SUMF1 Common Variant rs793391 Is Associated with Response to Inhaled Corticosteroids in Patients with COPD
Abstract
1. Introduction
2. Results
2.1. Patients’ Characteristics, Genotype and Allele Frequencies
2.2. Association of SUMF1 Polymorphisms with Lung Function, Histological and Blood-Sampling Parameters at Baseline
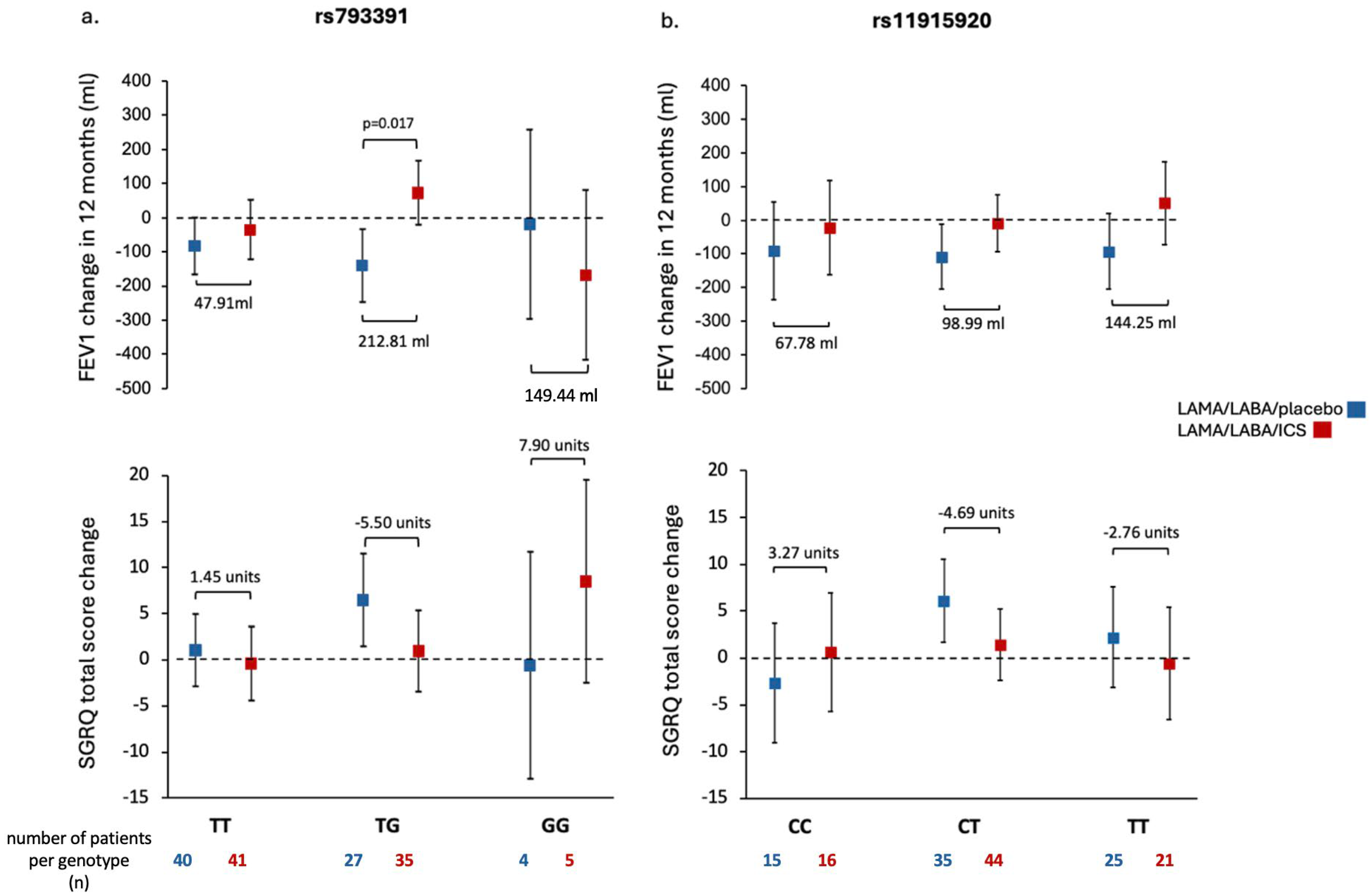
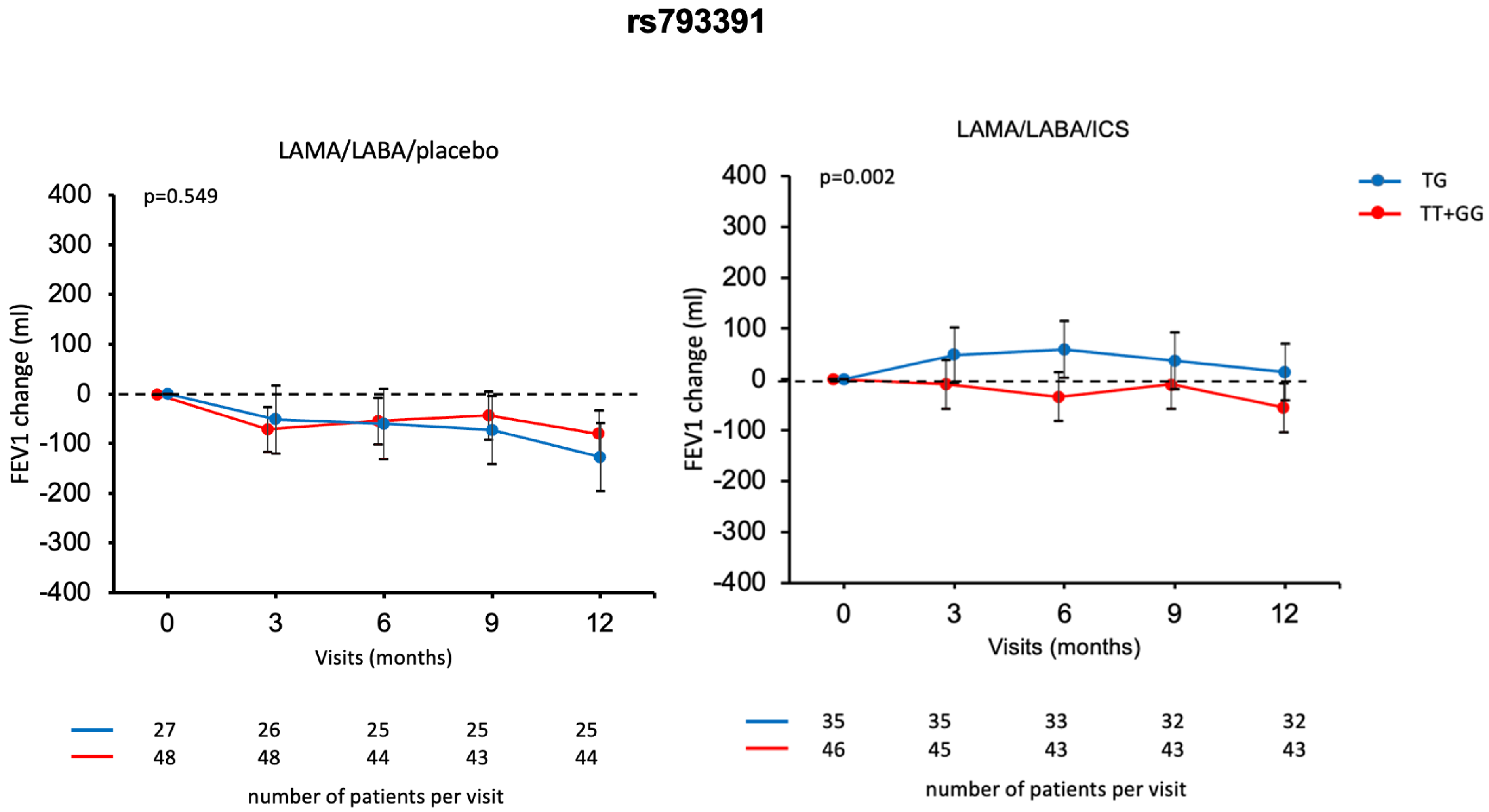
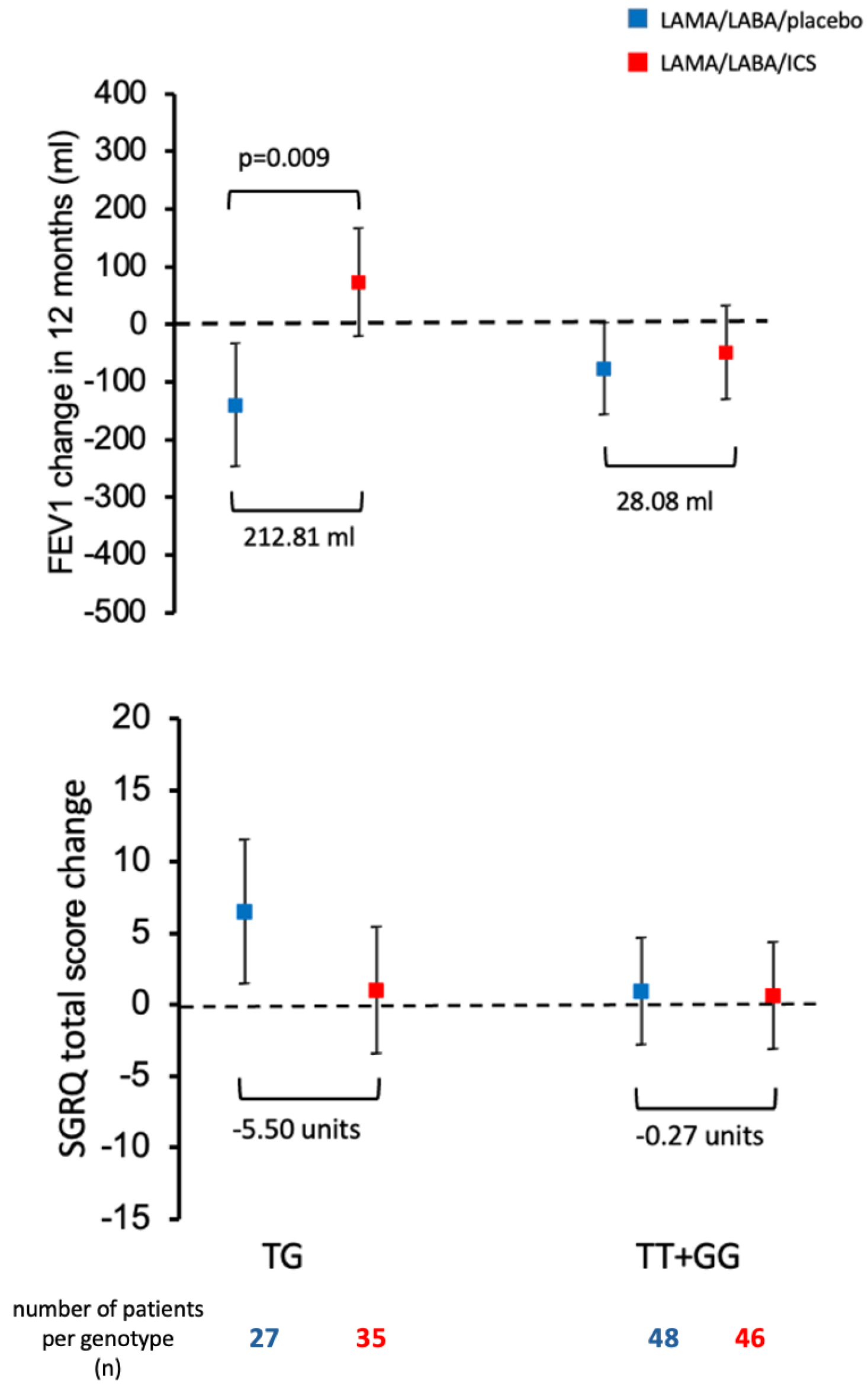
2.3. Association of SUMF1 Polymorphisms with Response to Treatment with ICS
3. Discussion
4. Materials and Methods
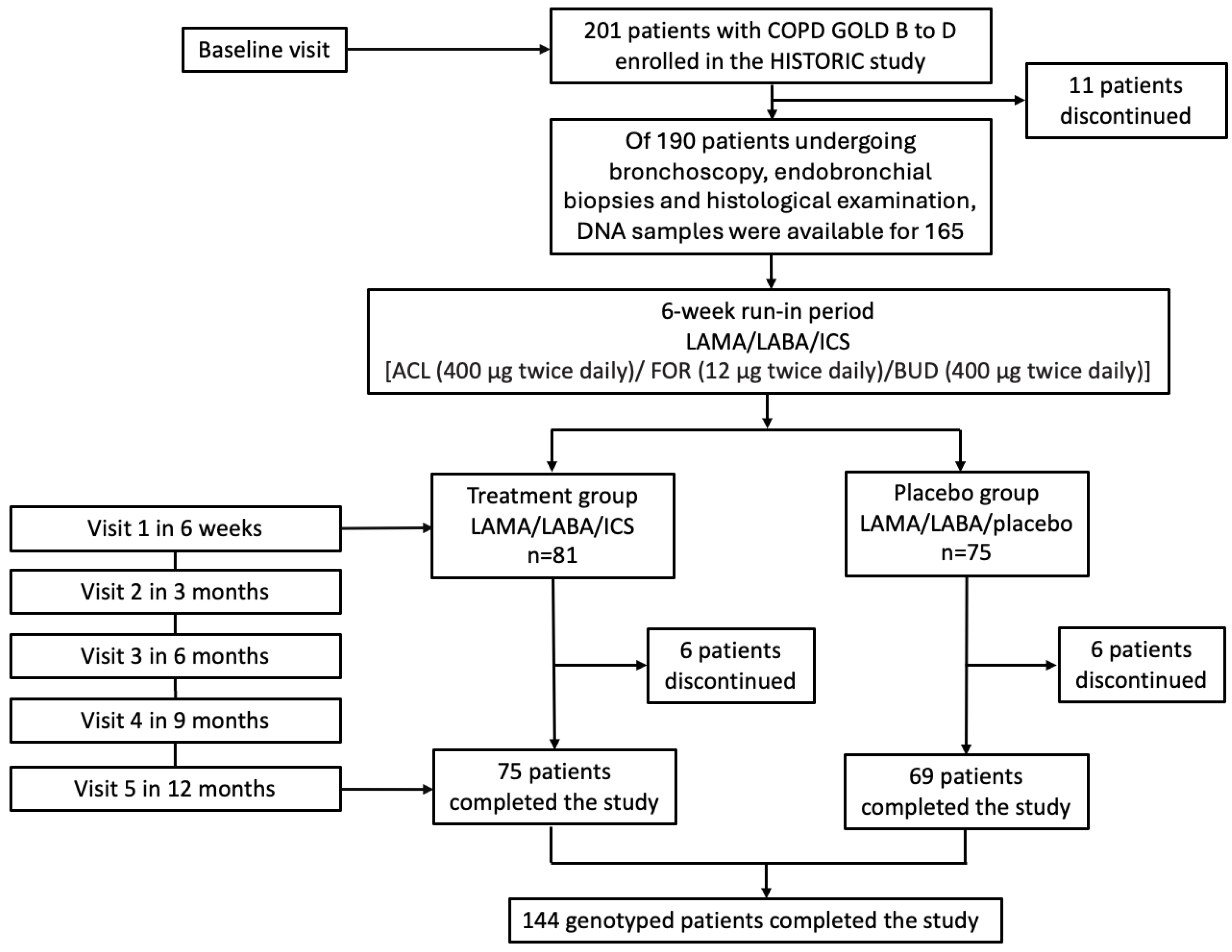
4.1. Study Population
4.2. Sample Size and Power Justification
4.3. DNA Extraction and Genotyping
4.4. Linkage Disequilibrium Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACL | Aclidinium |
| AECOPD | Acute Exacerbations of COPD |
| ASMC | Airway Smooth Muscle Cells |
| BAL | Bronchoalveolar Lavage |
| BUD | Budesonide |
| CAT | COPD Assessment Test |
| COPD | Chronic Obstructive Pulmonary Disease |
| CT | Computed Tomography |
| DLCO | Diffusing Capacity for Carbon Monoxide |
| ECM | Extracellular Matrix |
| FEV1 | Forced Expiratory Volume in 1 Second |
| FOR | Formoterol |
| GAGs | Glycosaminoglycans |
| GOLD | Global Initiative for Chronic Obstructive Lung Disease |
| GWAS | Genome-Wide Association Study |
| HWE | Hardy–Weinberg Equilibrium |
| ICS | Inhaled Corticosteroids |
| ISRCTN | International Standard Randomized Controlled Trial Number |
| LABA | Long-Acting Beta-2 Agonist |
| LAMA | Long-Acting Muscarinic Antagonist |
| LD | Linkage Disequilibrium Analysis |
| MAF | Minor Allele Frequency |
| MCID | Minimal Clinically Important Difference |
| MMRC | Modified Medical Research Council (Dyspnea Scale) |
| NCBI | National Center for Biotechnology Information |
| NO | Nitric Oxide |
| QoL | Quality of Life |
| RCT | Randomized Controlled Trial |
| RNA | Ribonucleic Acid |
| RV | Residual Volume |
| SABA | Short-Acting Beta-2 Agonist |
| SAMA | Short-Acting Muscarinic Antagonist |
| SAS® | Statistical Analysis System |
| SGRQ | St. George’s Respiratory Questionnaire |
| SF-36 | Short Form-36 (General Health Questionnaire) |
| SNP | Single Nucleotide Polymorphism |
| SUMF1 | Sulfatase Modifying Factor 1 |
| TLC | Total Lung Capacity |
References
- Pillai, S.G.; Ge, D.; Zhu, G.; Kong, X.; Shianna, K.V.; Need, A.C.; Feng, S.; Hersh, C.P.; Bakke, P.; Gulsvik, A.; et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): Identification of two major susceptibility loci. PLoS Genet. 2009, 5, e1000421. [Google Scholar] [CrossRef]
- Cho, M.H.; McDonald, M.L.; Zhou, X.; Mattheisen, M.; Castaldi, P.J.; Hersh, C.P.; Demeo, D.L.; Sylvia, J.S.; Ziniti, J.; Laird, N.M.; et al. Risk loci for chronic obstructive pulmonary disease: A genome-wide association study and meta-analysis. Lancet Respir. Med. 2014, 2, 214–225. [Google Scholar] [CrossRef]
- Cho, M.H.; Castaldi, P.J.; Hersh, C.P.; Hobbs, B.D.; Barr, R.G.; Tal Singer, R.; Bakke, P.; Gulsvik, A.; San Jose Estepar, R.; Van Beek, E.J.; et al. A Genome-Wide Association Study of Emphysema and Airway Quantitative Imaging Phenotypes. Am. J. Respir. Crit. Care Med. 2015, 192, 559–569. [Google Scholar] [CrossRef]
- Barnes, P.J. Inhaled corticosteroids are not beneficial in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2000, 161, 342–344. [Google Scholar]
- Yang, I.A.; Ferry, O.R.; Clarke, M.S.; Sim, E.H.; Fong, K.M. Inhaled corticosteroids versus placebo for stable chronic obstructive pulmonary disease. Cochrane Database Syst. Rev. 2023, 3, CD002991. [Google Scholar] [CrossRef]
- Anzueto, A.R.; Kostikas, K.; Mezzi, K.; Shen, S.; Larbig, M.; Patalano, F.; Fogel, R.; Banerji, D.; Wedzicha, J.A. Indacaterol/glycopyrronium versus salmeterol/fluticasone in the prevention of clinically important deterioration in COPD: Results from the FLAME study. Respir. Res. 2018, 19, 121. [Google Scholar] [CrossRef]
- Bhatt, S.P.; Anderson, J.A.; Brook, R.D.; Calverley, P.M.A.; Celli, B.R.; Cowans, N.J.; Crim, C.; Martinez, F.J.; Newby, D.E.; Vestbo, J.; et al. Cigarette smoking and response to inhaled corticosteroids in COPD. Eur. Respir. J. 2018, 51, 1701393. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J.; Adcock, I.M. Glucocorticoid resistance in inflammatory diseases. Lancet 2009, 342, 1905–1917. [Google Scholar] [CrossRef] [PubMed]
- Vestbo, J.; Anderson, J.A.; Brook, R.D.; Calverley, P.M.; Celli, B.R.; Crim, C.; Martinez, F.; Yates, J.; Newby, D.E.; SUMMIT Investigators. Fluticasone furoate and vilanterol and survival in chronic obstructive pulmonary disease with heightened cardiovascular risk (SUMMIT): A double-blind randomised controlled trial. Lancet 2016, 387, 1817–1826. [Google Scholar] [CrossRef] [PubMed]
- Miravitlles, M.; Auladell-Rispau, A.; Monteagudo, M.; Vázquez-Niebla, J.C.; Mohammed, J.; Nuñez, A.; Urrútia, G. Systematic review on long-term adverse effects of inhaled corticosteroids in the treatment of COPD. Eur. Respir. Rev. 2021, 30, 210075. [Google Scholar] [CrossRef]
- Cosma, M.P.; Pepe, S.; Annunziata, I.; Newbold, R.F.; Grompe, M.; Parenti, G.; Ballabio, A. The Multiple Sulfatase Deficiency Gene Encodes an Essential and Limiting Factor for the Activity of Sulfatases. Cell 2003, 113, 445–456. [Google Scholar] [CrossRef]
- Dierks, T.; Schmidt, B.; Borissenko, L.V.; Peng, J.; Preusser, A.; Mariappan, M.; von Figura, K. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C(alpha)-formylglycine generating enzyme. Cell 2003, 113, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Fraldi, A.; Biffi, A.; Lombardi, A.; Visigalli, I.; Pepe, S.; Settembre, C.; Nusco, E.; Auricchio, A.; Naldini, L.; Ballabio, A.; et al. SUMF1 enhances sulfatase activities in vivo in five sulfatase deficiencies. Biochem. J. 2007, 403, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Ntenti, C.; Papakonstantinou, E.; Fidani, L.; Stolz, D.; Goulas, A. The Genetics behind Sulfation: Impact on Airway Remodeling. J. Pers. Med. 2024, 14, 248. [Google Scholar] [CrossRef]
- Jarenbäck, L.; Frantz, S.; Weidner, J.; Ankerst, J.; Nihlén, U.; Bjermer, L.; Wollmer, P.; Tufvesson, E. Single-nucleotide polymorphisms in the sulfatase-modifying factor 1 gene are associated with lung function and COPD. ERJ Open Res. 2022, 8, 00668–02021. [Google Scholar] [CrossRef]
- Weidner, J.; Jarenbäck, L.; de Jong, K.; Vonk, J.M.; van den Berge, M.; Brandsma, C.A.; Boezen, H.M.; Sin, D.; Bossé, Y.; Nickle, D.; et al. Sulfatase modifying factor 1 (SUMF1) is associated with Chronic Obstructive Pulmonary Disease. Respir. Res. 2017, 18, 77. [Google Scholar] [CrossRef]
- Donohue, J.F. Minimal clinically important differences in COPD lung function. COPD 2005, 2, 111–124. [Google Scholar] [CrossRef]
- Lei, Y.; Gao, Y.; Chen, J.; Li, M.; Wu, X.; Ning, Q.; Zhao, J.; Xiong, W.; Xu, Y.; Xie, J. GLCCI1 rs37973: A potential genetic predictor of therapeutic response to inhaled corticosteroids in Chinese chronic obstructive pulmonary disease patients. Sci. Rep. 2017, 7, 42552. [Google Scholar] [CrossRef]
- Russo, P.; Tomino, C.; Santoro, A.; Prinzi, G.; Proietti, S.; Kisialiou, A.; Cardaci, V.; Fini, M.; Magnani, M.; Collacchi, F.; et al. FKBP5 rs4713916: A Potential Genetic Predictor of Interindividual Different Response to Inhaled Corticosteroids in Patients with Chronic Obstructive Pulmonary Disease in a Real-Life Setting. Int. J. Mol. Sci. 2019, 20, 2024. [Google Scholar] [CrossRef]
- Ntenti, C.; Misirlis, T.N.; Goulas, A. Pharmacogenetic Factors Shaping Treatment Outcomes in Chronic Obstructive Pulmonary Disease. Genes 2025, 16, 314. [Google Scholar] [CrossRef]
- Stolz, D.; Papakonstantinou, E.; Pascarella, M.; Jahn, K.; Siebeneichler, A.; Darie, A.M.; Herrmann, M.J.; Strobel, W.; Salina, A.; Grize, L.; et al. Airway smooth muscle area to predict steroid responsiveness in COPD patients receiving triple therapy (HISTORIC): A randomised, placebo-controlled, double-blind, investigator-initiated trial. Eur. Respir. J. 2023, 62, 2300218. [Google Scholar] [CrossRef]
- ISRCTN. International Standard Randomised Controlled Trial Number (ISRCTN11017699); BioMed Central: London, UK, 2016. Available online: https://www.isrctn.com/ISRCTN11017699 (accessed on 14 October 2025).

| Sex | |
|---|---|
| Male | 114 (69.1) |
| Female | 51 (30.9) |
| Age in years | 67 (59–72) |
| Ethnic origin | |
| White | 163 (100) |
| Smoking status | |
| Former smokers | 103 (62.4) |
| Current smokers | 62 (37.6) |
| Pack Years | 50 (37–70) |
| Disease duration (months) | 60 (23–120) |
| Disease characteristics at baseline | |
| Post-bronchodilator FEV1 (liters) | 1.59 (1.2–2.0) |
| Post-bronchodilator FEV1 (% of predicted value) | 59 (49–67) |
| Post-bronchodilator DLCO_SB (% of predicted value) | 61.5 (48.4–78.2) |
| Post-bronchodilator TLC (% of predicted value) | 109 (98–122) |
| Post-bronchodilator RV (% of predicted value) | 136 (111–162) |
| Post-bronchodilator ratio RV/TLC (% of predicted value) | 116 (104–133) |
| Fractional exhaled NO (ppb) | 15 (8–23) |
| 6 min walking distance (meters) | 460 (390–524) |
| MMRC dyspnea scale | 1 (0–2) |
| CAT score, total sum | 14 (9–19) |
| SGRQ, total score | 32.8 (20.0–48.7) |
| ASMC (%) * | 17.4 (12.4–22.2) |
| SNP | Chromosome Position | Genotype Frequencies n (%) | Minor Allele | MAF % | HWE (p) |
|---|---|---|---|---|---|
| rs11915920 C > T | chr3:4368850 | CC = 33 (20) CT = 83 (50.3) TT = 49 (29.7) | C | 45.2 | 0.84 |
| Intron Variant | |||||
| rs793391 T > G | chr3:4427508 | TT = 91 (55.2) TG = 65 (39.4) GG = 9 (5.5) | G | 25.1 | 0.55 |
| Intron Variant |
| SNP | Genotype | Treatment Group (n) | Mean Change Score (Min–Max) in 12 Months | Difference Between Treatment Groups (Units) | p-Value |
|---|---|---|---|---|---|
| rs793391 | TT | LAMA/LABA/placebo (40) | −2.05 | 1.50 | 0.66 |
| LAMA/LABA/ICS (41) | −0.55 | ||||
| TG | LAMA/LABA/placebo (27) | 0.70 | −2.68 | 0.17 | |
| LAMA/LABA/ICS (35) | −1.97 | ||||
| GG | LAMA/LABA/placebo (4) | 3.75 | −0.35 | 0.92 | |
| LAMA/LABA/ICS (5) | 3.40 | ||||
| rs11915920 | CC | LAMA/LABA/placebo (15) | 1.40 | −1.15 | 0.89 |
| LAMA/LABA/ICS (16) | 0.25 | ||||
| CT | LAMA/LABA/placebo (35) | −1.03 | 0.27 | 0.76 | |
| LAMA/LABA/ICS (44) | −0.76 | ||||
| TT | LAMA/LABA/placebo (25) | −1.64 | −0.50 | 0.58 | |
| LAMA/LABA/ICS (21) | −2.14 |
| SNP | Genotype | Treatment Group (n) | Mean Change Score (Min–Max) in 12 Months | Difference Between Treatment Groups (Units) | p-Value |
|---|---|---|---|---|---|
| rs793391 | TT | LAMA/LABA/placebo (40) | −6.38 (−40–30) | 2.94 | 0.75 |
| LAMA/LABA/ICS (41) | −3.44 (−95–27.5) | ||||
| TG | LAMA/LABA/placebo (27) | −11.40 (−45–30) | 8.12 | 0.39 | |
| LAMA/LABA/ICS (35) | −3.28 (−40–30) | ||||
| GG | LAMA/LABA/placebo (4) | 5.00 (−5–20) | −24.00 | 0.018 | |
| LAMA/LABA/ICS (5) | −19.00 (−40–5) | ||||
| rs11915920 | CC | LAMA/LABA/placebo (15) | −5.67 (−45–30) | 9.00 | 0.34 |
| LAMA/LABA/ICS (16) | 3.33 (−35–30) | ||||
| CT | LAMA/LABA/placebo (35) | −5.78 (−40–30) | −0.85 | 0.93 | |
| LAMA/LABA/ICS (44) | −6.63 (−95–25) | ||||
| TT | LAMA/LABA/placebo (25) | −11.36 (−40–20) | 5.97 | 0.52 | |
| LAMA/LABA/ICS (21) | −5.40 (−40–30) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ntenti, C.; Papakonstantinou, E.; Grize, L.; Pascarella, M.; Frye, B.C.; Fähndrich, S.; Ioannidou, D.; Savic Prince, S.; Goulas, A.; Stolz, D. SUMF1 Common Variant rs793391 Is Associated with Response to Inhaled Corticosteroids in Patients with COPD. Int. J. Mol. Sci. 2025, 26, 10225. https://doi.org/10.3390/ijms262010225
Ntenti C, Papakonstantinou E, Grize L, Pascarella M, Frye BC, Fähndrich S, Ioannidou D, Savic Prince S, Goulas A, Stolz D. SUMF1 Common Variant rs793391 Is Associated with Response to Inhaled Corticosteroids in Patients with COPD. International Journal of Molecular Sciences. 2025; 26(20):10225. https://doi.org/10.3390/ijms262010225
Chicago/Turabian StyleNtenti, Charikleia, Eleni Papakonstantinou, Leticia Grize, Maria Pascarella, Björn C. Frye, Sebastian Fähndrich, Despoina Ioannidou, Spasenija Savic Prince, Antonis Goulas, and Daiana Stolz. 2025. "SUMF1 Common Variant rs793391 Is Associated with Response to Inhaled Corticosteroids in Patients with COPD" International Journal of Molecular Sciences 26, no. 20: 10225. https://doi.org/10.3390/ijms262010225
APA StyleNtenti, C., Papakonstantinou, E., Grize, L., Pascarella, M., Frye, B. C., Fähndrich, S., Ioannidou, D., Savic Prince, S., Goulas, A., & Stolz, D. (2025). SUMF1 Common Variant rs793391 Is Associated with Response to Inhaled Corticosteroids in Patients with COPD. International Journal of Molecular Sciences, 26(20), 10225. https://doi.org/10.3390/ijms262010225






