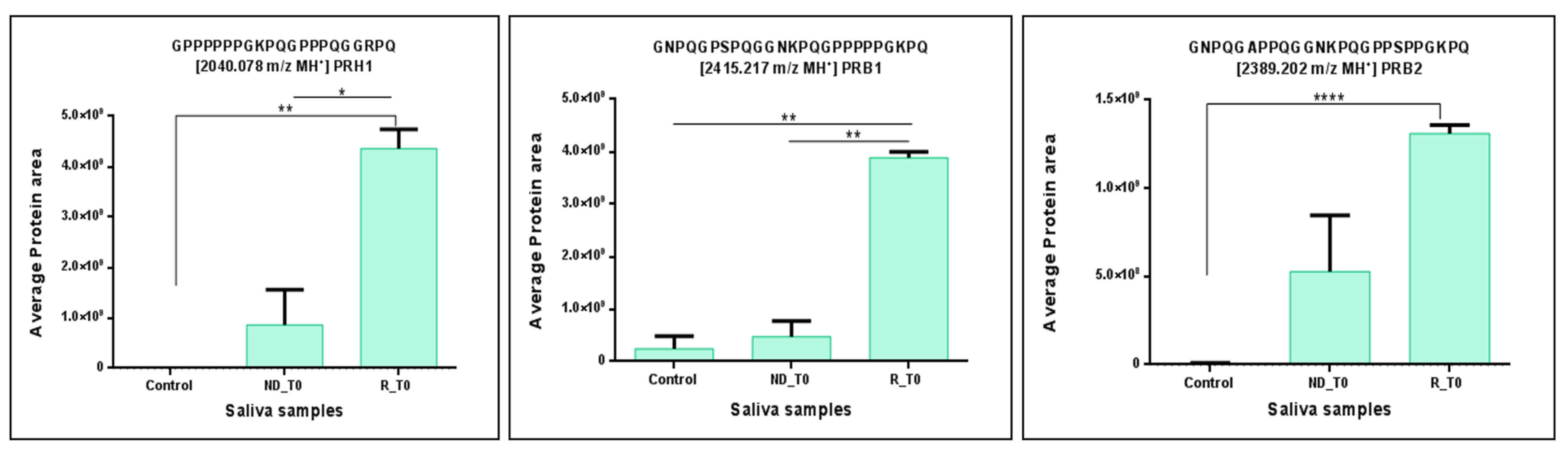
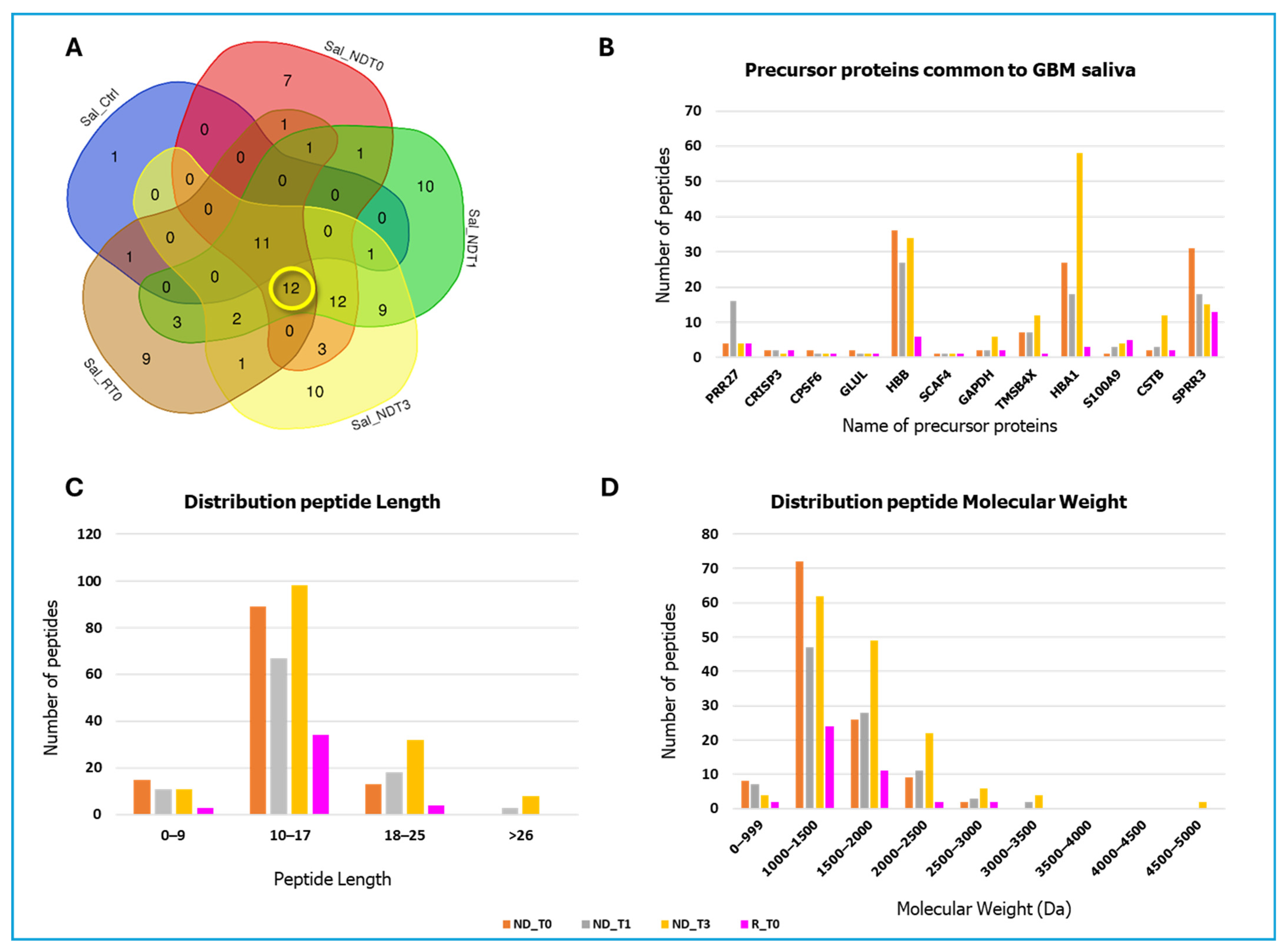
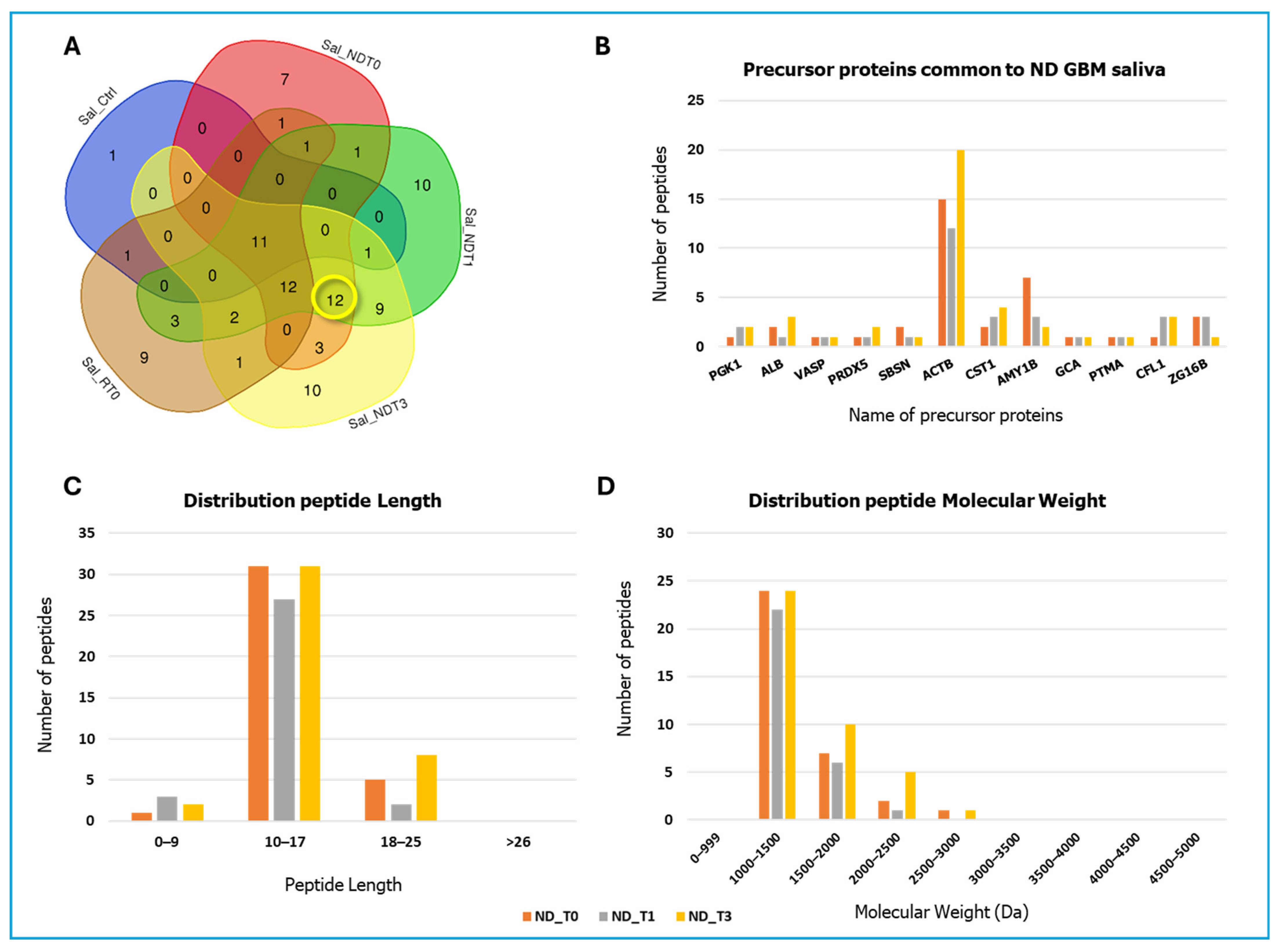
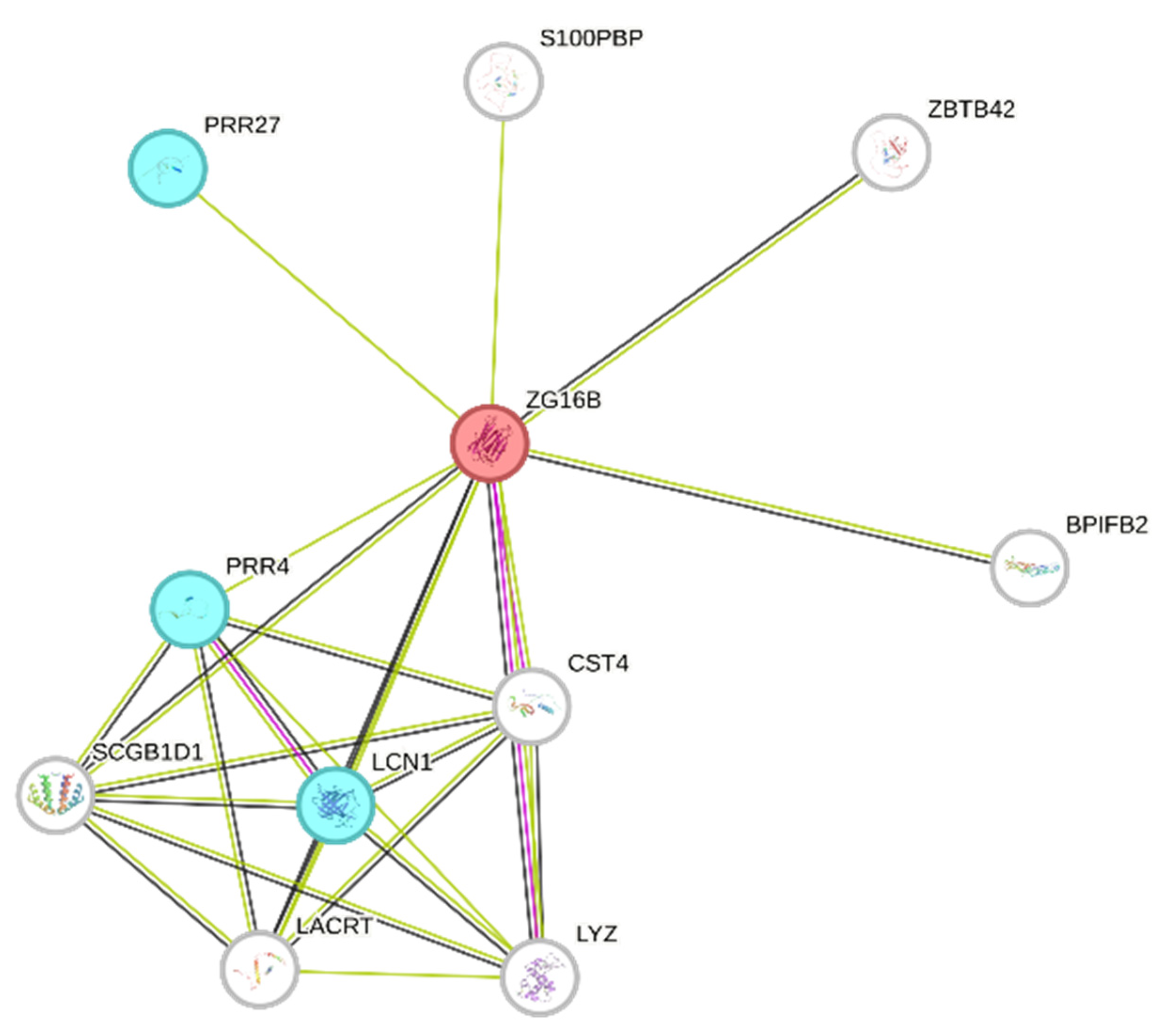
| Gene Name | Peptide Sequence | PTMs | m/z MH+ | Peptide Length | Sequence Position N/C-Terminal | Sequence Trait |
|---|---|---|---|---|---|---|
| PRR27 | FIGEDDNDDGHPLHPS | - | 1764.745 | 16 | - | 22–37 |
| KRRFPFIGEDDNDDGHPLHPS | - | 2449.160 | 21 | - | 17–37 | |
| CRISP3 | NEDKDPAFTAL | - | 1220.580 | 11 | - | 21–31 |
| CPSF6 | PQQGPPPPPG | - | 971.494 | 10 | - | 312–321 |
| GLUL | LNETGDEPFQYKN | - | 1554.709 | 13 | C-term | 361–373 |
| HBB | VHLTPEEKSAV | - | 1209.649 | 11 | N-term | 2–12 |
| DEVGGEALGRL | - | 1115.568 | 11 | - | 22–32 | |
| FESFGDLSTPD | - | 1214.524 | 11 | - | 43–53 | |
| SCAF4 | FGPGVPPPPPPP | - | 1155.618 | 12 | - | 712–722 |
| TMSB4X | SDKPDMAEIEKF | Acetylation (N-term) Oxidation (M6) | 1451.675 | 12 | N-term | 2–13 |
| HBA1 | VDPVNFKLL | - | 1044.612 | 9 | - | 94–102 |
| S100A9 | DTNADKQLSFEEF | - | 1543.692 | 13 | - | 67–79 |
| SPRR3 | KQTFTPPPQLQQQ | - | 1540.810 | 13 | - | 7–20 |
| TFTPPPQLQQQ | - | 1284.659 | 11 | - | 9–20 | |
| TFTPPPQLQ | - | 1028.541 | 9 | - | 9–17 | |
| VKQPSQPPPQEIFVPT | - | 1791.966 | 16 | - | 21–36 | |
| AIKVPEQGYT | - | 1105.589 | 10 | - | 130–139 | |
| FTPPPQLQ | - | 927.492 | 8 | - | 10–17 |
| Gene Name | Peptide Sequence | PTMs | m/z MH+ | Peptide Length | Sequence Position N/C-Terminal | Sequence Trait |
|---|---|---|---|---|---|---|
| ALB | GEYKFQNAL | - | 1069.529 | 9 | - | 423–431 |
| VASP | GPPAPPAGGPPPPP | - | 1205.631 | 14 | - | 161–174 |
| PRDX5 | APIKVGDAIPAVEV | - | 1378.795 | 14 | - | 54–67 |
| SBSN | ASDDPIEKVIE | - | 1215.611 | 11 | - | 24–34 |
| ACTB | FYNELRVAPEEHPV | - | 1699.844 | 14 | - | 90–103 |
| LLTEAPLNPKAN | - | 1280.723 | 12 | - | 104–115 | |
| GFAGDDAPRAVFPS | - | 1406.668 | 14 | - | 20–33 | |
| VAPEEHPVLLTEAPLNPK | - | 1954.066 | 18 | - | 96–113 | |
| FAGDDAPRAVFPS | - | 1349.651 | 13 | - | 21–33 | |
| NELRVAPEEHPV | - | 1389.713 | 12 | - | 92–103 | |
| GCA | AYPGYGGGFGNF | Acetylation (N-term) | 1248.532 | 12 | N-term | 2–13 |
| PTMA | SDAAVDTSSEITTK | Acetylation (N-term) | 1466.685 | 14 | N-term | 2–15 |
| CFL1 | ASGVAVSDGVIKVF | Acetylation (N-term) | 1390.759 | 14 | N-term | 2–15 |
| Gene Name (Sample Pool of Identification) | Peptide Sequence | PTMs | m/z MH+ | Peptide Length | Sequence Position N/C-Terminal | Sequence Trait |
|---|---|---|---|---|---|---|
| ZG16B (NDT0) | GPGGGKYFSTTEDY | - | 1478.641 | 14 | - | 21–34 |
| KLGALGGNTQEV | - | 1186.641 | 12 | - | 64–75 | |
| FEWNYPLEEPT | - | 1424.638 | 11 | - | 145–155 | |
| GDSWDVKLG | - | 976.472 | 9 | - | 58–66 | |
| ZG16B (NDT1) | TLQPGEYITKVF | - | 1395.753 | 12 | - | 76–87 |
| EDYDHEITGL | - | 1191.514 | 10 | - | 32–41 | |
| ZG16B (NDT3) | TLQPGEYITKVF | - | 1395.753 | 12 | - | 76–87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muntiu, A.; Vincenzoni, F.; Rossetti, D.V.; Castagnola, M.; Messana, I.; Iavarone, F.; Urbani, A.; La Rocca, G.; Albanese, A.; Olivi, A.; et al. Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients II: A Preliminary Investigation of the Complementary Low Molecular Mass Fraction. Int. J. Mol. Sci. 2025, 26, 9995. https://doi.org/10.3390/ijms26209995
Muntiu A, Vincenzoni F, Rossetti DV, Castagnola M, Messana I, Iavarone F, Urbani A, La Rocca G, Albanese A, Olivi A, et al. Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients II: A Preliminary Investigation of the Complementary Low Molecular Mass Fraction. International Journal of Molecular Sciences. 2025; 26(20):9995. https://doi.org/10.3390/ijms26209995
Chicago/Turabian StyleMuntiu, Alexandra, Federica Vincenzoni, Diana Valeria Rossetti, Massimo Castagnola, Irene Messana, Federica Iavarone, Andrea Urbani, Giuseppe La Rocca, Alessio Albanese, Alessandro Olivi, and et al. 2025. "Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients II: A Preliminary Investigation of the Complementary Low Molecular Mass Fraction" International Journal of Molecular Sciences 26, no. 20: 9995. https://doi.org/10.3390/ijms26209995
APA StyleMuntiu, A., Vincenzoni, F., Rossetti, D. V., Castagnola, M., Messana, I., Iavarone, F., Urbani, A., La Rocca, G., Albanese, A., Olivi, A., Sabatino, G., & Desiderio, C. (2025). Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients II: A Preliminary Investigation of the Complementary Low Molecular Mass Fraction. International Journal of Molecular Sciences, 26(20), 9995. https://doi.org/10.3390/ijms26209995









