FLAG Immunoprecipitation-Based Mapping of the In Vivo Assembled Spliceosomal C* Complex
Abstract
1. Introduction
2. Results
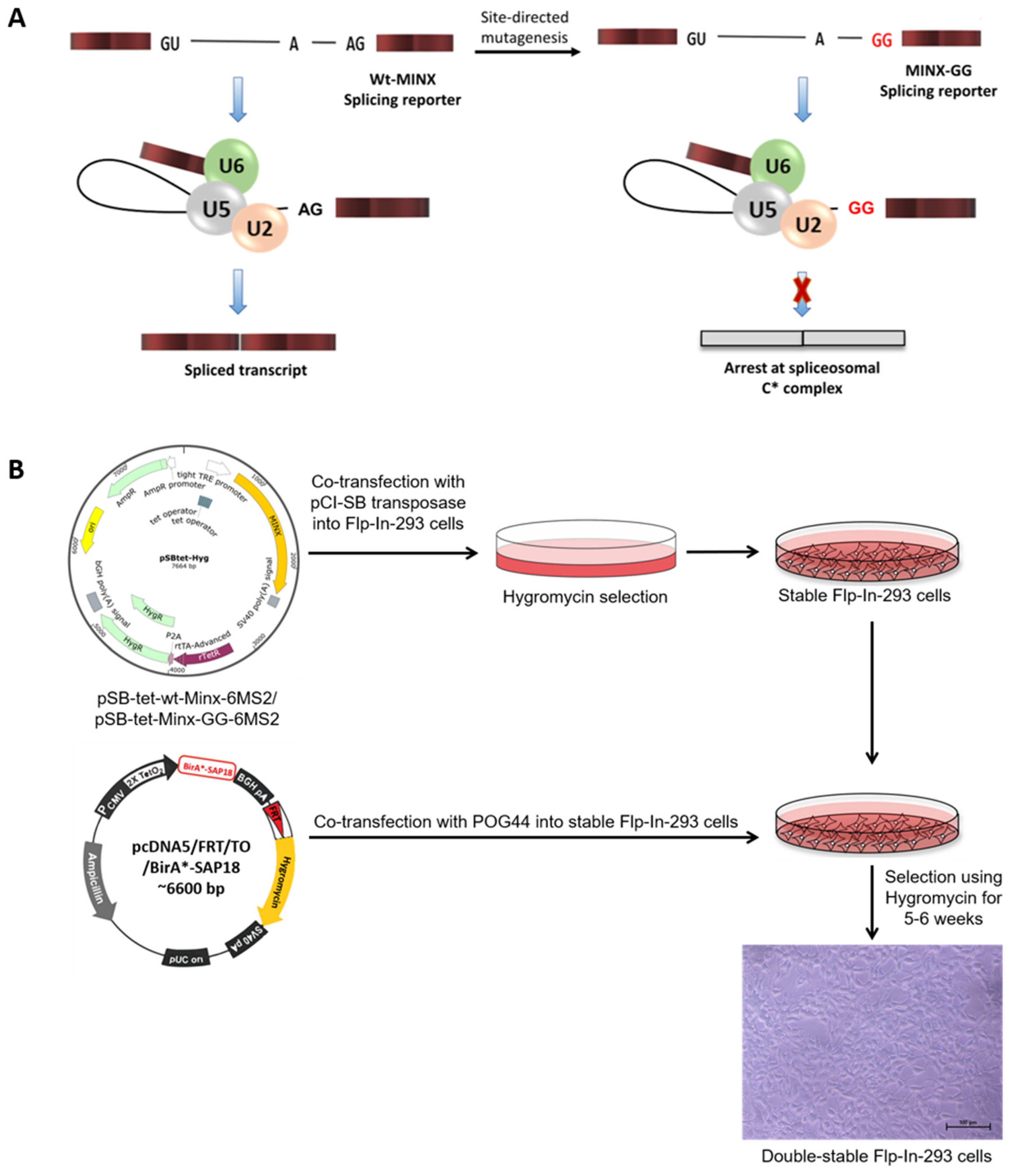
2.1. Design of the MS2-Based In Vivo Splicing Reporter System
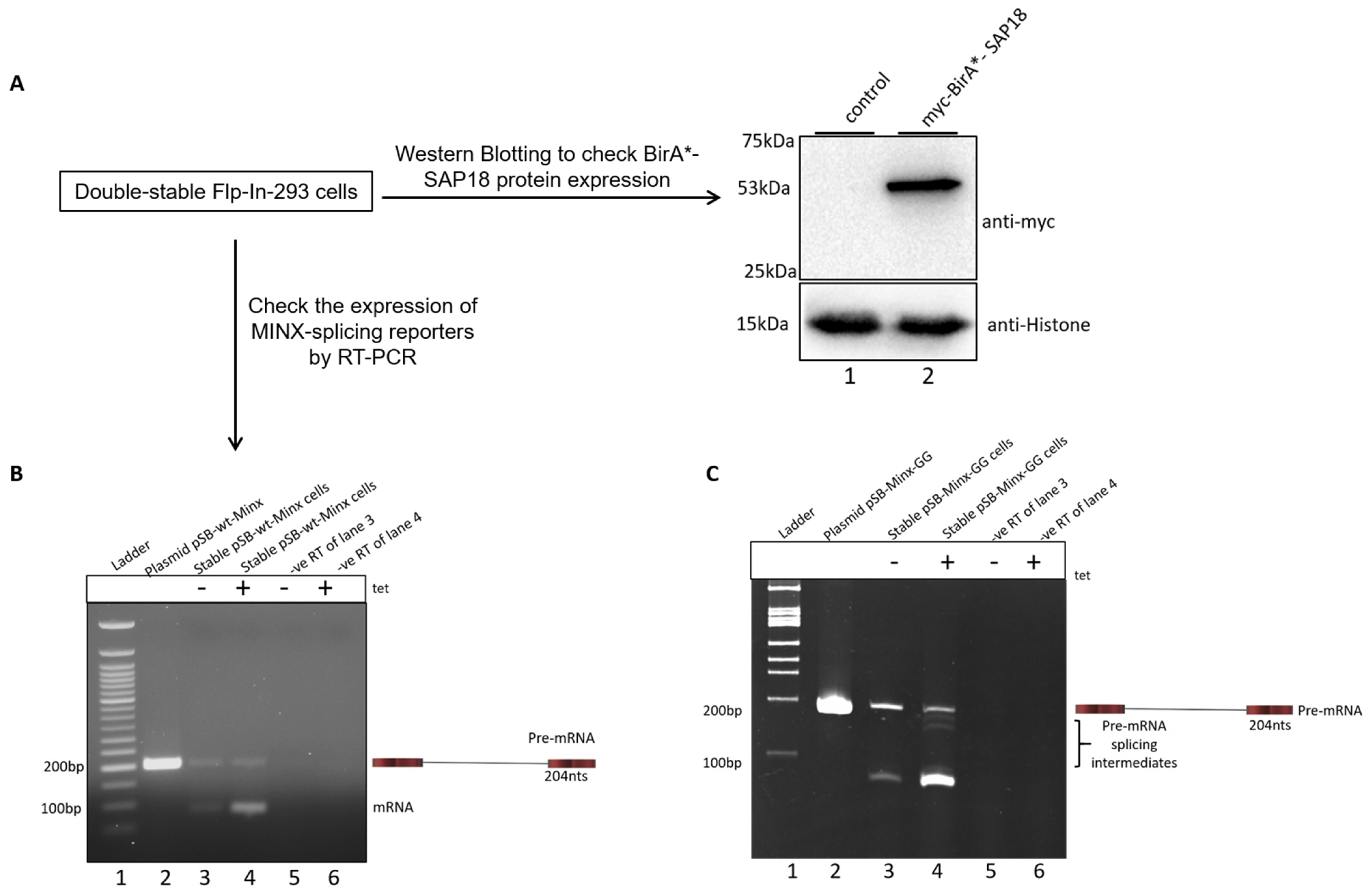
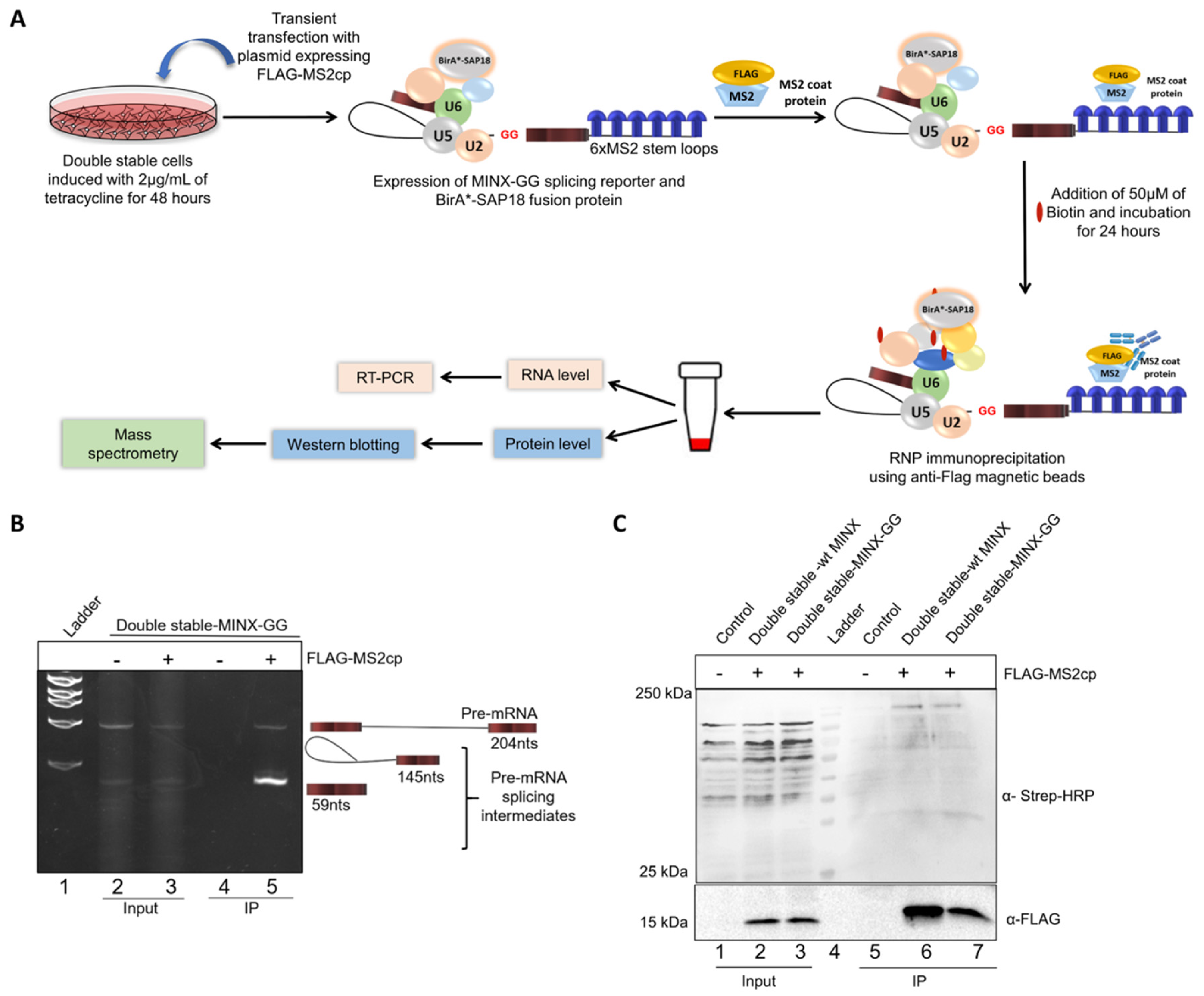
2.2. Mapping the Spliceosomal Complex Assembled on MINX Reporters by RNP IP
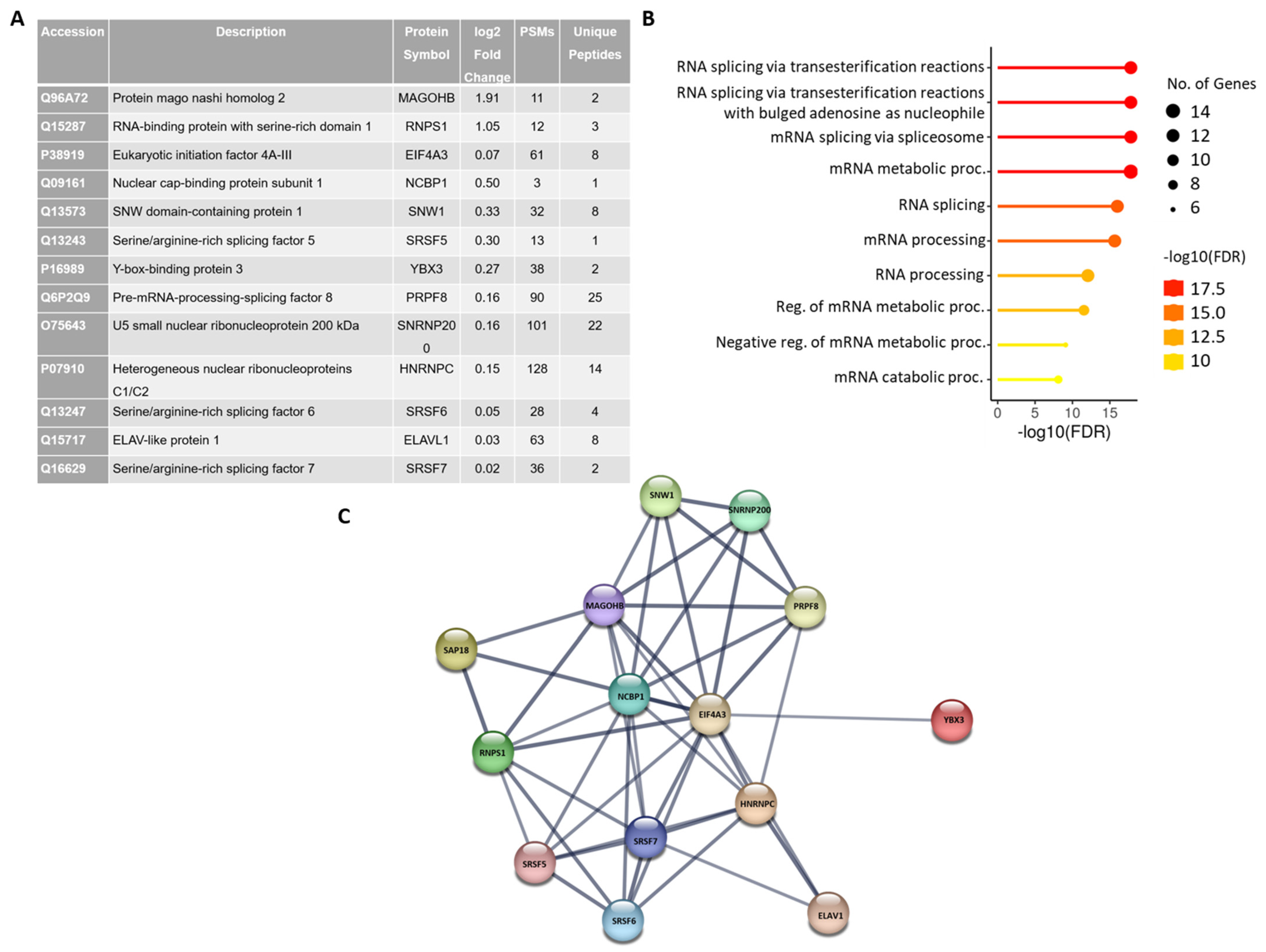
2.3. Identification of the Spliceosomal Proteins Enriched on the MINX-GG Splicing Reporter by Mass Spectrometry
3. Discussion
4. Materials and Methods
4.1. Mammalian Cell Culture
4.2. Plasmids
4.3. Generation of Double-Stable Flp-InTM-293 Cells Expressing BioID Fusion Proteins and Splicing Reporter
4.4. RNA Isolation and cDNA Synthesis
4.5. Reverse Transcriptase PCR (RT-PCR)
4.6. RNP Immunoprecipitation (RNP IP)
4.7. Mass Spectrometric Analysis and Data Processing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Will, C.L.; Lührmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef]
- Brody, E.; Abelson, J. The “spliceosome”: Yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science 1985, 228, 963–967. [Google Scholar] [CrossRef]
- Kastner, B.; Will, C.L.; Stark, H.; Lührmann, R. Structural Insights into Nuclear pre-mRNA Splicing in Higher Eukaryotes. Cold Spring Harb. Perspect. Biol. 2019, 11, a032417. [Google Scholar] [CrossRef]
- Wilkinson, M.E.; Charenton, C.; Nagai, K. RNA Splicing by the Spliceosome. Annu. Rev. Biochem. 2020, 89, 359–388. [Google Scholar] [CrossRef]
- Wahl, M.C.; Will, C.L.; Lührmann, R. The Spliceosome: Design Principles of a Dynamic RNP Machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Kucukural, A.; Cenik, C.; Leszyk, J.D.; Shaffer, S.A.; Weng, Z.; Moore, M.J. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 2012, 151, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Saulière, J.; Murigneux, V.; Wang, Z.; Marquenet, E.; Barbosa, I.; Le Tonquèze, O.; Audic, Y.; Paillard, L.; Crollius, H.R.; Le Hir, H. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat. Struct. Mol. Biol. 2012, 19, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, H.; Izaurralde, E.; Maquat, L.E.; Moore, M.J. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000, 19, 6860–6869. [Google Scholar] [CrossRef]
- Kataoka, N.; Diem, M.D.; Kim, V.N.; Yong, J.; Dreyfuss, G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon–exon junction complex. EMBO J. 2001, 20, 6424–6433. [Google Scholar] [CrossRef]
- Tange, T.; Shibuya, T.; Jurica, M.S.; Moore, M.J. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 2005, 11, 1869–1883. [Google Scholar] [CrossRef]
- Fribourg, S.; Gatfield, D.; Izaurralde, E.; Conti, E. A novel mode of RBD-protein recognition in the Y14–Mago complex. Nat. Struct. Mol. Biol. 2003, 10, 433–439. [Google Scholar] [CrossRef]
- Dybkov, O.; Preussner, M.; El Ayoubi, L.; Feng, V.-Y.; Harnisch, C.; Merz, K.; Leupold, P.; Yudichev, P.; Agafonov, D.E.; Will, C.L.; et al. Regulation of 3′ splice site selection after step 1 of splicing by spliceosomal C* proteins. Sci. Adv. 2023, 9, eadf1785. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, C.; Hang, J.; Finci, L.I.; Lei, J.; Shi, Y. An Atomic Structure of the Human Spliceosome. Cell 2017, 169, 918–929.e14. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Adhikary, A.; Singh, K.K. BioID Proximity Mapping Reveals Novel Interactions of SAP18 in the Prespliceosomal Complex. Biochem. Biophys. Res. Commun. 2024, 738, 150944. [Google Scholar] [CrossRef] [PubMed]
- Kowarz, E.; Löscher, D.; Marschalek, R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 2015, 10, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Busetto, V.; Barbosa, I.; Basquin, J.; Marquenet, É.; Hocq, R.; Hennion, M.; Paternina, J.A.; Namane, A.; Conti, E.; Bensaude, O.; et al. Structural and functional insights into CWC27/CWC22 heterodimer linking the exon junction complex to spliceosomes. Nucleic Acids Res. 2020, 48, 5670–5683. [Google Scholar] [CrossRef]
- Steckelberg, A.L.; Altmueller, J.; Dieterich, C.; Gehring, N.H. CWC22-dependent pre-mRNA splicing and eIF4A3 binding enables global deposition of exon junction complexes. Nucleic Acids Res. 2015, 43, 4687–4700. [Google Scholar] [CrossRef]
- Steckelberg, A.L.; Boehm, V.; Gromadzka, A.M.; Gehring, N.H. CWC22 connects pre-mRNA splicing and exon junction complex assembly. Cell Rep. 2012, 2, 454–461. [Google Scholar] [CrossRef]
- Barbosa, I.; Haque, N.; Fiorini, F.; Barrandon, C.; Tomasetto, C.; Blanchette, M.; Le Hir, H. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat. Struct. Mol. Biol. 2012, 19, 983–990. [Google Scholar] [CrossRef]
- Alexandrov, A.; Colognori, D.; Shu, M.-D.; Steitz, J.A. Human spliceosomal protein CWC22 plays a role in coupling splicing to exon junction complex deposition and nonsense-mediated decay. Proc. Natl. Acad. Sci. USA 2012, 109, 21313–21318. [Google Scholar] [CrossRef]
- Bertram, K.; Agafonov, D.E.; Liu, W.-T.; Dybkov, O.; Will, C.L.; Hartmuth, K.; Urlaub, H.; Kastner, B.; Stark, H.; Lührmann, R. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 2017, 542, 318–323. [Google Scholar] [CrossRef]
- Schwerk, C.; Prasad, J.; Degenhardt, K.; Erdjument-Bromage, H.; White, E.; Tempst, P.; Kidd, V.J.; Manley, J.L.; Lahti, J.M.; Reinberg, D. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol. Cell. Biol. 2003, 23, 2981–2990. [Google Scholar] [CrossRef]
- Boehm, V.; Britto-Borges, T.; Steckelberg, A.-L.; Singh, K.K.; Gerbracht, J.V.; Gueney, E.; Blazquez, L.; Altmüller, J.; Dieterich, C.; Gehring, N.H. Exon Junction Complexes Suppress Spurious Splice Sites to Safeguard Transcriptome Integrity. Mol. Cell 2018, 72, 482–495.e7. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumari, S.; Singh, K.K. FLAG Immunoprecipitation-Based Mapping of the In Vivo Assembled Spliceosomal C* Complex. Int. J. Mol. Sci. 2025, 26, 9914. https://doi.org/10.3390/ijms26209914
Kumari S, Singh KK. FLAG Immunoprecipitation-Based Mapping of the In Vivo Assembled Spliceosomal C* Complex. International Journal of Molecular Sciences. 2025; 26(20):9914. https://doi.org/10.3390/ijms26209914
Chicago/Turabian StyleKumari, Sweta, and Kusum K. Singh. 2025. "FLAG Immunoprecipitation-Based Mapping of the In Vivo Assembled Spliceosomal C* Complex" International Journal of Molecular Sciences 26, no. 20: 9914. https://doi.org/10.3390/ijms26209914
APA StyleKumari, S., & Singh, K. K. (2025). FLAG Immunoprecipitation-Based Mapping of the In Vivo Assembled Spliceosomal C* Complex. International Journal of Molecular Sciences, 26(20), 9914. https://doi.org/10.3390/ijms26209914




