Next-Generation Sequencing in Congenital Eye Malformations: Identification of Genetic Causes and Comparison of Different Panel-Based Diagnostic Strategies
Abstract
1. Introduction
2. Results
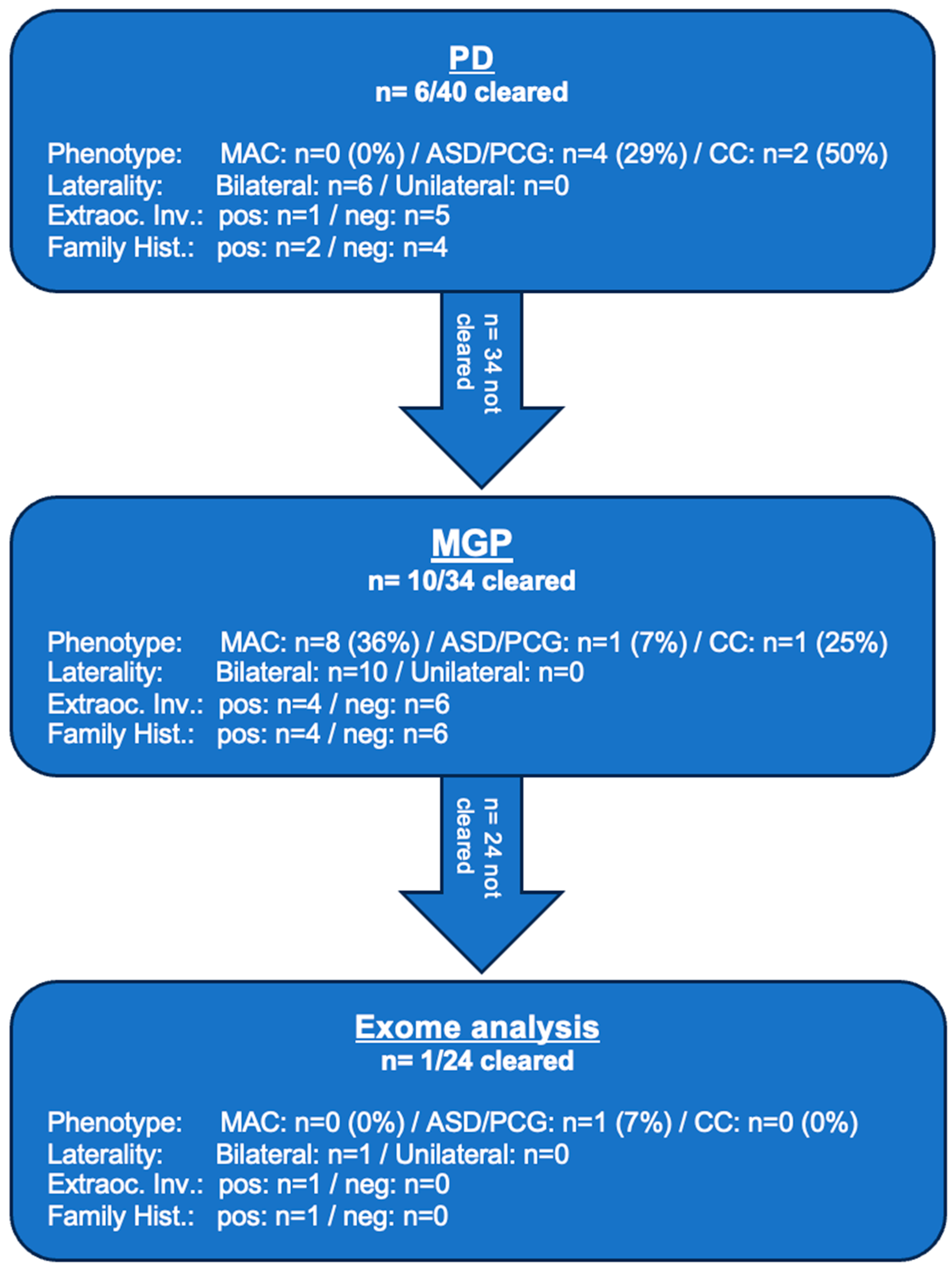
2.1. Study Population Characteristics and Sequencing Results
2.2. Phenotype-Subgroup Results
2.2.1. MAC
2.2.2. ASD/PCG
2.2.3. CC
2.3. Diagnostic Yield
2.4. Variants of Unknown Significance and Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Patient Selection and Clinical Data Collection
4.2. Genetic Testing Workflow
4.3. Bioinformatic Analysis, Variant Classification and Interpretation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, Z.; Yu, C.; Li, J.; Wang, Y.; Liu, D.; Xiang, X.; Su, W.; Pan, Q.; Xie, L.; Xia, K. A Novel Locus for Congenital Simple Microphthalmia Family Mapping to 17p12-q12. Investig. Ophthalmol. Vis. Sci. 2011, 52, 3425–3429. [Google Scholar] [CrossRef][Green Version]
- Haer-Wigman, L.; van Zelst-Stams, W.A.; Pfundt, R.; Born, L.I.v.D.; Klaver, C.C.; Verheij, J.B.; Hoyng, C.B.; Breuning, M.H.; Boon, C.J.; Kievit, A.J.; et al. Diagnostic exome sequencing in 266 Dutch patients with visual impairment. Eur. J. Hum. Genet. 2017, 25, 591–599. [Google Scholar] [CrossRef]
- Sun, W.; Xiao, X.; Li, S.; Guo, X.; Zhang, Q.; Leung, Y.F. Exome Sequencing of 18 Chinese Families with Congenital Cataracts: A New Sight of the NHS Gene. PLoS ONE 2014, 9, e100455. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, S.; Sun, W.; Xiao, X.; Jia, X.; Liu, M.; Xu, L.; Long, Y.; Zhang, Q. An Ophthalmic Targeted Exome Sequencing Panel as a Powerful Tool to Identify Causative Mutations in Patients Suspected of Hereditary Eye Diseases. Transl. Vis. Sci. Technol. 2019, 8, 21. [Google Scholar] [CrossRef]
- Chassaing, N.; Causse, A.; Vigouroux, A.; Delahaye, A.; Alessandri, J.; Boespflug-Tanguy, O.; Boute-Benejean, O.; Dollfus, H.; Duban-Bedu, B.; Gilbert-Dussardier, B.; et al. Molecular findings and clinical data in a cohort of 150 patients with anophthalmia/microphthalmia. Clin. Genet. 2013, 86, 326–334. [Google Scholar] [CrossRef]
- Gillespie, R.L.; O’Sullivan, J.; Ashworth, J.; Bhaskar, S.; Williams, S.; Biswas, S.; Kehdi, E.; Ramsden, S.C.; Clayton-Smith, J.; Black, G.C.; et al. Personalized Diagnosis and Management of Congenital Cataract by Next-Generation Sequencing. Ophthalmology 2014, 121, 2124–2137. [Google Scholar] [CrossRef] [PubMed]
- Magnin, E.; Ayrignac, X.; Berger, E.; Mine, M.; Tournier-Lasserve, E.; Labauge, P. Late Diagnosis of COL4A1 Mutation and Problematic Vascular Risk Fac-tor Management. Eur. Neurol. 2014, 72, 150–152. [Google Scholar] [CrossRef]
- Méjécase, C.; Malka, S.; Guan, Z.; Slater, A.; Arno, G.; Moosajee, M. Practical guide to genetic screening for inherited eye diseases. Ther. Adv. Ophthalmol. 2020, 12, 2515841420954592. [Google Scholar] [CrossRef]
- Deng, H.; Yuan, L. Molecular genetics of congenital nuclear cataract. Eur. J. Med. Genet. 2014, 57, 113–122. [Google Scholar] [CrossRef]
- Patel, A.; Hayward, J.D.; Tailor, V.; Nyanhete, R.; Ahlfors, H.; Gabriel, C.; Jannini, T.B.; Abbou-Rayyah, Y.; Henderson, R.; Nischal, K.K. The Oculome Panel Test: Next-Generation Sequencing to Diagnose a Di-verse Range of Genetic Developmental Eye Disorders. Ophthalmology 2019, 126, 888–907. [Google Scholar] [CrossRef]
- Harding, P.; Gore, S.; Malka, S.; Rajkumar, J.; Oluonye, N.; Moosajee, M. Real-world clinical and molecular management of 50 prospective patients with microphthalmia, anophthalmia and/or ocular coloboma. Br. J. Ophthalmol. 2022, 107, 1925–1935. [Google Scholar] [CrossRef] [PubMed]
- Hilal, L.; Boutayeb, S.; Serrou, A.; Refass-Buret, L.; Shisseh, H.; Bencherifa, F.; El Mzibri, M.; Benazzouz, B.; Berraho, A. Screening of CYP1B1 and MYOC in Moroccan families with primary congenital glaucoma: Three novel mutations in CYP1B1. Mol. Vis. 2010, 16, 1215–1226. [Google Scholar] [PubMed]
- Zhang, Y.; Chen, X.; Wang, L.; Sun, X.; Chen, Y. Heterogeneity of Axenfeld–Rieger Syndrome: Molecular and Clinical Findings in Chinese Patients. Front. Genet. 2021, 12, 732170. [Google Scholar] [CrossRef]
- Deml, B.; Reis, L.; Maheshwari, M.; Griffis, C.; Bick, D.; Semina, E. Whole exome analysis identifies dominant COL4A1 mutations in patients with complex ocular phenotypes involving microphthalmia. Clin. Genet. 2014, 86, 475–481. [Google Scholar] [CrossRef]
- Li, J.; Leng, Y.; Han, S.; Yan, L.; Lu, C.; Luo, Y.; Zhang, X.; Cao, L. Clinical and genetic characteristics of Chinese patients with familial or sporadic pe-diatric cataract. Orphanet J. Rare Dis. 2018, 13, 94. [Google Scholar] [CrossRef]
- Ng, D.; Thakker, N.; Corcoran, C.M.; Donnai, D.; Perveen, R.; Schneider, A.; Hadley, D.W.; Tifft, C.; Zhang, L.; Wilkie, A.O.M.; et al. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat. Genet. 2004, 36, 411–416. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Sergouniotis, P.I.; Mackay, D.S.; Day, A.C.; Wright, G.; Devery, S.; Leroy, B.P.; Robson, A.G.; Holder, G.E.; Li, Z.; et al. A detailed phenotypic assessment of individuals affected by MFRP-related oculopathy. Mol. Vis. 2010, 16, 540–548. [Google Scholar]
- Rainger, J.; Pehlivan, D.; Johansson, S.; Bengani, H.; Sanchez-Pulido, L.; Williamson, K.A.; Ture, M.; Barker, H.; Rosendahl, K.; Spranger, J.; et al. Monoallelic and Biallelic Mutations in MAB21L2 Cause a Spectrum of Major Eye Malformations. Am. J. Hum. Genet. 2014, 94, 915–923. [Google Scholar] [CrossRef]
- Gal, A.; Rau, I.; El Matri, L.; Kreienkamp, H.-J.; Fehr, S.; Baklouti, K.; Chouchane, I.; Li, Y.; Rehbein, M.; Fuchs, J.; et al. Autosomal-Recessive Posterior Microphthalmos Is Caused by Mutations in PRSS56, a Gene Encoding a Trypsin-Like Serine Protease. Am. J. Hum. Genet. 2011, 88, 382–390. [Google Scholar] [CrossRef]
- Reis, L.M.; Tyler, R.C.; Weh, E.; Hendee, K.E.; Kariminejad, A.; Abdul-Rahman, O.; Ben-Omran, T.; Manning, M.A.; Yesilyurt, A.; McCarty, C.A.; et al. Analysis of CYP1B1 in pediatric and adult glaucoma and other ocular pheno-types. Mol. Vis. 2016, 22, 1229–1238. [Google Scholar] [PubMed]
- Assia Batzir, N.; Posey, J.E.; Song, X.; Akdemir, Z.C.; Rosenfeld, J.A.; Brown, C.W.; Chen, E.; Holtrop, S.G.; Mizerik, E.; Nieto Moreno, M.; et al. Phenotypic expansion of POGZ-related intellectual disability syndrome (White-Sutton syndrome). Am. J. Med. Genet. A 2020, 182, 38–52. [Google Scholar] [CrossRef]
- Kamenarova, K.; Mihova, K.; Veleva, N.; Mermeklieva, E.; Mihaylova, B.; Dimitrova, G.; Oscar, A.; Shandurkov, I.; Cherninkova, S.; Kaneva, R. Panel-based next-generation sequencing identifies novel mutations in Bulgarian patients with inherited retinal dystrophies. Mol. Genet. Genom. Med. 2022, 10, e1997. [Google Scholar] [CrossRef]
- Jackson, D.; Malka, S.; Harding, P.; Palma, J.; Dunbar, H.; Moosajee, M. Molecular diagnostic challenges for non-retinal developmental eye disorders in the United Kingdom. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 578–589. [Google Scholar] [CrossRef]
- Chacon Fonseca, I.; Wong, J.; Mireskandari, K.; Chitayat, D. Newborn with bilateral congenital cataracts: Never forget congenital rubella syndrome. Paediatr. Child Health 2020, 25, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Prontera, P.; Stangoni, G.; Ardisia, C.; Rogaia, D.; Mencarelli, A.; Donti, E. Trisomy 2 mosaicism with caudal dysgenesis, Hirschsprung disease, and micro-anophthalmia. Am. J. Med. Genet. A 2011, 155, 928–930. [Google Scholar] [CrossRef] [PubMed]
- Coêlho, R.E.; Sena, D.R.; Cruz, F.S.; Moura, B.C.; Han, C.C.; Andrade, F.N.; Lira, R.P. CYP1B1 Gene and Phenotypic Correlation in Patients From Northeastern Brazil With Primary Congenital Glaucoma. Eur. J. Gastroenterol. Hepatol. 2019, 28, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Nischal, K.K. A new approach to the classification of neonatal corneal opacities. Curr. Opin. Ophthalmol. 2012, 23, 344–354. [Google Scholar] [CrossRef]
- Chandra, A.; Arno, G.; Williamson, K.; Sergouniotis, P.I.; Preising, M.N.; Charteris, D.G.; Thompson, D.A.; Holder, G.E.; Borman, A.D.; Davagnanam, I.; et al. Expansion of Ocular Phenotypic Features Associated With Mutations in ADAMTS18. JAMA Ophthalmol. 2014, 132, 996–1001, Erratum in JAMA Ophthalmol. 2014, 132, 1153. [Google Scholar] [CrossRef][Green Version]
- Williamson, K.A.; Hall, H.N.; Owen, L.J.; Livesey, B.J.; Hanson, I.M.; Adams, G.; Bodek, S.; Calvas, P.; Castle, B.; Clarke, M.; et al. Recurrent heterozygous PAX6 missense variants cause severe bilateral microphthalmia via predictable effects on DNA–protein interaction. Anesth. Analg. 2019, 22, 598–609. [Google Scholar] [CrossRef]
- Lahrouchi, N.; George, A.; Ratbi, I.; Schneider, R.; Elalaoui, S.C.; Moosa, S.; Bharti, S.; Sharma, R.; Abu-Asab, M.; Onojafe, F.; et al. Homozygous frameshift mutations in FAT1 cause a syndrome characterized by colobomatous-microphthalmia, ptosis, nephropathy and syndactyly. Nat. Commun. 2019, 10, 1180. [Google Scholar] [CrossRef]
- Murch, O.; Jain, V.; Benneche, A.; Metcalfe, K.; Hobson, E.; Prescott, K.; Chandler, K.; Ghali, N.; Carmichael, J.; Foulds, N.C.; et al. Further delineation of the clinical spectrum of White–Sutton syndrome: 12 new individuals and a review of the literature. Eur. J. Hum. Genet. 2021, 30, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Williamson, K.A.; FitzPatrick, D.R. The genetic architecture of microphthalmia, anophthalmia and coloboma. Eur. J. Med. Genet. 2014, 57, 369–380. [Google Scholar] [CrossRef]
- Moon, D.; Park, H.W.; Surl, D.; Won, D.; Lee, S.-T.; Shin, S.; Choi, J.R.; Han, J. Precision Medicine through Next-Generation Sequencing in Inherited Eye Diseases in a Korean Cohort. Genes 2021, 13, 27. [Google Scholar] [CrossRef]
- Rechsteiner, D.; Issler, L.; Koller, S.; Lang, E.; Bähr, L.; Feil, S.; Rüegger, C.M.; Kottke, R.; Toelle, S.P.; Zweifel, N.; et al. Genetic Analysis in a Swiss Cohort of Bilateral Congenital Cataract. JAMA Ophthalmol. 2021, 139, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]


| Diagnostic Level | Patient ID | Described Ocular Phenotype | Unilateral/Bilateral | Systemic Features | Family History (Phenotype) | Gene | Sequence Variant | Amino Acid Change | Variant Type | ACMG Class | Variant Known/Novel | Segregation/Variant in Family Members? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PD | ASD/PCG-8 | Congenital glaucoma | Bilateral | - | Negative | CYP1B1 | c.1168C>A; het/c.1076_1082dup; het | p.Arg390Ser/p.Asp361Glufs*16 | Missense/Frameshift | Class 5/Class 5 | Known [12]/Novel | ø |
| PD | ASD/PCG-22 | ARS, early childhood glaucoma | Bilateral | - | Positive (mother, aunt, grandmother, daughter) | FOXC1 | Deletion 3’ Probe Exon 1; het | - | Deletion | Class 4 | - | ø |
| PD | ASD/PCG-23 | Anterior segment dysgenesis (Rieger-anomaly) | Bilateral | Teeth anomalies, excessive umbilical skin | Positive (daughter) | PITX2 | Deletion of all Probes (Intron 3, Exon 4, Exon 5); het | - | Deletion | Class 4 | - | ø |
| PD | ASD/PCG-34 | Anterior segment dysgenesis/congenital glaucoma | Bilateral | - | Negative | FOXC1 | c.247T>C; het | p.Tyr83His | Missense | Class 4 | Known [13] | de novo |
| PD | CC-30 | Congenital cataract, microcornea | Bilateral | Cerebral hemorrhages | Negative | COL4A1 | c.2317G>A; het | p.Gly773Arg | Missense | Class 5 | Known [14] | de novo |
| PD | CC-33 | Congenital cataract, microcornea | Bilateral | - | Negative | PAX6 | c.113G>A; het | p.Arg38Gln | Missense | Class 4 | Known [15] | ø |
| MGP | MAC-9 | Microphthalmia, congenital cataract | Bilateral | Radiculomegaly, oligodontia, toe syndactyly, broad nasal tip | Positive (mother, sister) | BCOR | c.4038_4039del; het | p.Glu1348Ilefs*26 | Frameshift | Class 4 | Known [16] | Mother affected |
| MGP | MAC-10 | Microphthalmia, congenital cataract | Bilateral | - | Positive (sister) | NHS | c.320del; hem | p.Gly107Alafs*89 | Frameshift | Class 5 | Novel | ø |
| MGP | MAC-11 | Iris/retinal/choroidal coloboma | Bilateral | - | Negative | MAB21L2 | c.58del; het | p.Cys20Valfs*37 | Frameshift | Class 4 | Novel | ø |
| MGP | MAC-15 | Iris/retinal/choroidal coloboma | Bilateral | - | Negative | MAB21L2 | c.58del; het | p.Cys20Valfs*37 | Frameshift | Class 4 | Novel | ø |
| MGP | MAC-19 | Microphthalmia, glaucoma, extreme hyperopia | Bilateral | - | Positive (sister) | MFRP | c.201G>A; het/c.1534T>G; het | p.Trp67*/p.Cys512Gly | Nonsense/Missense | Class 4/Class 4 | Known [17]/ Novel | ø |
| MGP | MAC-37 | Microphthalmia, congenital cataract | Bilateral | Ureteral stricture, psoriasis, dental duplication | Negative | NHS | c.3659dup; hem | p.Asn1220Lysfs*5 | Frameshift | Class 4 | Novel | ø |
| MGP | MAC-39 | Iris/retinal/choroidal/optic disk coloboma | Bilateral | Sandal gap deformity | Negative | MAB21L2 | c.145G>A; het | p.Glu49Lys | Missense | Class 5 | Known [18] | de novo |
| MGP | MAC-40 | Microphthalmia | Bilateral | - | Negative | PRSS56 | c.1066dup; hom | p.Gln356Profs*152 | Frameshift | Class 5 | Known [19] | ø |
| MGP | ASD/PCG-27 | Corneal opacity, iris hypoplasia, optic disk changes, congenital glaucoma | Bilateral | - | Negative | CYP1B1 | c.1159G>A; het/c.1064_1076del; het | p.Glu387Lys/p.Arg355Hisfs*69 | Missense/Frameshift | Class 5/Class 5 | Known [20] | ø |
| MGP | CC-4 | Congenital cataract | Bilateral | Language-related developmental delay/microcephaly | Positive (brother) | GCNT2 | c.1148G>A; hom | p.Arg383His | Missense | Class 4 | Known [15] | ø |
| Exome | ASD/PCG-20 | ARS, high myopia, microcornea, maculopathy, glaucoma | Bilateral | Taurodontism, conspicuous auricle | Positive (sister, brother) | ADAMTS18 | c.1399G>T; hom | p.Gly467* | Nonsense | Class 5 | Novel | Sister affected |
| Patient ID | Described Ocular Phenotype | Unilateral/Bilateral | Systemic Features | Family History (Phenotype) | Gene | Sequence Variant | Amino Acid Change | Variant Type | ACMG Class | Variant Known/Novel | Segregation/Variant in Family Members? |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MAC-1 | Iris/choroidal coloboma OD, optic disk coloboma OS | Bilateral | Motor development delay | Negative | POGZ | c.3271C>T; het | p.His1091Tyr | Missense | Class 3 | Novel | ø |
| MAC-5 | Iris/choroidal coloboma | Unilateral | Developmental delay | Negative | POGZ | c.4086A>C; het | p.Glu1362Asp | Missense | Class 3 | Known [21] * | ø |
| MAC-26 | Iris/retinal/choroidal coloboma | Bilateral | - | Negative | FAT1 | c.1025G>C; het | p.Gly342Ala | Missense | Class 3 | Novel | ø |
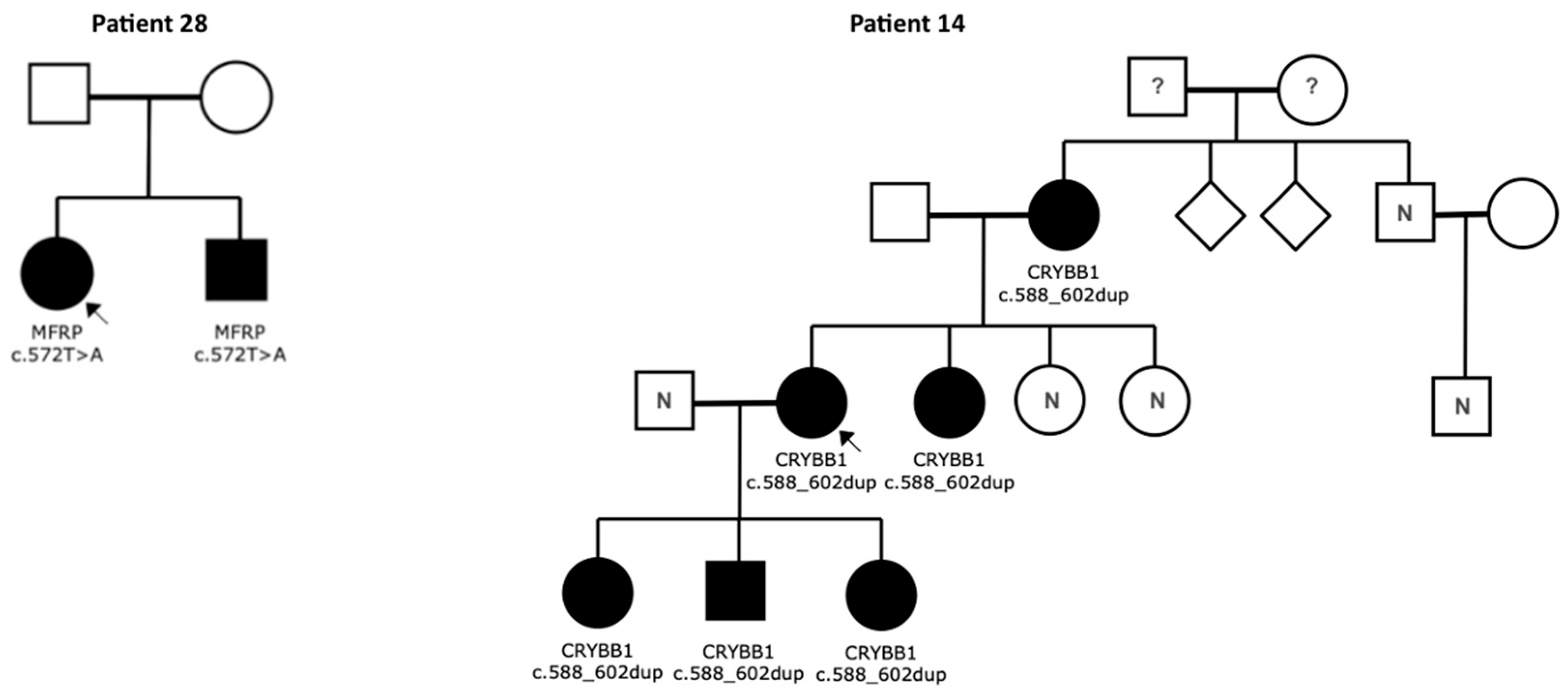
| MAC-28 | Microphthalmia, extreme hyperopia, glaucoma | Bilateral | - | Positive (brother) | MFRP | c.572T>A; hom | p.Ile191Lys | Missense | Class 3 | Novel | Brother affected |
| ASD/PCG-12 | Early childhood glaucoma | Unilateral | - | Negative | COL18A1 | c.2156C>T; het | p.Thr719Met | Missense | Class 3 | Novel | ø |
| CC-14 | Congenital cataract | Bilateral | - | Positive (mother, sister, 2 daughters, son) | CRYBB1 | c.588_602dup; het | p.Gln197_Tyr201dup | Intragenic Duplication | Class 3 | Novel | Mother, Sister, 2 Daughters, Son affected |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neuhann, L.; Laner, A.; Holinski-Feder, E.; Neuhann, T. Next-Generation Sequencing in Congenital Eye Malformations: Identification of Genetic Causes and Comparison of Different Panel-Based Diagnostic Strategies. Int. J. Mol. Sci. 2025, 26, 9854. https://doi.org/10.3390/ijms26209854
Neuhann L, Laner A, Holinski-Feder E, Neuhann T. Next-Generation Sequencing in Congenital Eye Malformations: Identification of Genetic Causes and Comparison of Different Panel-Based Diagnostic Strategies. International Journal of Molecular Sciences. 2025; 26(20):9854. https://doi.org/10.3390/ijms26209854
Chicago/Turabian StyleNeuhann, Lukas, Andreas Laner, Elke Holinski-Feder, and Teresa Neuhann. 2025. "Next-Generation Sequencing in Congenital Eye Malformations: Identification of Genetic Causes and Comparison of Different Panel-Based Diagnostic Strategies" International Journal of Molecular Sciences 26, no. 20: 9854. https://doi.org/10.3390/ijms26209854
APA StyleNeuhann, L., Laner, A., Holinski-Feder, E., & Neuhann, T. (2025). Next-Generation Sequencing in Congenital Eye Malformations: Identification of Genetic Causes and Comparison of Different Panel-Based Diagnostic Strategies. International Journal of Molecular Sciences, 26(20), 9854. https://doi.org/10.3390/ijms26209854






