Transcriptomic Profiling of the Tumor Microenvironment in High-Grade Serous Carcinoma: A Pilot Study of Morphologic and Molecular Distinctions Between Classic and SET Patterns
Abstract
1. Introduction
2. Results
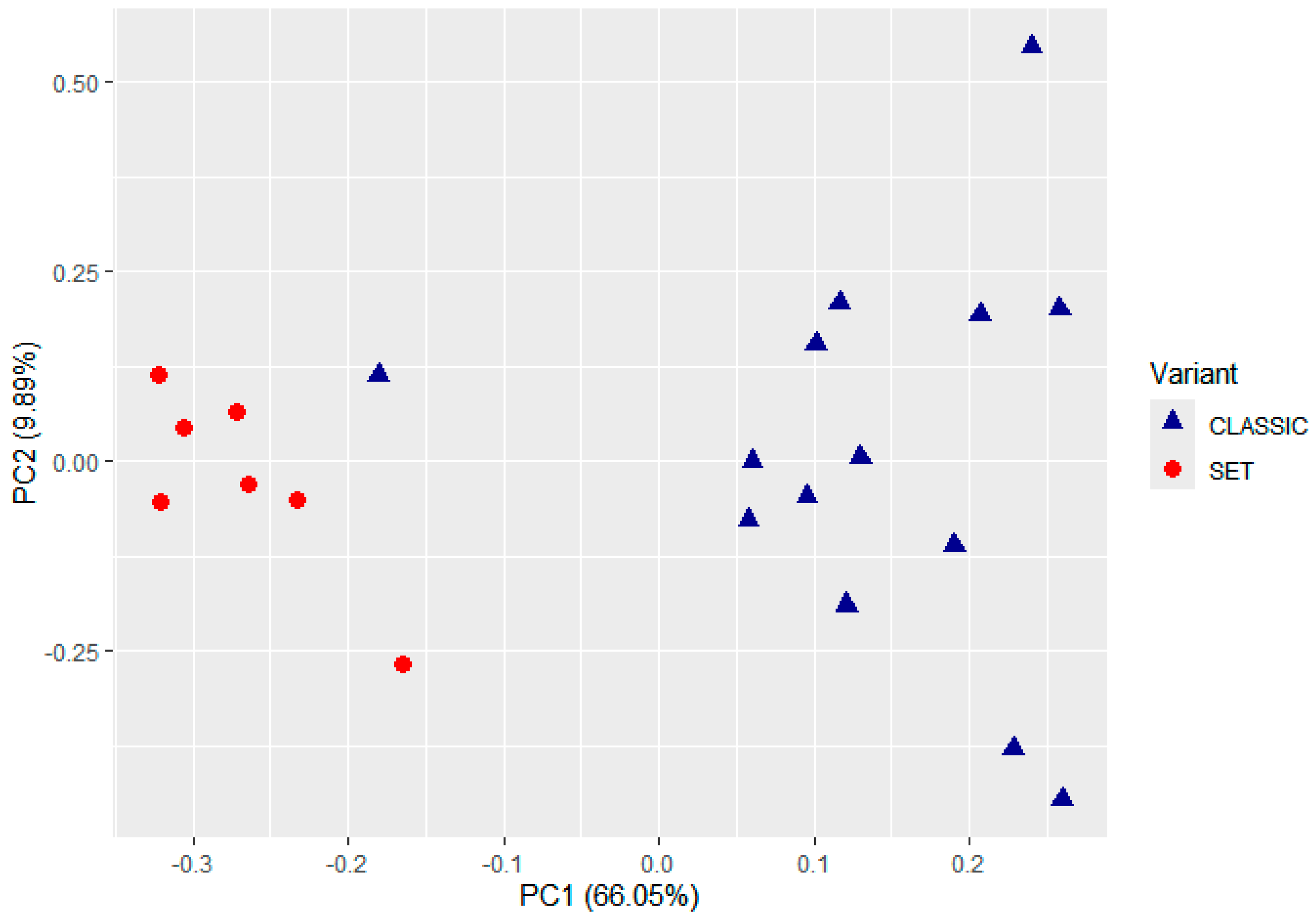
2.1. Unsupervised Classification of TME Profiles
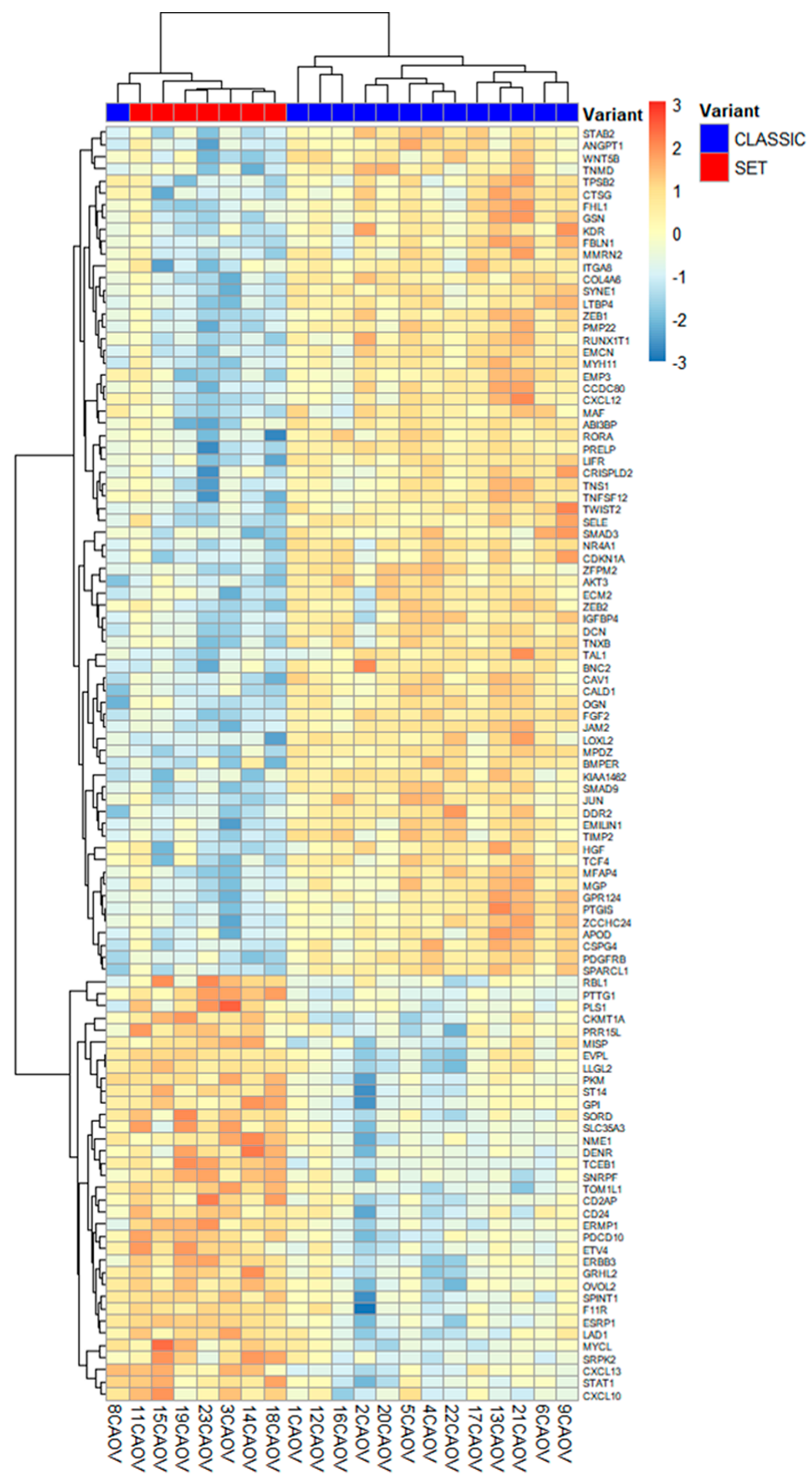
2.2. Differential TME Gene Expression Between SET and Classic HGSC
2.3. Pathway-Level and Microenvironment Insights
3. Discussion
4. Materials and Methods
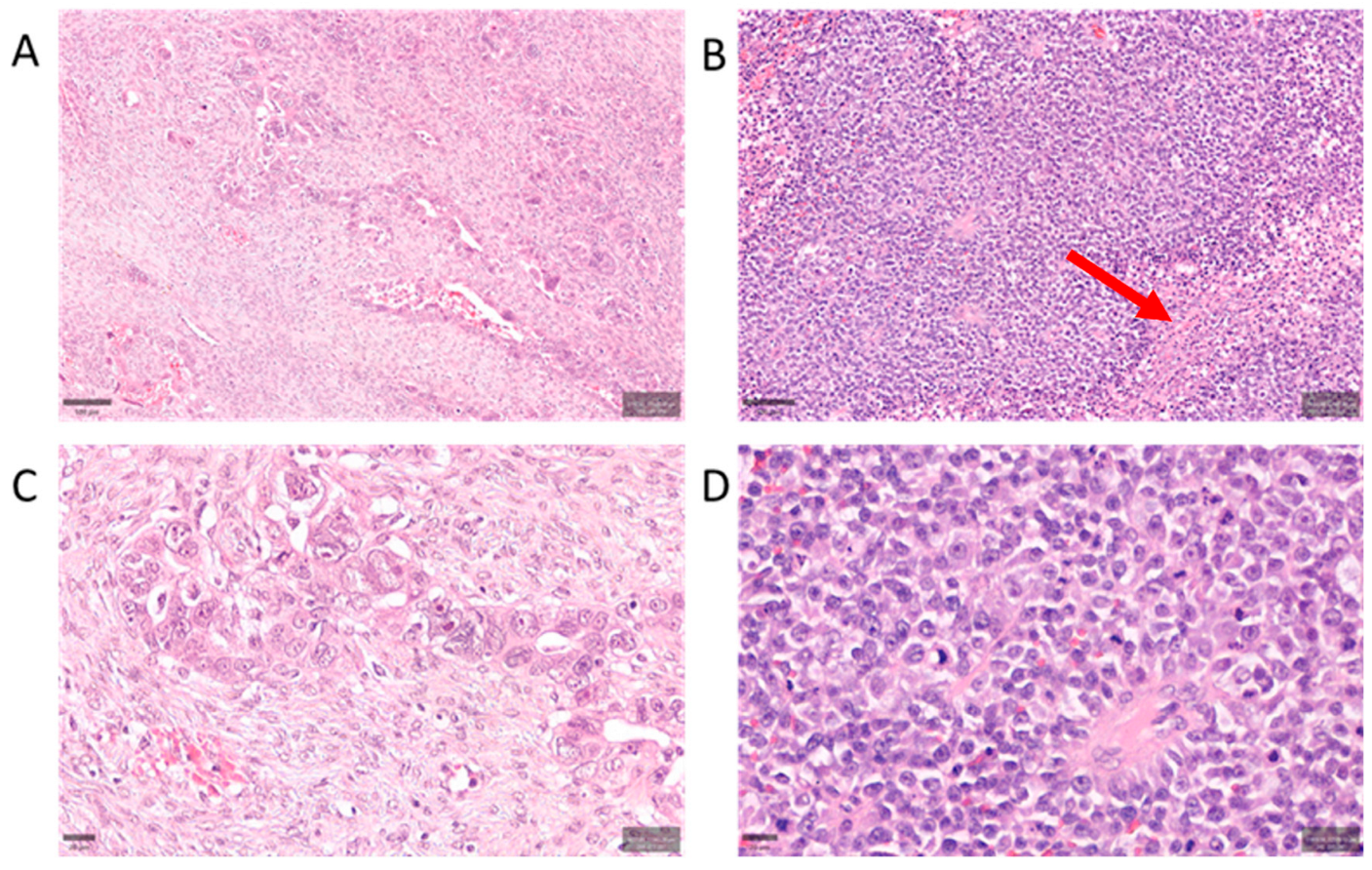
4.1. Case Selection and Morphology Classification
4.2. RNA Extraction and Gene Expression Profiling
4.3. Data Processing and Differential Expression Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AOUP | Azienda Ospedaliero-Universitaria Pisana |
| CAFs | cancer-associated fibroblasts |
| DEGs | differentially expressed genes |
| ECM | extracellular matrix |
| EGFR | epidermal growth factor receptor |
| EMT | epithelial–mesenchymal transition |
| ERBB3 | erb-b2 receptor tyrosine kinase 3 (HER3) |
| FFPE | formalin-fixed, paraffin-embedded |
| FIGO | International Federation of Gynecology and Obstetrics |
| GLM | generalized linear model |
| H&E | hematoxylin and eosin |
| HGSC | high-grade serous carcinoma |
| HRD | homologous recombination deficiency |
| IHC | immunohistochemistry |
| MET | mesenchymal-to-epithelial transition |
| NK | natural killer (cell) |
| PARP | poly(ADP-ribose) polymerase |
| PAX8 | Paired box gene 8 |
| PCA | principal component analysis |
| PI3K | phosphatidylinositol 3-kinase |
| QC | quality control |
| SET | solid, pseudo-endometrioid, transitional-like (morphology) |
| STIC | serous tubal intraepithelial carcinoma |
| TGF-β | transforming growth factor beta |
| TILs | tumor-infiltrating lymphocytes |
| TME | tumor microenvironment |
| WT1 | Wilms tumor 1 |
| TP53 | tumor protein p53 |
| HER2 | human epidermal growth factor receptor 2 |
| HER3 | human epidermal growth factor receptor 3 (ERBB3) |
References
- Bell, D.; Berchuck, A.; Birrer, M.; Chien, J.; Cramer, D.W.; Dao, F.; Dhir, R.; DiSaia, P.; Gabra, H.; Glenn, P.; et al. Integrated Genomic Analyses of Ovarian Carcinoma. Nature 2011, 474, 609–615, Erratum in Nature 2012, 490, 292. [Google Scholar] [CrossRef]
- Bromley, A.B.; Altman, A.D.; Chu, P.; Nation, J.G.; Nelson, G.S.; Ghatage, P.; Kalloger, S.E.; Han, G.; Köbel, M. Architectural Patterns of Ovarian/Pelvic High-Grade Serous Carcinoma. Int. J. Gynecol. Pathol. 2012, 31, 397–404. [Google Scholar] [CrossRef]
- Kurman, R.J.; Shih, I.-M. Molecular Pathogenesis and Extraovarian Origin of Epithelial Ovarian Cancer–Shifting the Paradigm. Hum. Pathol. 2011, 42, 918–931. [Google Scholar] [CrossRef]
- Uner, H.; Demir, M.; Goksuluk, D.; Kars, A.; Uner, M.; Usubutun, A. Evidence for Diverse Prognosis in High-Grade Serous Ovarian Carcinoma: Solid, Pseudoendometrioid, and Transitional-Like; So-Called “SET Morphology” and Progesterone Receptor Status. Turk. Patoloji Derg. 2022, 38, 240–250. [Google Scholar] [CrossRef]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.H.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual Microdissection Identifies Distinct Tumor- and Stroma-Specific Subtypes of Pancreatic Ductal Adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Sergina, N.V.; Rausch, M.; Wang, D.; Blair, J.; Hann, B.; Shokat, K.M.; Moasser, M.M. Escape from HER-Family Tyrosine Kinase Inhibitor Therapy by the Kinase-Inactive HER3. Nature 2007, 445, 437–441. [Google Scholar] [CrossRef]
- Ashkenazi, A. Targeting the Extrinsic Apoptotic Pathway in Cancer: Lessons Learned and Future Directions. J. Clin. Investig. 2015, 125, 487. [Google Scholar] [CrossRef]
- Tothill, R.W.; Tinker, A.V.; George, J.; Brown, R.; Fox, S.B.; Lade, S.; Johnson, D.S.; Trivett, M.K.; Etemadmoghadam, D.; Locandro, B.; et al. Novel Molecular Subtypes of Serous and Endometrioid Ovarian Cancer Linked to Clinical Outcome. Clin. Cancer Res. 2008, 14, 5198–5208. [Google Scholar] [CrossRef]
- de Kruijf, E.M.; van Nes, J.G.H.; van de Velde, C.J.H.; Putter, H.; Smit, V.T.H.B.M.; Liefers, G.J.; Kuppen, P.J.K.; Tollenaar, R.A.E.M.; Mesker, W.E. Tumor-Stroma Ratio in the Primary Tumor Is a Prognostic Factor in Early Breast Cancer Patients, Especially in Triple-Negative Carcinoma Patients. Breast Cancer Res. Treat. 2011, 125, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.H.; Zhang, B.; McDermott, J.E.; Zhou, J.Y.; Petyuk, V.A.; Chen, L.; Ray, D.; et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.J.; Takeda, K.; Hayakawa, Y.; Peschon, J.J.; van den Brink, M.R.M.; Yagita, H. Nature’s TRAIL—On a Path to Cancer Immunotherapy. Immunity 2003, 18, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Yamaguchi, N.; Akiba, H.; Kojima, Y.; Hayakawa, Y.; Tanner, J.E.; Sayers, T.J.; Seki, N.; Okumura, K.; Yagita, H.; et al. Induction of Tumor-Specific T Cell Immunity by Anti-DR5 Antibody Therapy. J. Exp. Med. 2004, 199, 437–448. [Google Scholar] [CrossRef]
- Sheng, Q.; Liu, X.; Fleming, E.; Yuan, K.; Piao, H.; Chen, J.; Moustafa, Z.; Thomas, R.K.; Greulich, H.; Schinzel, A.; et al. An Activated ErbB3/NRG1 Autocrine Loop Supports in Vivo Proliferation in Ovarian Cancer Cells. Cancer Cell 2010, 17, 298–310. [Google Scholar] [CrossRef]
- Garrett, J.T.; Olivares, M.G.; Rinehart, C.; Granja-Ingram, N.D.; Sánchez, V.; Chakrabarty, A.; Dave, B.; Cook, R.S.; Pao, W.; McKinely, E.; et al. Transcriptional and Posttranslational Up-Regulation of HER3 (ErbB3) Compensates for Inhibition of the HER2 Tyrosine Kinase. Proc. Natl. Acad. Sci. USA 2011, 108, 5021–5026. [Google Scholar] [CrossRef]
- Yang, J.; Mani, S.A.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; Weinberg, R.A. Twist, a Master Regulator of Morphogenesis, Plays an Essential Role in Tumor Metastasis. Cell 2004, 117, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular Mechanisms of Epithelial–Mesenchymal Transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. XCell: Digitally Portraying the Tissue Cellular Heterogeneity Landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed]
- Shield, K.; Ackland, M.L.; Ahmed, N.; Rice, G.E. Multicellular Spheroids in Ovarian Cancer Metastases: Biology and Pathology. Gynecol. Oncol. 2009, 113, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Burger, R.A.; Brady, M.F.; Bookman, M.A.; Fleming, G.F.; Monk, B.J.; Huang, H.; Mannel, R.S.; Homesley, H.D.; Fowler, J.; Greer, B.E.; et al. Incorporation of Bevacizumab in the Primary Treatment of Ovarian Cancer. N. Engl. J. Med. 2011, 365, 2473–2483. [Google Scholar] [CrossRef]
- Ahmed, N.; Abubaker, K.; Findlay, J.; Quinn, M. Epithelial Mesenchymal Transition and Cancer Stem Cell-like Phenotypes Facilitate Chemoresistance in Recurrent Ovarian Cancer. Curr. Cancer Drug Targets 2010, 10, 268–278. [Google Scholar] [CrossRef]
- Kroeger, P.T.; Drapkin, R. Pathogenesis and Heterogeneity of Ovarian Cancer. Curr. Opin. Obstet. Gynecol. 2017, 29, 26–34. [Google Scholar] [CrossRef]
- Lengyel, E. Ovarian Cancer Development and Metastasis. Am. J. Pathol. 2010, 177, 1053–1064. [Google Scholar] [CrossRef]
- Cheon, D.-J.; Tong, Y.; Sim, M.-S.; Dering, J.; Berel, D.; Cui, X.; Lester, J.; Beach, J.A.; Tighiouart, M.; Walts, A.E.; et al. A Collagen-Remodeling Gene Signature Regulated by TGF-β Signaling Is Associated with Metastasis and Poor Survival in Serous Ovarian Cancer. Clin. Cancer Res. 2014, 20, 711–723. [Google Scholar] [CrossRef]
- Jeong, H.M.; Han, J.; Lee, S.H.; Park, H.J.; Lee, H.J.; Choi, J.S.; Lee, Y.M.; Choi, Y.L.; Shin, Y.K.; Kwon, M.J. ESRP1 Is Overexpressed in Ovarian Cancer and Promotes Switching from Mesenchymal to Epithelial Phenotype in Ovarian Cancer Cells. Oncogenesis 2017, 6, e389, Erratum in Exp. Hematol. Oncol. 2017, 6, e391; Correction in J. Biomed. Sci. 2019, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Kojima, Y.; Yonemori, K.; Yoshida, H.; Sugiura, Y.; Ohtake, Y.; Okuma, H.S.; Nishikawa, T.; Tanioka, M.; Sudo, K.; et al. Neoadjuvant Chemotherapy Promotes the Expression of HER3 in Patients with Ovarian Cancer. Oncol. Lett. 2020, 20, 336. [Google Scholar] [CrossRef]
- De Leo, A.; Santini, D.; Ceccarelli, C.; Santandrea, G.; Palicelli, A.; Acquaviva, G.; Chiarucci, F.; Rosini, F.; Ravegnini, G.; Pession, A.; et al. What Is New on Ovarian Carcinoma: Integrated Morphologic and Molecular Analysis Following the New 2020 World Health Organization Classification of Female Genital Tumors. Diagnostics 2021, 11, 697. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhang, Y.; Feng, M.; Shen, M.; Yang, L.; Wei, B.; Zhou, Y.; Zhang, Z. HER3: Updates and Current Biology Function, Targeted Therapy and Pathologic Detecting Methods. Life Sci. 2024, 357, 123087. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiu, J.G.; Wang, W.; Sun, F.L.; Wang, X.; Liu, W.J.; Jia, X.Y.; Ji, H.; Wang, L.; Jiang, B.H. Suppression of CYLD by HER3 Confers Ovarian Cancer Platinum Resistance via Inhibiting Apoptosis and by Inducing Drug Efflux. Exp. Hematol. Oncol. 2025, 14, 21. [Google Scholar] [CrossRef]
- Mujoo, K.; Choi, B.-K.; Huang, Z.; Zhang, N.; An, Z.; Mujoo, K.; Choi, B.-K.; Huang, Z.; Zhang, N.; An, Z. Regulation of ERBB3/HER3 Signaling in Cancer. Oncotarget 2014, 5, 10222–10236. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.; Holmes, A.; Lomo, L.; Steinkamp, M.P.; Kang, H.; Muller, C.Y.; Wilson, B.S. High Incidence of ErbB3, ErbB4 and MET Expression In Ovarian Cancer. Int. J. Gynecol. Pathol. 2014, 33, 402. [Google Scholar] [CrossRef]
- Dasari, S.; Fang, Y.; Mitra, A.K. Cancer Associated Fibroblasts: Naughty Neighbors That Drive Ovarian Cancer Progression. Cancers 2018, 10, 406. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.C.; Liao, Y.C.; Chen, P.M.; Yang, Y.Y.; Wang, Y.H.; Tung, S.L.; Chuang, C.M.; Sung, Y.W.; Jang, T.H.; Chuang, S.E.; et al. Periostin Promotes Ovarian Cancer Metastasis by Enhancing M2 Macrophages and Cancer-Associated Fibroblasts via Integrin-Mediated NF-ΚB and TGF-Β2 Signaling. J. Biomed. Sci. 2022, 29, 109, Erratum in J. Biomed. Sci. 2023, 30, 54. [Google Scholar] [CrossRef] [PubMed]
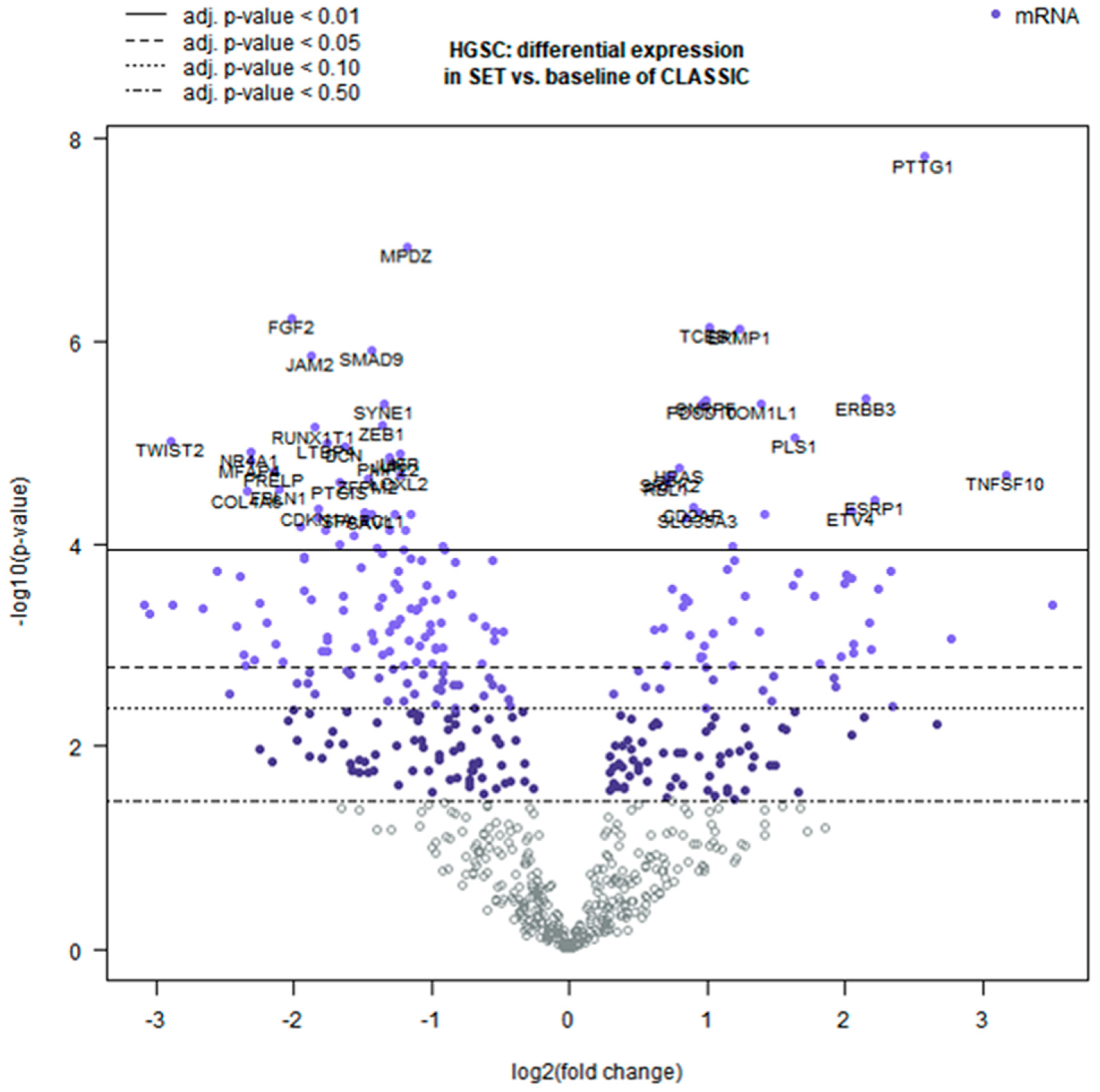
| Gene | Log2FC (SET vs. Classic) | 95% CI (Log2) | q (BY) | Higher in |
|---|---|---|---|---|
| PTTG1 | +2.57 | [2.03, 3.10] | 7.21 × 10−5 | SET (cell cycle regulator) |
| TNFSF10 | +3.17 | [2.06, 4.27] | 0.0037 | SET (immune apoptosis ligand) |
| ESRP1 | +2.22 | [1.41, 3.04] | 0.0052 | SET (epithelial splicing factor) |
| ERBB3 | +2.15 | [1.49, 2.81] | 0.00166 | SET (growth factor receptor) |
| PLS1 | +1.64 | [1.11, 2.18] | 0.0028 | SET (actin-bundling protein) |
| HRAS | +0.80 | [0.52, 1.08] | 0.0036 | SET (RAS oncogene) |
| TOP2A | +1.10 | [0.70, 1.50] | <0.01 | SET (topoisomerase, proliferation) |
| CDC20 | +1.05 | [0.65, 1.45] | <0.01 | SET (cell cycle checkpoint) |
| UBE2C | +1.00 | [0.60, 1.40] | <0.01 | SET (ubiquitin ligase) |
| TWIST2 | −2.89 | [−3.84, −1.94] | 0.0028 | Classic (EMT transcription factor) |
| FGF2 | −2.02 | [−2.56, −1.48] | 0.00072 | Classic (angiogenic factor) |
| FBLN1 | −2.11 | [−2.86, −1.35] | 0.0044 | Classic (ECM glycoprotein) |
| OGN | −1.95 | [−2.70, −1.19] | 0.0068 | Classic (stromal proteoglycan) |
| LTBP4 | −1.75 | [−2.32, −1.17] | 0.0028 | Classic (TGF-β binding protein) |
| ZEB1 | −1.36 | [−1.80, −0.93] | 0.0025 | Classic (EMT transcription factor) |
| DCN | −1.63 | [−2.17, −1.09] | 0.0029 | Classic (ECM proteoglycan) |
| TGFBI | −1.20 | [−1.66, −0.74] | <0.01 | Classic (TGF-β–induced protein) |
| POSTN | −1.10 | [−1.54, −0.66] | <0.01 | Classic (matricellular protein) |
| ANGPTL4 | −1.05 | [−1.48, −0.62] | <0.01 | Classic (angiogenesis factor) |
| THBS2 | −1.00 | [−1.40, −0.60] | <0.01 | Classic (angiogenesis inhibitor) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannini, R.; Bartoli, F.; Ieso, K.D.; Camacci, T.; Bertolucci, A.; Piccini, L.; Rreka, E.; Volterrani, D.; Gemignani, F.; Landi, S.; et al. Transcriptomic Profiling of the Tumor Microenvironment in High-Grade Serous Carcinoma: A Pilot Study of Morphologic and Molecular Distinctions Between Classic and SET Patterns. Int. J. Mol. Sci. 2025, 26, 10229. https://doi.org/10.3390/ijms262010229
Giannini R, Bartoli F, Ieso KD, Camacci T, Bertolucci A, Piccini L, Rreka E, Volterrani D, Gemignani F, Landi S, et al. Transcriptomic Profiling of the Tumor Microenvironment in High-Grade Serous Carcinoma: A Pilot Study of Morphologic and Molecular Distinctions Between Classic and SET Patterns. International Journal of Molecular Sciences. 2025; 26(20):10229. https://doi.org/10.3390/ijms262010229
Chicago/Turabian StyleGiannini, Riccardo, Francesco Bartoli, Katia De Ieso, Tiziano Camacci, Andrea Bertolucci, Lorenzo Piccini, Erion Rreka, Duccio Volterrani, Federica Gemignani, Stefano Landi, and et al. 2025. "Transcriptomic Profiling of the Tumor Microenvironment in High-Grade Serous Carcinoma: A Pilot Study of Morphologic and Molecular Distinctions Between Classic and SET Patterns" International Journal of Molecular Sciences 26, no. 20: 10229. https://doi.org/10.3390/ijms262010229
APA StyleGiannini, R., Bartoli, F., Ieso, K. D., Camacci, T., Bertolucci, A., Piccini, L., Rreka, E., Volterrani, D., Gemignani, F., Landi, S., Ugolini, C., Lippolis, P. V., & Faviana, P. (2025). Transcriptomic Profiling of the Tumor Microenvironment in High-Grade Serous Carcinoma: A Pilot Study of Morphologic and Molecular Distinctions Between Classic and SET Patterns. International Journal of Molecular Sciences, 26(20), 10229. https://doi.org/10.3390/ijms262010229






