A Semi-Automatic Tool for the Standardized Analysis of Fluorescent Intensity Changes in Polarized Cells
Abstract
1. Introduction
2. Results
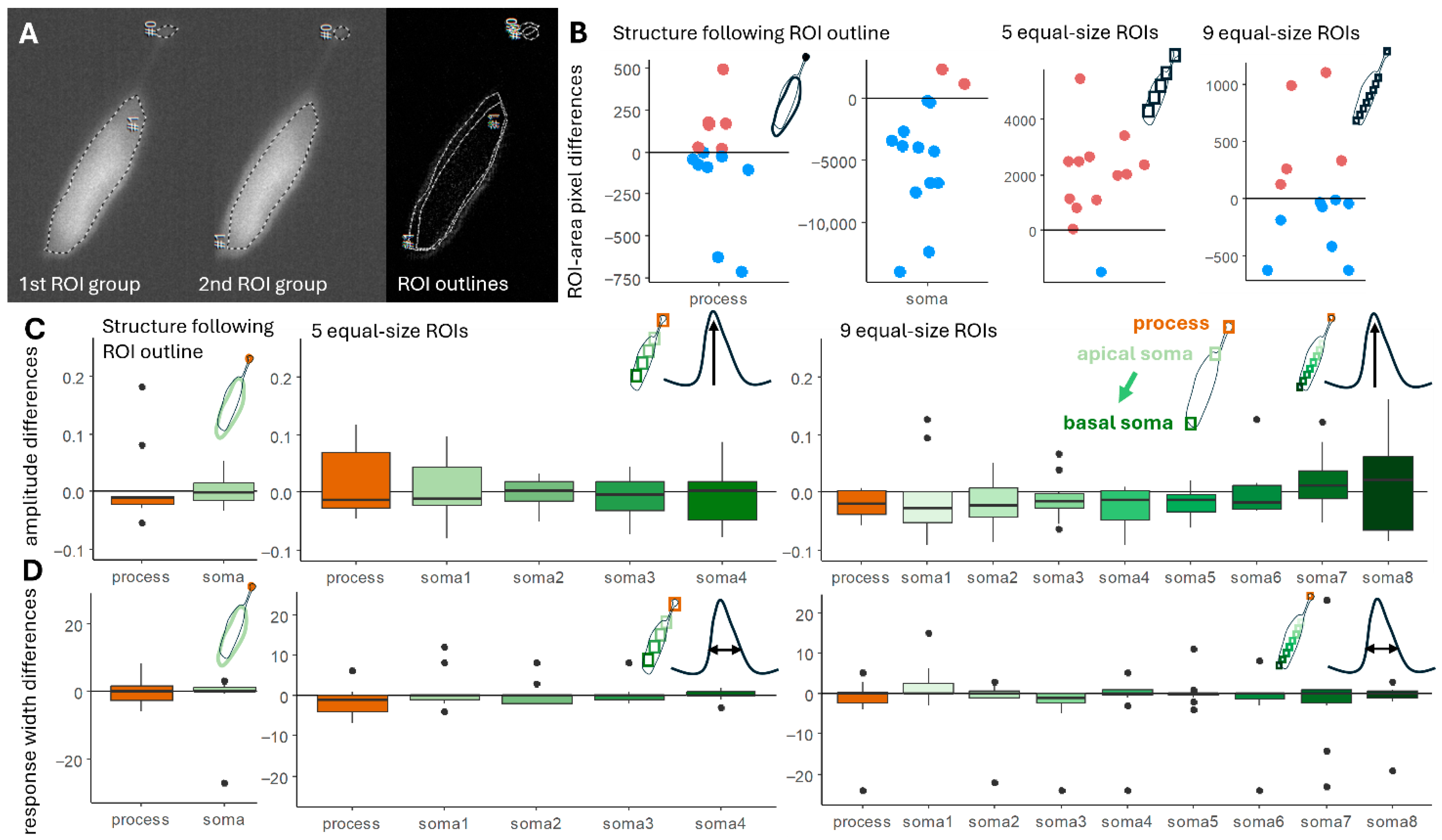
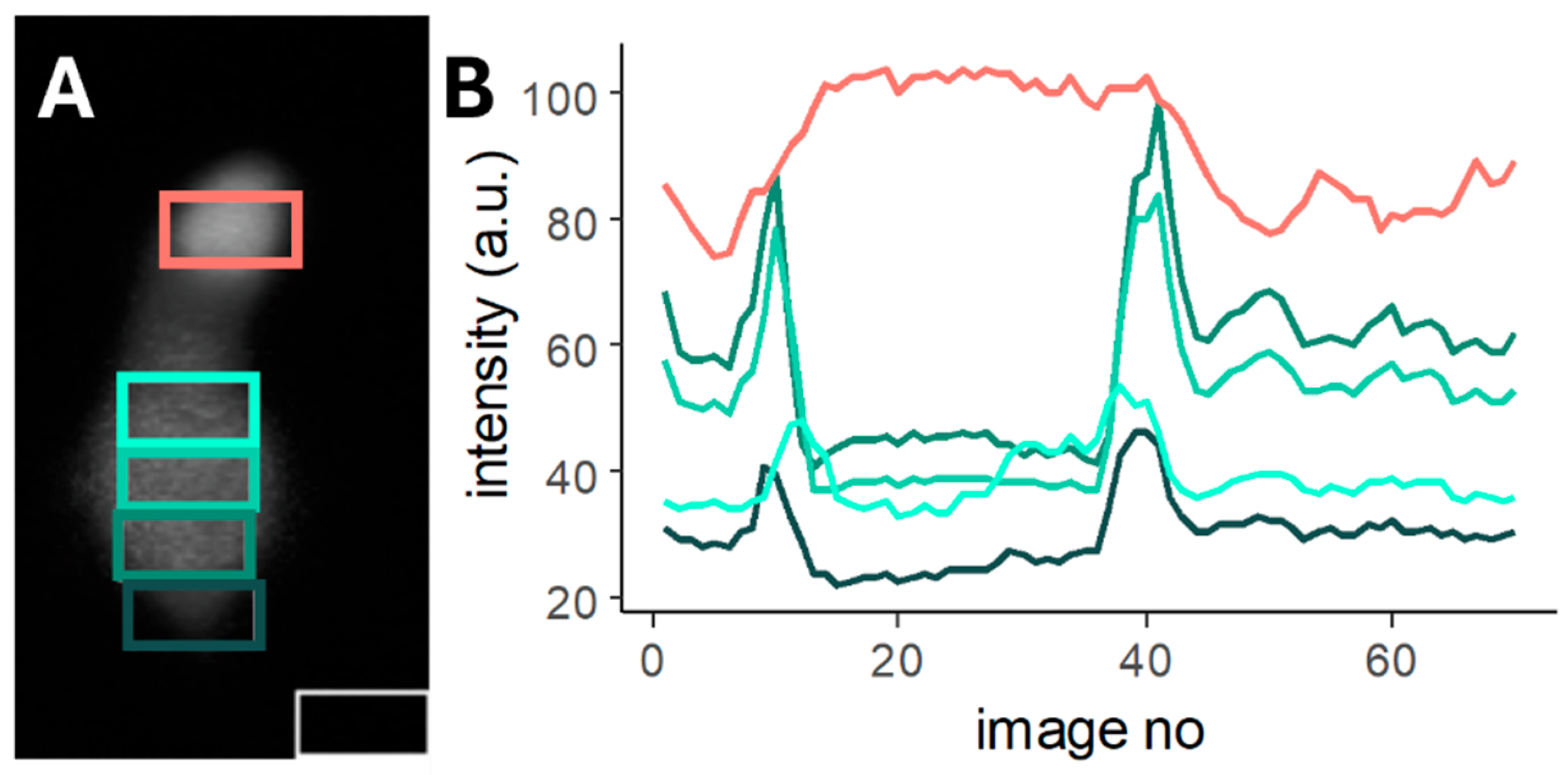
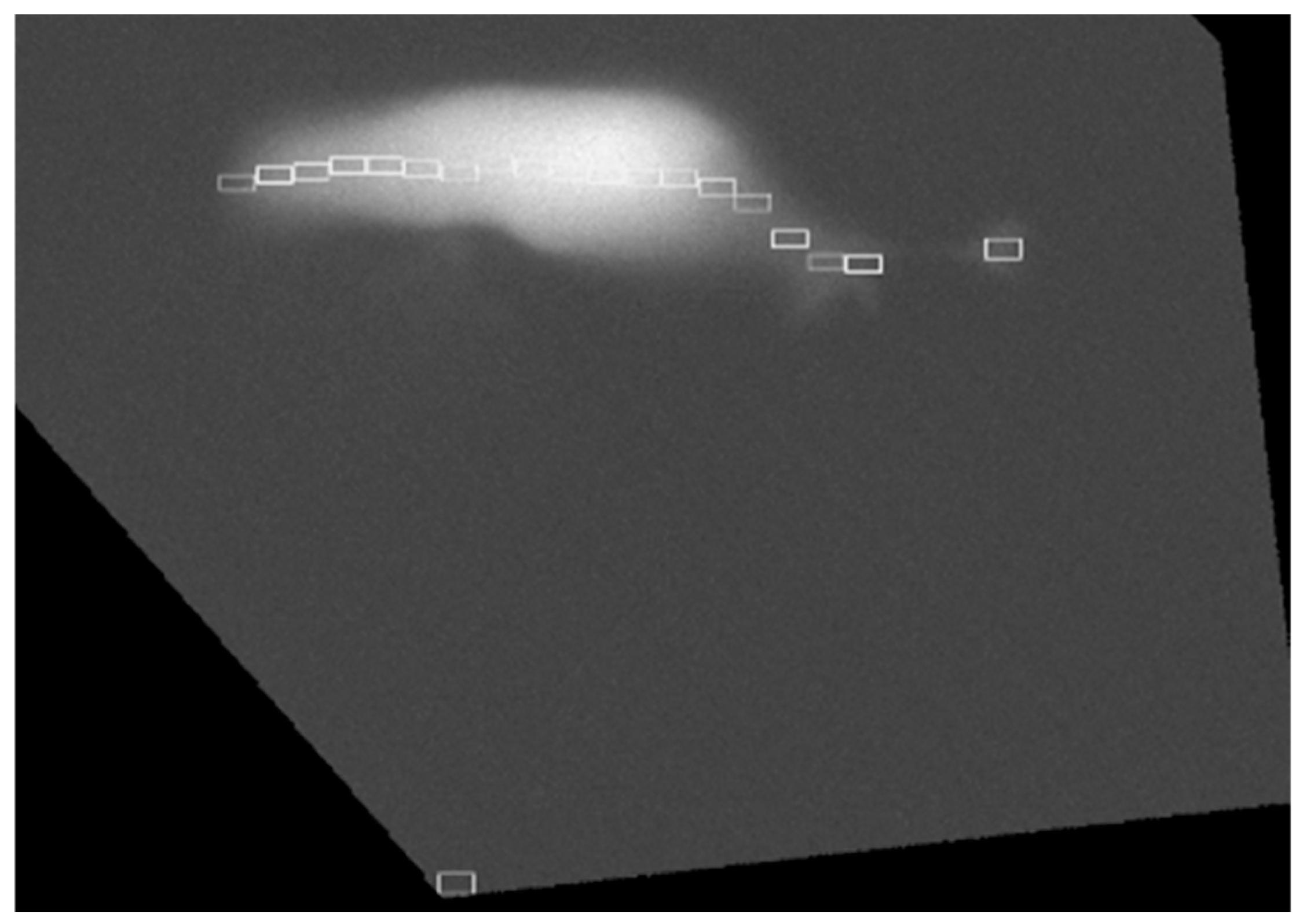
2.1. Region of Interest Identification for Subcellular Ca2+ Imaging in Deiters’ Cells
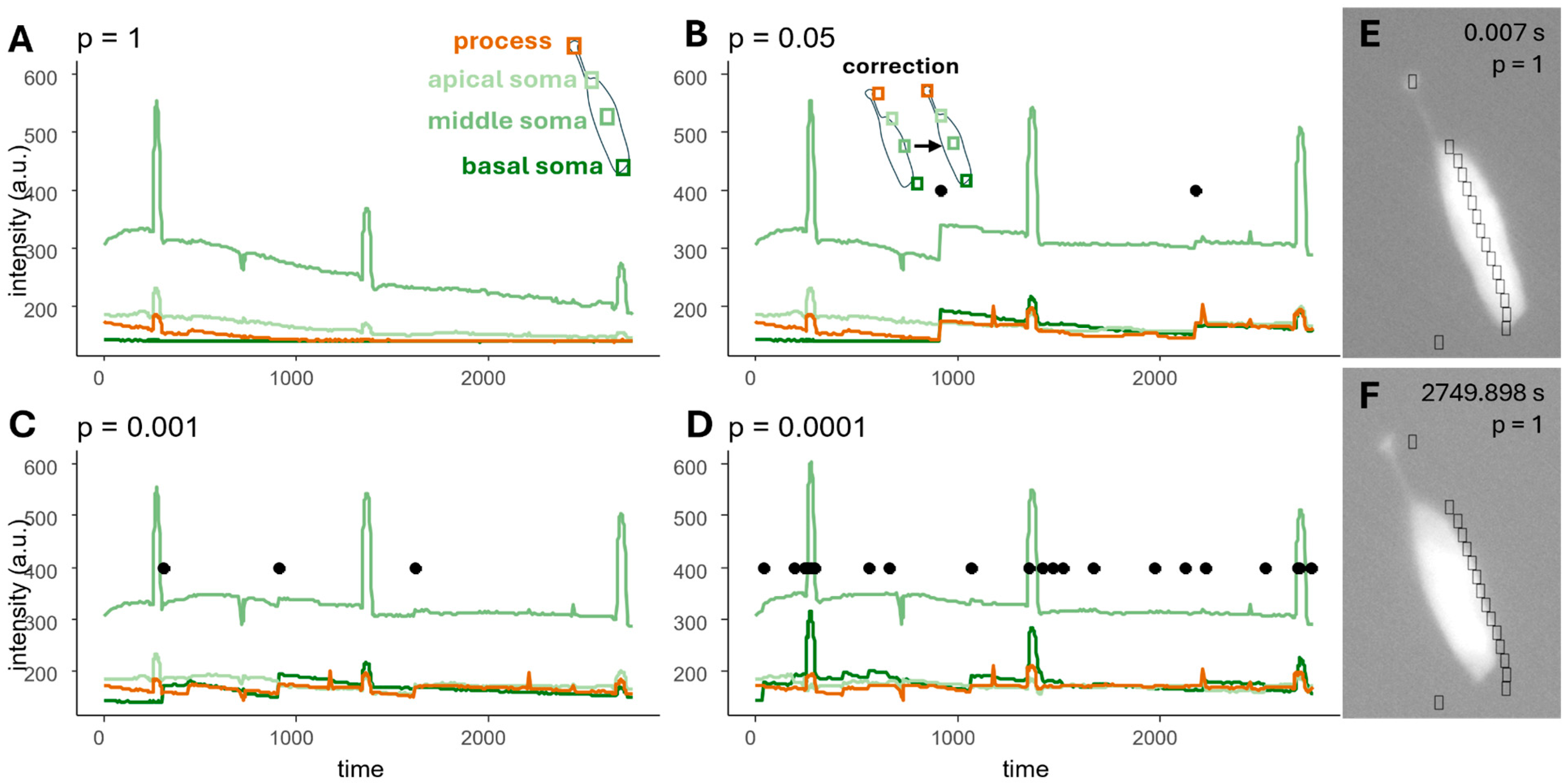
2.2. Motion Detection and Correction
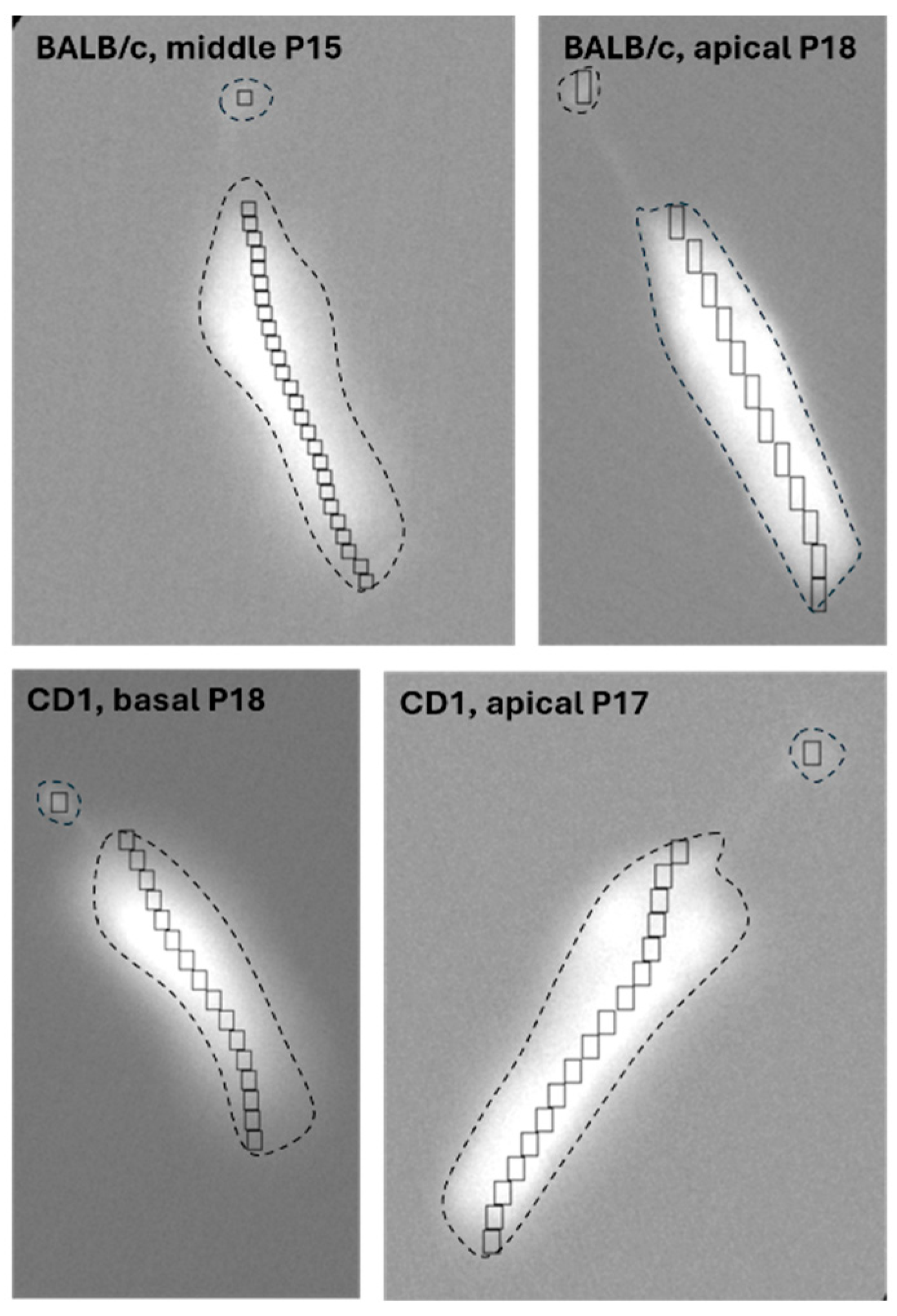
2.3. Other Cell Types
3. Discussion
4. Materials and Methods
4.1. Collection of Experimental Data from Ca2+ Imaging of Deiters’ Cells
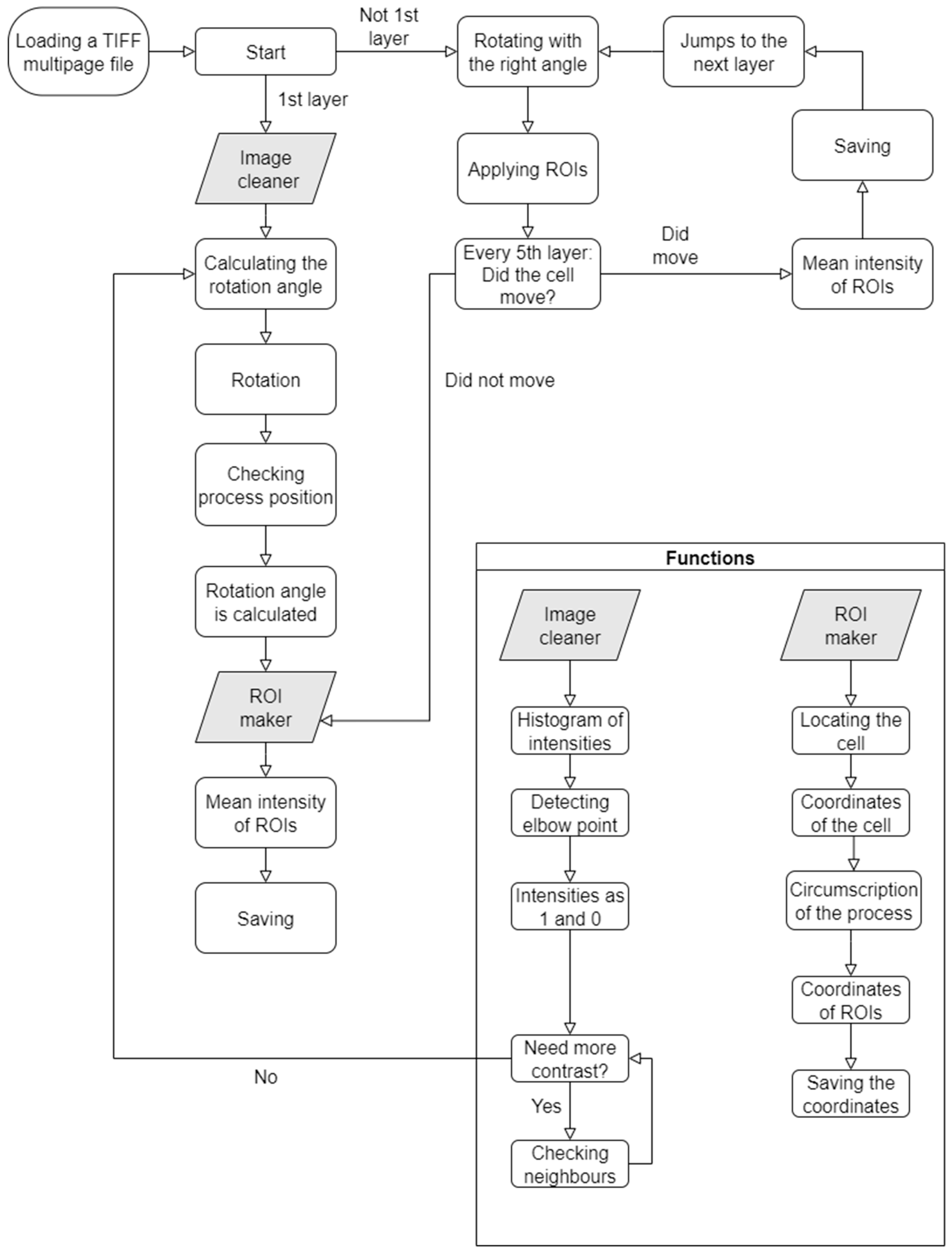
4.2. Structure and Operation of the Algorithm
4.2.1. Start
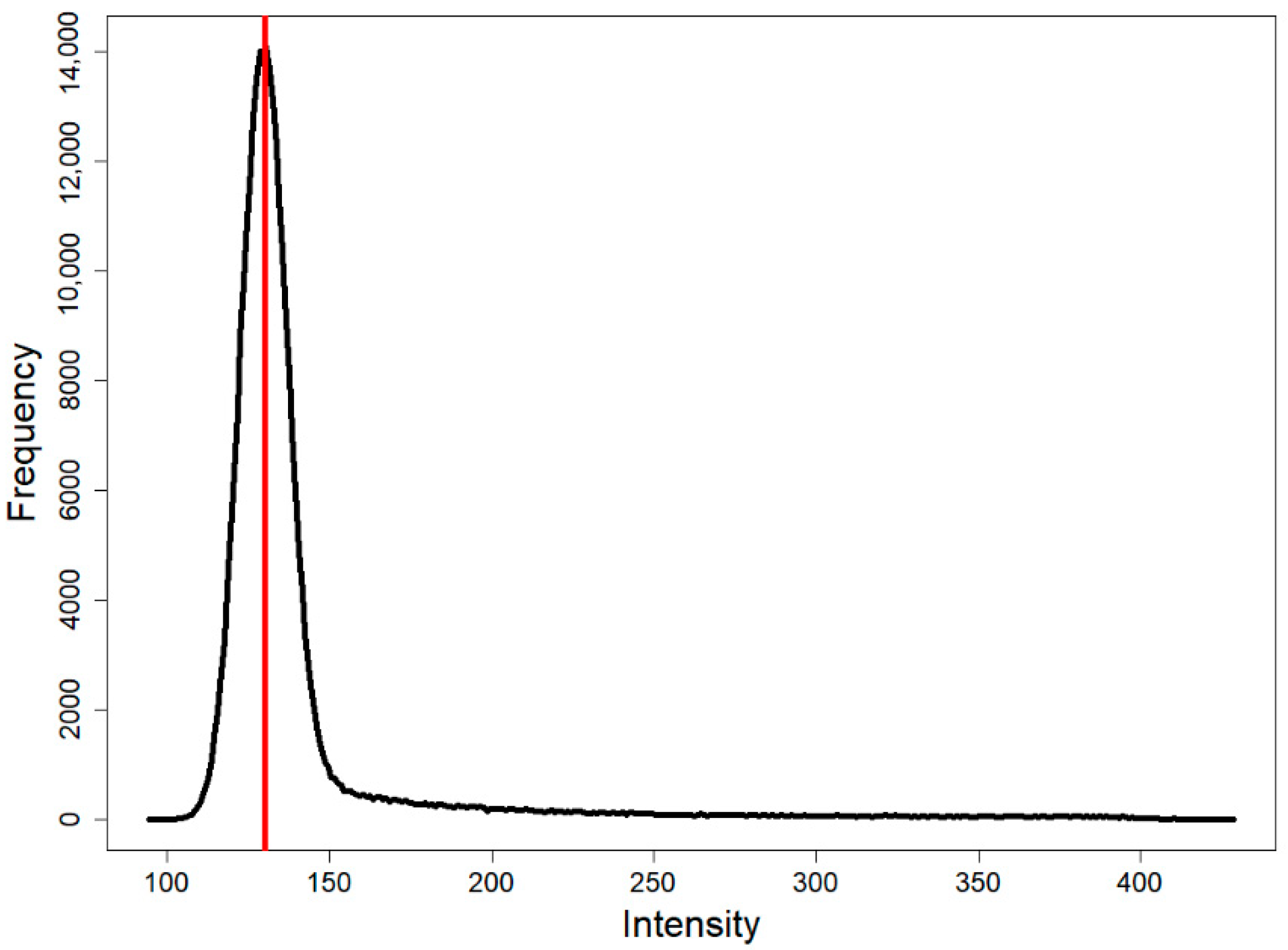
4.2.2. Kneedle
4.2.3. Image Cleaner
4.2.4. Calculation of the Rotation Angle
4.2.5. Rotation
4.2.6. Examination of Process Orientation
4.2.7. ROI Maker
4.2.8. ROI Calculator
4.2.9. Motion Detection
4.2.10. Saving
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ROI | Region of interest |
| OGB | Oregon Green BAPTA 488 |
References
- Canepari, M.; Ross, W.N. Spatial and temporal aspects of neuronal calcium and sodium signals measured with low-affinity fluorescent indicators. Pflug. Arch. Eur. J. Physiol. 2024, 476, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Yokoyama, T. Probing neuronal activity with genetically encoded calcium and voltage fluorescent indicators. Neurosci. Res. 2025, 215, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Mackey, C.; Feng, Y.; Liang, C.; Liang, A.; Tian, H.; Narayan, O.P.; Dong, J.; Tai, Y.; Hu, J.; Mu, Y.; et al. Mechanical Modulation, Physiological Roles, and Imaging Innovations of Intercellular Calcium Waves in Living Systems. Cancers 2025, 17, 1851. [Google Scholar] [CrossRef]
- Zhang, Y.; Rózsa, M.; Liang, Y.; Bushey, D.; Wei, Z.; Zheng, J.; Reep, D.; Broussard, G.J.; Tsang, A.; Tsegaye, G.; et al. Fast and sensitive GCaMP calcium indicators for imaging neural populations. Nature 2023, 615, 884–891. [Google Scholar] [CrossRef]
- Suzuki, J.; Kanemaru, K.; Iino, M. Genetically Encoded Fluorescent Indicators for Organellar Calcium Imaging. Biophys. J. 2016, 111, 1119–1131. [Google Scholar] [CrossRef]
- Zhang, Y.; Looger, L.L. Fast and sensitive GCaMP calcium indicators for neuronal imaging. J. Physiol. 2024, 602, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Shannonhouse, J.; Zhang, Y.; Son, H.; Kim, E.; Han, D.; Park, J.T.; Kim, Y.S. Lessons from the use of in vivo cellular calcium imaging in primary sensory neurons and spinal cord. Neuroscientist 2025. [Google Scholar] [CrossRef]
- Cea Salazar, V.I.; Perez, M.; Robison, A.J.; Trainor, B.C. Impacts of sex differences on optogenetic, chemogenetic, and calcium-imaging tools. Curr. Opin. Neurobiol. 2024, 84, 102817. [Google Scholar] [CrossRef]
- Pnevmatikakis, E.A. Analysis pipelines for calcium imaging data. Curr. Opin. Neurobiol. 2019, 55, 15–21. [Google Scholar] [CrossRef]
- de Melo Reis, R.A.; Freitas, H.R.; de Mello, F.G. Cell Calcium Imaging as a Reliable Method to Study Neuron–Glial Circuits. Front. Neurosci. 2020, 14, 569361. [Google Scholar] [CrossRef]
- Desai, N.S.; Zhong, C.; Kim, R.; Talmage, D.A.; Role, L.W. A simple MATLAB toolbox for analyzing calcium imaging data in vitro and in vivo. J. Neurosci. Methods 2024, 409, 110202. [Google Scholar] [CrossRef]
- Mackay, L.; Mikolajewicz, N.; Komarova, S.V.; Khadra, A. Systematic characterization of dynamic parameters of intracellular calcium signals. Front. Physiol. 2016, 7, 525. [Google Scholar] [CrossRef]
- Sità, L.; Brondi, M.; Lagomarsino de Leon Roig, P.; Curreli, S.; Panniello, M.; Vecchia, D.; Fellin, T. A deep-learning approach for online cell identification and trace extraction in functional two-photon calcium imaging. Nat. Commun. 2022, 13, 1529. [Google Scholar] [CrossRef] [PubMed]
- Hattori, R.; Komiyama, T. PatchWarp: Corrections of non-uniform image distortions in two-photon calcium imaging data by patchwork affine transformations. Cell Rep. Methods 2022, 2, 100205. [Google Scholar] [CrossRef] [PubMed]
- Bjørnstad, D.M.; Åbjørsbråten, K.S.; Hennestad, E.; Cunen, C.; Hermansen, G.H.; Bojarskaite, L.; Pettersen, K.H.; Vervaeke, K.; Enger, R. Begonia—A Two-Photon Imaging Analysis Pipeline for Astrocytic Ca2+ Signals. Front. Cell. Neurosci. 2021, 15, 681066. [Google Scholar] [CrossRef] [PubMed]
- Alsup, A.M.; Fowlds, K.; Cho, M.; Luber, J.M. BetaBuddy: An automated end-to-end computer vision pipeline for analysis of calcium fluorescence dynamics in β-cells. PLoS ONE 2024, 19, e0299549. [Google Scholar] [CrossRef]
- de Kraker, L.; Seignette, K.; Thamizharasu, P.; van den Boom, B.J.G.; Ferreira Pica, I.; Willuhn, I.; Levelt, C.N.; Togt, C. van der SpecSeg is a versatile toolbox that segments neurons and neurites in chronic calcium imaging datasets based on low-frequency cross-spectral power. Cell Rep. Methods 2022, 2, 100299. [Google Scholar] [CrossRef]
- Lagache, T.; Hanson, A.; Pérez-Ortega, J.E.; Fairhall, A.; Yuste, R. Tracking calcium dynamics from individual neurons in behaving animals. PLoS Comput. Biol. 2021, 17, e1009432. [Google Scholar] [CrossRef]
- Liu, W.; Pan, J.; Xu, Y.; Wang, M.; Jia, H.; Zhang, K.; Chen, X.; Li, X.; Liao, X. Fast and Accurate Motion Correction for Two-Photon Ca2+ Imaging in Behaving Mice. Front. Neuroinform. 2022, 16, 851188. [Google Scholar] [CrossRef]
- Cantu, D.A.; Wang, B.; Gongwer, M.W.; He, C.X.; Goel, A.; Suresh, A.; Kourdougli, N.; Arroyo, E.D.; Zeiger, W.; Portera-Cailliau, C. EZcalcium: Open-Source Toolbox for Analysis of Calcium Imaging Data. Front. Neural Circuits 2020, 14, 00025. [Google Scholar] [CrossRef]
- Aghayee, S.; Winkowski, D.E.; Bowen, Z.; Marshall, E.E.; Harrington, M.J.; Kanold, P.O.; Losert, W. Particle tracking facilitates real time capable motion correction in 2D or 3D two-photon imaging of neuronal activity. Front. Neural Circuits 2017, 11, 00056. [Google Scholar] [CrossRef]
- Li, M.; Liu, C.; Cui, X.; Jung, H.; You, H.; Feng, L.; Zhang, S. An Open-Source Real-Time Motion Correction Plug-In for Single-Photon Calcium Imaging of Head-Mounted Microscopy. Front. Neural Circuits 2022, 16, 891825. [Google Scholar] [CrossRef]
- Karavitaki, K. Measurements and Models of Electrically-Evoked Motion in the Gerbil Organ of Corti. Ph.D. Thesis, Massachusetts Institute of Thechnology, Cambridge, MA, USA, 2002. [Google Scholar]
- Tippani, M.; Pattie, E.A.; Davis, B.A.; Nguyen, C.V.; Wang, Y.; Sripathy, S.R.; Maher, B.J.; Martinowich, K.; Jaffe, A.E.; Page, S.C. CaPTure: Calcium PeakToolbox for analysis of in vitro calcium imaging data. BMC Neurosci. 2022, 23, 71. [Google Scholar] [CrossRef]
- Giovannucci, A.; Friedrich, J.; Gunn, P.; Kalfon, J.; Brown, B.L.; Koay, S.A.; Taxidis, J.; Najafi, F.; Gauthier, J.L.; Zhou, P.; et al. Caiman an open source tool for scalable calcium imaging data analysis. eLife 2019, 8, e38173. [Google Scholar] [CrossRef]
- Velez Rueda, A.J.; Gonano, L.A.; Smith, A.G.; Parisi, G.; Fornasari, M.S.; Sommese, L.M. CardIAP: Calcium transients confocal image analysis tool. Front. Bioinform. 2023, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Szikra, T.; Barabas, P.; Bartoletti, T.M.; Huang, W.; Akopian, A.; Thoreson, W.B.; Krizaj, D. Calcium homeostasis and cone signaling are regulated by interactions between calcium stores and plasma membrane ion channels. PLoS ONE 2009, 4, 11–13. [Google Scholar] [CrossRef]
- Kulkarni, M.; Trifunović, D.; Schubert, T.; Euler, T.; Paquet-Durand, F. Calcium dynamics change in degenerating cone photoreceptors. Hum. Mol. Genet. 2016, 25, 3729–3740. [Google Scholar] [CrossRef] [PubMed]
- Harpring, A.L.; Pradeep, S.; Zangle, T.A. Comparison of automated and manual intracellular particle tracking using quantitative phase imaging. J. Opt. Soc. Am. A 2024, 41, C49. [Google Scholar] [CrossRef]
- Dolenšek, J.; Pohorec, V.; Skelin Klemen, M.; Gosak, M.; Stožer, A. Ultrafast multicellular calcium imaging of calcium spikes in mouse beta cells in tissue slices. Acta Physiol. 2025, 241, e14261. [Google Scholar] [CrossRef] [PubMed]
- Gee, J.M.; Smith, N.A.; Fernandez, F.R.; Economo, M.N.; Brunert, D.; Rothermel, M.; Morris, S.C.; Talbot, A.; Palumbos, S.; Ichida, J.M.; et al. Imaging Activity in Neurons and Glia with a Polr2a-Based and Cre-Dependent GCaMP5G-IRES-tdTomato Reporter Mouse. Neuron 2014, 83, 1058–1072. [Google Scholar] [CrossRef]
- Li, F.; Yang, C.; Yuan, F.; Liao, D.; Li, T.; Guilak, F.; Zhong, P. Dynamics and mechanisms of intracellular calcium waves elicited by tandem bubble-induced jetting flow. Proc. Natl. Acad. Sci. USA 2018, 115, E353–E362. [Google Scholar] [CrossRef]
- Guisoni, N.; Ferrero, P.; Layana, C.; Diambra, L. Abortive and propagating intracellular calcium waves: Analysis from a hybrid model. PLoS ONE 2015, 10, e0115187. [Google Scholar] [CrossRef]
- Keiler, S.; Richter, C.-P. Cochlear dimensions obtained in hemicochleae of four different strains of mice: CBA/CaJ, 129/CD1, 129/SvEv and C57BL/6J. Hear. Res. 2001, 162, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Berekméri, E.; Fekete, Á.; Köles, L.; Zelles, T. Postnatal Development of the Subcellular Structures and Purinergic Signaling of Deiters’ Cells along the Tonotopic Axis of the Cochlea. Cells 2019, 8, 1266. [Google Scholar] [CrossRef] [PubMed]
- Edge, R.M.; Evans, B.N.; Pearce, M.; Richter, C.P.; Hu, X.; Dallos, P. Morphology of the unfixed cochlea. Hear. Res. 1998, 124, 1–16. [Google Scholar] [CrossRef]
- Waxman, S.; Quinn, M.; Donahue, C.; Falo, L.D.; Sun, D.; Jakobs, T.C.; Sigal, I.A. Individual astrocyte morphology in the collagenous lamina cribrosa revealed by multicolor DiOlistic labeling. Exp. Eye Res. 2023, 230, 109458. [Google Scholar] [CrossRef]
- Levy, A.F.; Zayats, M.; Guerrero-Cazares, H.; Quiñones-Hinojosa, A.; Searson, P.C. Influence of basement membrane proteins and endothelial cell-derived factors on the morphology of human fetal-derived astrocytes in 2D. PLoS ONE 2014, 9, e92165. [Google Scholar] [CrossRef]
- Domingos, C.; Müller, F.E.; Passlick, S.; Wachten, D.; Ponimaskin, E.; Schwarz, M.K.; Schoch, S.; Zeug, A.; Henneberger, C. Induced Remodelling of Astrocytes In Vitro and In Vivo by Manipulation of Astrocytic RhoA Activity. Cells 2023, 12, 331. [Google Scholar] [CrossRef]
- Cuffaro, H.M.; Pääkkönen, V.; Tjäderhane, L. Enzymatic isolation of viable human odontoblasts. Int. Endod. J. 2016, 49, 454–461. [Google Scholar] [CrossRef]
- Kasahara, K.; Muramatsu, J.; Kurashina, Y.; Miura, S.; Miyata, S.; Onoe, H. Spatiotemporal single-cell tracking analysis in 3D tissues to reveal heterogeneous cellular response to mechanical stimuli. Sci. Adv. 2023, 9, eadf9917. [Google Scholar] [CrossRef]
- Grienberger, C.; Giovannucci, A.; Zeiger, W.; Portera-Cailliau, C. Two-photon calcium imaging of neuronal activity. Nat. Rev. Methods Prim. 2022, 2, 67. [Google Scholar] [CrossRef] [PubMed]
- Nietz, A.K.; Popa, L.S.; Streng, M.L.; Carter, R.E.; Kodandaramaiah, S.B.; Ebner, T.J. Wide-Field Calcium Imaging of Neuronal Network Dynamics In Vivo. Biology 2022, 11, 1601. [Google Scholar] [CrossRef]
- Olivo-Marin, J.C. Extraction of spots in biological images using multiscale products. Pattern Recognit. 2002, 35, 1989–1996. [Google Scholar] [CrossRef]
- Kamran, S.A.; Hossain, K.F.; Moghnieh, H.; Riar, S.; Bartlett, A.; Tavakkoli, A.; Sanders, K.M.; Baker, S.A. New open-source software for subcellular segmentation and analysis of spatiotemporal fluorescence signals using deep learning. iScience 2022, 25, 104277. [Google Scholar] [CrossRef]
- Mandracchia, B.; Zheng, C.; Rajendran, S.; Liu, W.; Forghani, P.; Xu, C.; Jia, S. High-speed optical imaging with sCMOS pixel reassignment. Nat. Commun. 2024, 15, 4598. [Google Scholar] [CrossRef]
- Lu, Z.; Zuo, S.; Shi, M.; Fan, J.; Xie, J.; Xiao, G.; Yu, L.; Wu, J.; Dai, Q. Long-term intravital subcellular imaging with confocal scanning light-field microscopy. Nat. Biotechnol. 2025, 43, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Lines, J.; Baraibar, A.; Nanclares, C.; Martin, E.D.; Aguilar, J.; Kofuji, P.; Navarrete, M.; Araque, A. A spatial threshold for astrocyte calcium surge. eLife 2024, 12, RP90046. [Google Scholar] [CrossRef]
- Bonato, J.; Curreli, S.; Romanzi, S.; Panzeri, S.; Fellin, T. ASTRA: A deep learning algorithm for fast semantic segmentation of large-scale astrocytic networks. bioRxiv 2023. [Google Scholar] [CrossRef]
- Zhou, W.; Jabeen, T.; Sabha, S.; Becker, J.; Nam, J.H. Deiters Cells Act as Mechanical Equalizers for Outer Hair Cells. J. Neurosci. 2022, 42, 8361–8372. [Google Scholar] [CrossRef]
- Morton-Jones, R.T.; Vlajkovic, S.M.; Thorne, P.R.; Cockayne, D.A.; Ryan, A.F.; Housley, G.D. Properties of ATP-gated ion channels assembled from P2X2 subunits in mouse cochlear Reissner’s membrane epithelial cells. Purinergic Signal. 2015, 11, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Dubbs, A.; Guevara, J.; Yuste, R. moco: Fast motion correction for calcium imaging. Front. Neuroinform. 2016, 10, 00006. [Google Scholar] [CrossRef]
- Guizar-Sicairos, M.; Thurman, S.T.; Fienup, J.R. Efficient subpixel image registration algorithms. Opt. Lett. 2008, 33, 156. [Google Scholar] [CrossRef]
- Kaifosh, P.; Zaremba, J.D.; Danielson, N.B.; Losonczy, A. SIMA: Python software for analysis of dynamic fluorescence imaging data. Front. Neuroinform. 2014, 8, 00080. [Google Scholar] [CrossRef]
- Greenberg, D.S.; Kerr, J.N.D. Automated correction of fast motion artifacts for two-photon imaging of awake animals. J. Neurosci. Methods 2009, 176, 1–15. [Google Scholar] [CrossRef]
- Li, K. Image Stabilizer. 2009. Available online: https://www.cs.cmu.edu/~kangli/code/Image_Stabilizer.html (accessed on 8 September 2025).
- Pnevmatikakis, E.A.; Giovannucci, A. NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods 2017, 291, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Mitani, A.; Komiyama, T. Real-time processing of two-photon calcium imaging data including lateral motion artifact correction. Front. Neuroinform. 2018, 12, 00098. [Google Scholar] [CrossRef] [PubMed]
- Berekméri, E.; Deák, O.; Téglás, T.; Sághy, É.; Horváth, T.; Aller, M.; Fekete, Á.; Köles, L.; Zelles, T. Targeted single-cell electroporation loading of Ca2+ indicators in the mature hemicochlea preparation. Hear. Res. 2019, 371, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Tam, E. Kneedle. 2022. Available online: https://github.com/etam4260/kneedle (accessed on 8 October 2025).
- Satopää, V.; Albrecht, J.; Irwin, D.; Raghavan, B. Finding a “kneedle” in a haystack: Detecting knee points in system behavior. In Proceedings of the 31st International Conference on Distributed Computing Systems Workshops, Minneapolis, MN, USA, 20–24 June 2011; pp. 166–171. [Google Scholar]
- Conway, J.H. An enumeration of knots and links, and some of their algebraic properties. In Computational Problems in Abstract Algebra; Elsevier: Amsterdam, The Netherlands, 1970; pp. 329–358. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fazekas, F.; Zelles, T.; Berekméri, E. A Semi-Automatic Tool for the Standardized Analysis of Fluorescent Intensity Changes in Polarized Cells. Int. J. Mol. Sci. 2025, 26, 9987. https://doi.org/10.3390/ijms26209987
Fazekas F, Zelles T, Berekméri E. A Semi-Automatic Tool for the Standardized Analysis of Fluorescent Intensity Changes in Polarized Cells. International Journal of Molecular Sciences. 2025; 26(20):9987. https://doi.org/10.3390/ijms26209987
Chicago/Turabian StyleFazekas, Fruzsina, Tibor Zelles, and Eszter Berekméri. 2025. "A Semi-Automatic Tool for the Standardized Analysis of Fluorescent Intensity Changes in Polarized Cells" International Journal of Molecular Sciences 26, no. 20: 9987. https://doi.org/10.3390/ijms26209987
APA StyleFazekas, F., Zelles, T., & Berekméri, E. (2025). A Semi-Automatic Tool for the Standardized Analysis of Fluorescent Intensity Changes in Polarized Cells. International Journal of Molecular Sciences, 26(20), 9987. https://doi.org/10.3390/ijms26209987





