Does Genetic Variation in Detoxification Capacity Influence Hepatic Biomarker Responses to a Liver Support Supplementation Regimen?
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
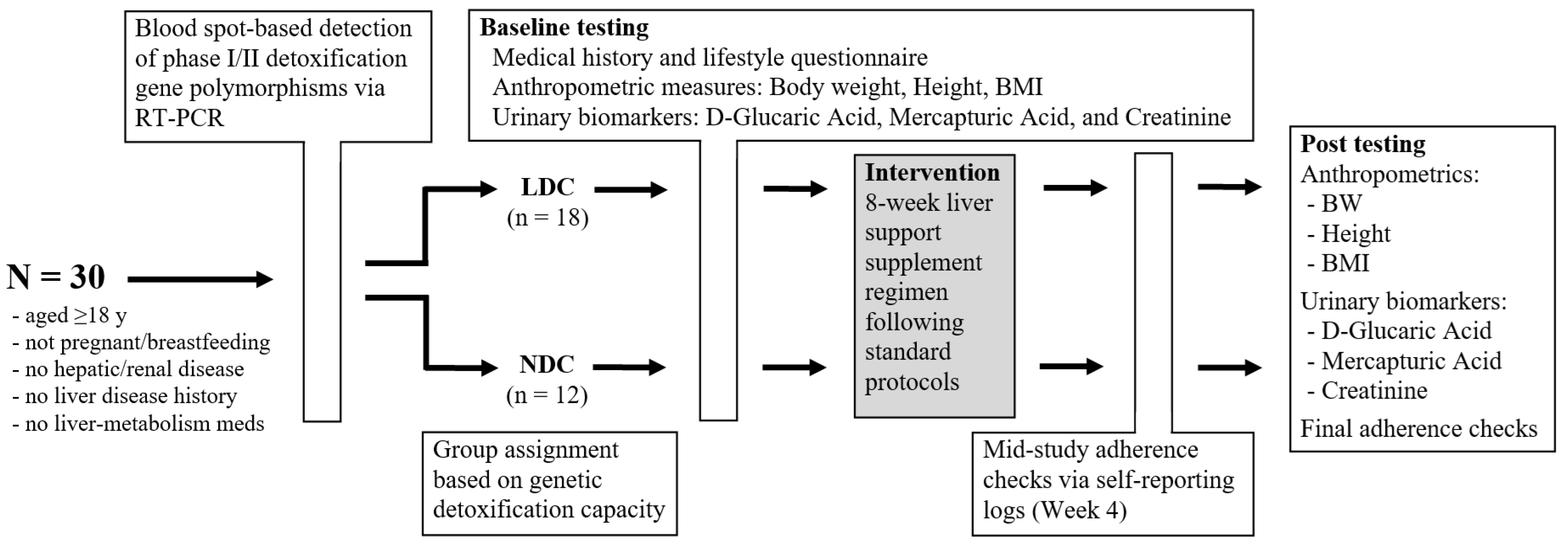
4.1. Study Design and Participants
4.2. Inclusion and Exclusion Criteria
4.3. Procedures and Group Allocation
4.4. Intervention and Protocol Compliance
4.5. Biospecimen Collection, Processing, and Laboratory Analysis
4.6. Outcome Measures
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAMA | Acrylamide Mercapturic Acid |
| BMI | Body Mass Index |
| CI | Confidence Interval |
| CV | Coefficient of Variation |
| CYP | Cytochrome P450 |
| CYP1A1 | Cytochrome P450 Family 1 Subfamily A Member 1 |
| DAO | Diamine Oxidase |
| DHGS | Deutsche Hochschule für Gesundheit & Sport |
| DIM | Diindolylmethane |
| DNA | Deoxyribonucleic Acid |
| DF | Degrees of Freedom |
| GAMA | Glycidamide Mercapturic Acid |
| GSH | Glutathione |
| GST | Glutathione S-transferase |
| GSTM1 | Glutathione S-transferase Mu 1 |
| GSTP1 | Glutathione S-transferase Pi 1 |
| GSTT1 | Glutathione S-transferase Theta 1 |
| HPLC-UV | High-Performance Liquid Chromatography with Ultraviolet Detection |
| HNMT | Histamine N-Methyltransferase |
| LC-MS/MS | Liquid Chromatography–Tandem Mass Spectrometry |
| LDC | Limited Detox Capacity |
| NAC | N-Acetylcysteine |
| NDC | Normal Detox Capacity |
| NQO1 | NAD(P)H Quinone Dehydrogenase 1 |
| OR | Odds Ratio |
| PCR | Polymerase Chain Reaction |
| PMA | Panaceo Micro Activation |
| R2 | Coefficient of Determination |
| RCT | Randomized Controlled Trial |
| RT-PCR | Real-Time Polymerase Chain Reaction |
| SBMA | Sulfobetaine Methacrylate |
| S-PMA | S-Phenylmercapturic Acid |
| SD | Standard Deviation |
| SE | Standard Error |
| UGT | UDP-Glucuronosyltransferase |
| UGT1A | UDP-Glucuronosyltransferase Family 1 Member A |
| UGT2B | UDP-Glucuronosyltransferase Family 2 Member B |
| UGT2B17 | UDP-Glucuronosyltransferase Family 2 Member B17 |
| UDP | Uridine Diphosphate |
References
- Grant, D.M. Detoxification pathways in the liver. J. Inherit. Metab. Dis. 1991, 14, 421–430. [Google Scholar] [CrossRef]
- Kalra, A.; Yetiskul, E.; Wehrle, C.J.; Tuma, F. Physiology, Liver. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK535438/ (accessed on 25 September 2025).
- Xu, C.; Li, C.Y.; Kong, A.N. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 2005, 28, 249–268. [Google Scholar] [CrossRef]
- Abdelmonem, H.B.; Abdelaal, N.M.; Anwer, E.K.E.; Rashwan, A.A.; Hussein, M.A.; Ahmed, Y.F.; Khashana, R.; Hanna, M.M.; Abdelnaser, A. Decoding the Role of CYP450 Enzymes in Metabolism and Disease: A Comprehensive Review. Biomedicines 2024, 12, 1467. [Google Scholar] [CrossRef]
- Sargis, R.M.; Heindel, J.J.; Padmanabhan, V. Interventions to Address Environmental Metabolism-Disrupting Chemicals: Changing the Narrative to Empower Action to Restore Metabolic Health. Front. Endocrinol. 2019, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Panda, C.; Komarnytsky, S.; Fleming, M.N.; Marsh, C.; Barron, K.; Le Brun-Blashka, S.; Metzger, B. Guided Metabolic Detoxification Program Supports Phase II Detoxification Enzymes and Antioxidant Balance in Healthy Participants. Nutrients 2023, 15, 2209. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.J.G.; Serrano, M.A.; Monte, M.J.; Sanchez-Martin, A.; Temprano, A.G.; Briz, O.; Romero, M.R. Role of Genetic Variations in the Hepatic Handling of Drugs. Int. J. Mol. Sci. 2020, 21, 2884. [Google Scholar] [CrossRef] [PubMed]
- Prysyazhnyuk, V.; Sydorchuk, L.; Sydorchuk, R.; Prysiazhniuk, I.; Bobkovych, K.; Buzdugan, I.; Dzuryak, V.; Prysyazhnyuk, P. Glutathione-S-transferases genes-promising predictors of hepatic dysfunction. World J. Hepatol. 2021, 13, 620–633. [Google Scholar] [CrossRef]
- Ross, D.; Siegel, D. The diverse functionality of NQO1 and its roles in redox control. Redox Biol. 2021, 41, 101950. [Google Scholar] [CrossRef]
- Włodarczyk, M.; Nowicka, G. Common polymorphisms in CYP1A1, GSTM1, GSTT1, GSTP1 and XPD genes and endogenous DNA damage. Mol. Biol. Rep. 2012, 39, 5699–5704. [Google Scholar] [CrossRef]
- Nakanishi, G.; Pita-Oliveira, M.; Bertagnolli, L.S.; Torres-Loureiro, S.; Scudeler, M.M.; Cirino, H.S.; Chaves, M.L.; Miwa, B.; Rodrigues-Soares, F. Worldwide Systematic Review of GSTM1 and GSTT1 Null Genotypes by Continent, Ethnicity, and Therapeutic Area. OMICS 2022, 26, 528–541. [Google Scholar] [CrossRef]
- Aronica, L.; Ordovas, J.M.; Volkov, A.; Lamb, J.J.; Stone, P.M.; Minich, D.; Leary, M.; Class, M.; Metti, D.; Larson, I.A.; et al. Genetic Biomarkers of Metabolic Detoxification for Personalized Lifestyle Medicine. Nutrients 2022, 14, 768. [Google Scholar] [CrossRef]
- Thakur, S.; Kumar, V.; Das, R.; Sharma, V.; Mehta, D.K. Biomarkers of Hepatic Toxicity: An Overview. Curr. Ther. Res. Clin. Exp. 2024, 100, 100737. [Google Scholar] [CrossRef]
- Ayyadurai, V.A.S.; Deonikar, P.; Fields, C. Mechanistic Understanding of D-Glucaric Acid to Support Liver Detoxification Essential to Muscle Health Using a Computational Systems Biology Approach. Nutrients 2023, 15, 733. [Google Scholar] [CrossRef]
- Mathias, P.I.; B’hymer, C. Mercapturic acids: Recent advances in their determination by liquid chromatography/mass spectrometry and their use in toxicant metabolism studies and in occupational and environmental exposure studies. Biomarkers 2016, 21, 293–315. [Google Scholar] [CrossRef]
- Pluym, N.; Gilch, G.; Scherer, G.; Scherer, M. Analysis of 18 urinary mercapturic acids by two high-throughput multiplex-LC-MS/MS methods. Anal. Bioanal. Chem. 2015, 407, 5463–5476. [Google Scholar] [CrossRef] [PubMed]
- Murray, K.J.; Mckeon, D.; Lecchi, C.; Maertens, L.; Villalta, P.W.; Balbo, S. Positive Ion Tandem Mass Spectrometry Offers Enhanced Structural Insights for the Discovery of Mercapturic Acids. Chem. Res. Toxicol. 2025, 38, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Diaper, A.M.; Law, F.D.; Melichar, J.K. Pharmacological strategies for detoxification. Br. J. Clin. Pharmacol. 2014, 77, 302–314. [Google Scholar] [CrossRef]
- Hodges, R.E.; Minich, D.M. Modulation of Metabolic Detoxification Pathways Using Foods and Food-Derived Components: A Scientific Review with Clinical Application. J. Nutr. Metab. 2015, 2015, 760689. [Google Scholar] [CrossRef] [PubMed]
- Hanje, A.J.; Fortune, B.; Song, M.; Hill, D.; McClain, C. The use of selected nutrition supplements and complementary and alternative medicine in liver disease. Nutr. Clin. Pract. 2006, 21, 255–272. [Google Scholar] [CrossRef]
- Mokhtari, V.; Afsharian, P.; Shahhoseini, M.; Kalantar, S.M.; Moini, A. A Review on Various Uses of N-Acetyl Cysteine. Cell J. 2017, 19, 11–17. [Google Scholar] [CrossRef]
- Jozefczak, M.; Remans, T.; Vangronsveld, J.; Cuypers, A. Glutathione is a key player in metal-induced oxidative stress defenses. Int. J. Mol. Sci. 2012, 13, 3145–3175. [Google Scholar] [CrossRef]
- Superti, F.; Russo, R. Alpha-Lipoic Acid: Biological Mechanisms and Health Benefits. Antioxidants 2024, 13, 1228. [Google Scholar] [CrossRef]
- Nho, C.W.; Jeffery, E. The synergistic upregulation of phase II detoxification enzymes by glucosinolate breakdown products in cruciferous vegetables. Toxicol. Appl. Pharmacol. 2001, 174, 146–152. [Google Scholar] [CrossRef] [PubMed]
- van Vugt-Lussenburg, B.M.A.; Capinha, L.; Reinen, J.; Rooseboom, M.; Kranendonk, M.; Onderwater, R.C.; Jennings, P. “Commandeuring” Xenobiotic Metabolism: Advances in Understanding Xenobiotic Metabolism. Chem. Res. Toxicol. 2022, 35, 1184–1201. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.J.L.; Hanigan, M.H. Metabolism of Glutathione S-Conjugates: Multiple Pathways. In Comprehensive Toxicology; Elsevier: Oxford, UK, 2018; pp. 363–406. [Google Scholar] [CrossRef]
- Rosting, C.; Olsen, R. Biomonitoring of the benzene metabolite S-phenylmercapturic acid and the toluene metabolite S-benzylmercapturic acid in urine from firefighters. Toxicol. Lett. 2020, 329, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Carbonari, D.; Proietto, A.; Fioretti, M.; Tranfo, G.; Paci, E.; Papacchini, M.; Mansi, A. Influence of genetic polymorphism on t,t-MA/S-PMA ratio in 301 benzene exposed subjects. Toxicol. Lett. 2014, 231, 205–212. [Google Scholar] [CrossRef]
- He, Y.; Qi, J.; He, F.; Zhang, Y.; Wang, Y.; Zhang, R.; Li, G. GSTM1 and GSTT1 Genes are Associated with DNA Damage of p53 Gene in Coke-oven Workers. J. Occup. Environ. Med. 2017, 59, 499–501. [Google Scholar] [CrossRef]
- Lodovici, M.; Luceri, C.; Guglielmi, F.; Bacci, C.; Akpan, V.; Fonnesu, M.L.; Boddi, V.; Dolara, P. Benzo(a)pyrene diolepoxide (BPDE)-DNA adduct levels in leukocytes of smokers in relation to polymorphism of CYP1A1, GSTM1, GSTP1, GSTT1, and mEH. Cancer Epidemiol. Biomark. Prev. 2004, 13, 1342–1348. [Google Scholar] [CrossRef]
- Wang, P.; Jia, Y.; Wu, R.; Chen, Z.; Yan, R. Human gut bacterial β-glucuronidase inhibition: An emerging approach to manage medication therapy. Biochem. Pharmacol. 2021, 190, 114566. [Google Scholar] [CrossRef]
- Gallagher, C.J.; Balliet, R.M.; Sun, D.; Chen, G.; Lazarus, P. Sex differences in UDP-glucuronosyltransferase 2B17 expression and activity. Drug Metab. Dispos. 2010, 38, 2204–2209. [Google Scholar] [CrossRef]
- Nakamura, A.; Nakajima, M.; Yamanaka, H.; Fujiwara, R.; Yokoi, T. Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines. Drug Metab. Dispos. 2008, 36, 1461–1464. [Google Scholar] [CrossRef]
- Meech, R.; Hu, D.G.; Hulin, J.A.; Mackenzie, P.I. Sex-specific UGT expression and function: Prevalence, potential mechanisms and significance. Expert Opin. Drug Metab. Toxicol. 2025, 21, 511–518. [Google Scholar] [CrossRef]
- Gonçalves-Dias, C.; Morello, J.; Semedo, V.; Correia, M.J.; Coelho, N.R.; Monteiro, E.C.; Antunes, A.M.M.; Pereira, S.A. The mercapturomic profile of health and non-communicable diseases. High Throughput 2019, 8, 10. [Google Scholar] [CrossRef]
- Laborde, E. Glutathione transferases as mediators of signaling pathways involved in cell proliferation and cell death. Cell Death Differ. 2010, 17, 1373–1380. [Google Scholar] [CrossRef]
- Nugrahaningsih, D.A.A.; Wihadmadyatami, H.; Widyarini, S.; Wijayaningsih, R.A. A Review of the GSTM1 Null Genotype Modifies the Association between Air Pollutant Exposure and Health Problems. Int. J. Genom. 2023, 2023, 4961487. [Google Scholar] [CrossRef]
- Simpson, J.B.; Walker, M.E.; Sekela, J.J.; Ivey, S.M.; Jariwala, P.B.; Storch, C.M.; Kowalewski, M.E.; Graboski, A.L.; Lietzan, A.D.; Walton, W.G.; et al. Gut microbial β-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics. Cell Host Microbe 2024, 32, 925–944.e10. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Ding, Q.; Zhang, W.; Kang, M.; Ma, J.; Zhao, L. Gut microbial beta-glucuronidase: A vital regulator in female estrogen metabolism. Gut Microbes 2023, 15, 2236749. [Google Scholar] [CrossRef] [PubMed]
- Walaszek, Z.; Szemraj, J.; Narog, M.; Adams, A.K.; Kilgore, J.; Sherman, U.; Hanausek, M. Metabolism, uptake, and excretion of a D-glucaric acid salt and its potential use in cancer prevention. Cancer Detect. Prev. 1997, 21, 178–190. [Google Scholar]
- Zoltaszek, R.; Kowalczyk, P.; Kowalczyk, M.C.; Hanausek, M.; Kilianska, Z.M.; Slaga, T.J.; Walaszek, Z. Dietary D-glucarate effects on the biomarkers of inflammation during early post-initiation stages of benzo[a]pyrene-induced lung tumorigenesis in A/J mice. Oncol. Lett. 2011, 2, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Saluk-Juszczak, J. A comparative study of antioxidative activity of calcium-D-glucarate, sodium-D-gluconate and D-glucono-1,4-lactone in a human blood platelet model. Platelets 2010, 21, 632–640. [Google Scholar] [CrossRef]
- Lizzo, G.; Migliavacca, E.; Lamers, D.; Frézal, A.; Corthesy, J.; Vinyes-Parès, G.; Bosco, N.; Karagounis, L.G.; Hövelmann, U.; Heise, T.; et al. A Randomized Controlled Clinical Trial in Healthy Older Adults to Determine Efficacy of Glycine and N-Acetylcysteine Supplementation on Glutathione Redox Status and Oxidative Damage. Front. Aging 2022, 3, 852569. [Google Scholar] [CrossRef]
- Schmitt, B.; Vicenzi, M.; Garrel, C.; Denis, F.M. Effects of N-acetylcysteine, oral glutathione (GSH) and a novel sublingual form of GSH on oxidative stress markers: A comparative crossover study. Redox Biol. 2015, 6, 198–205. [Google Scholar] [CrossRef]
- Goutzourelas, N.; Orfanou, M.; Charizanis, I.; Leon, G.; Spandidos, D.A.; Kouretas, D. GSH levels affect weight loss in individuals with metabolic syndrome and obesity following dietary therapy. Exp. Ther. Med. 2018, 16, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Abulmeaty, M.M.A.; Ghneim, H.K.; Alkhathaami, A.; Alnumair, K.; Al Zaben, M.; Razak, S.; Al-Sheikh, Y.A. Inflammatory Cytokines, Redox Status, and Cardiovascular Diseases Risk after Weight Loss via Bariatric Surgery and Lifestyle Intervention. Medicina 2023, 59, 751. [Google Scholar] [CrossRef]
- Qi, L. Gene-diet interaction and weight loss. Curr. Opin. Lipidol. 2014, 25, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Kosicka-Noworzyń, K.; Romaniuk-Drapała, A.; Sheng, Y.H.; Yohn, C.; Brunetti, L.; Kagan, L. Obesity-related drug-metabolizing enzyme expression alterations in the human liver. Biomed. Pharmacother. 2025, 187, 118155. [Google Scholar] [CrossRef] [PubMed]
- Donepudi, A.C.; Cheng, Q.; Lu, Z.J.; Cherrington, N.J.; Slitt, A.L. Hepatic transporter expression in metabolic syndrome: Phenotype, serum metabolic hormones, and transcription factor expression. Drug Metab. Dispos. 2016, 44, 518–526. [Google Scholar] [CrossRef]
- Almoshabek, H.A.; Mustafa, M.; Al-Asmari, M.M.; Alajmi, T.K.; Al-Asmari, A.K. Association of glutathione S-transferase GSTM1 and GSTT1 deletion polymorphisms with obesity and their relationship with body mass index, lipoprotein and hypertension among young age Saudis. JRSM Cardiovasc. Dis. 2016, 5, 2048004016669645. [Google Scholar] [CrossRef]
- Chen, Y.; Bai, Y.; Yuan, J.; Chen, W.; Sun, J.; Wang, H.; Liang, H.; Guo, L.; Yang, X.; Tan, H.; et al. Association of polymorphisms in AhR, CYP1A1, GSTM1, and GSTT1 genes with levels of DNA damage in peripheral blood lymphocytes among coke-oven workers. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1703–1707. [Google Scholar] [CrossRef]
- Sacco, R.; Eggenhoffner, R.; Giacomelli, L. Glutathione in the treatment of liver diseases: Insights from clinical practice. Minerva Gastroenterol. Dietol. 2016, 62, 316–324. [Google Scholar]
- Sinha, R.; Sinha, I.; Calcagnotto, A.; Trushin, N.; Haley, J.S.; Schell, T.D.; Richie, J.P. Oral supplementation with liposomal glutathione elevates body stores of glutathione and markers of immune function. Eur. J. Clin. Nutr. 2018, 72, 105–111. [Google Scholar] [CrossRef]
- Tenório, M.C.D.S.; Graciliano, N.G.; Moura, F.A.; Oliveira, A.C.M.; Goulart, M.O.F. N-Acetylcysteine (NAC): Impacts on Human Health. Antioxidants 2021, 10, 967. [Google Scholar] [CrossRef]
- Nikbaf-Shandiz, M.; Adeli, S.; Faghfouri, A.H.; Khademi, F.; Jamilian, P.; Zarezadeh, M.; Ebrahimi-Mamaghani, M. The efficacy of N-acetylcysteine in improving liver function: A systematic review and meta-analysis of controlled clinical trials. PharmaNutrition 2023, 24, 100343. [Google Scholar] [CrossRef]
- Salehi, B.; Berkay Yılmaz, Y.; Antika, G.; Boyunegmez Tumer, T.; Fawzi Mahomoodally, M.; Lobine, D.; Akram, M.; Riaz, M.; Capanoglu, E.; Sharopov, F.; et al. Insights on the Use of α-Lipoic Acid for Therapeutic Purposes. Biomolecules 2019, 9, 356. [Google Scholar] [CrossRef]
- Shay, K.P.; Moreau, R.F.; Smith, E.J.; Smith, A.R.; Hagen, T.M. Alpha-lipoic acid as a dietary supplement: Molecular mechanisms and therapeutic potential. Biochim. Biophys. Acta 2009, 1790, 1149–1160. [Google Scholar] [CrossRef]
- Banerjee, S.; Kong, D.; Wang, Z.; Bao, B.; Hillman, G.G.; Sarkar, F.H. Attenuation of multi-targeted proliferation-linked signaling by 3,3’-diindolylmethane (DIM): From bench to clinic. Mutat. Res. 2011, 728, 47–66. [Google Scholar] [CrossRef] [PubMed]
- Vermillion Maier, M.L.; Siddens, L.K.; Uesugi, S.L.; Choi, J.; Leonard, S.; Pennington, J.; Tilton, S.C.; Smith, J.N.; Ho, E.; Chow, S.; et al. 3,3’-Diindolylmethane Exhibits Significant Metabolism after Oral Dosing in Humans. Drug Metab. Dispos. 2021, 49, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Bulog, A.; Pavelic, K.; Šutić, I.; Kraljevic Pavelic, S. PMA-Zeolite: Chemistry and Diverse Medical Applications. J. Funct. Biomater. 2024, 15, 296. [Google Scholar] [CrossRef]
- Auerbach, S.M.; Kathleen, A.; Carrado, K.A.; Dutta, P.K. Handbook of Zeolite Science and Technology; CRC Press: New York, NY, USA, 2003. [Google Scholar] [CrossRef]
- Mancak, M.; Altintas, D.; Balaban, Y.; Caliskan, U.K. Evidence-based herbal treatments in liver diseases. Hepatol. Forum 2024, 5, 50–60. [Google Scholar] [CrossRef]
- Sunday Natural. Liv Ultra Liver Complex. Available online: https://www.sunday.de/en/liv-ultra-liver-complex.html (accessed on 15 October 2025).
- Poon, R.; Villeneuve, D.C.; Chu, I.; Kinach, R. HPLC determination of D-glucaric acid in human urine. J. Anal. Toxicol. 1993, 17, 146–150. [Google Scholar] [CrossRef]
- Laakso, E.I.; Tokola, R.A.; Hirvisalo, E.L. Determination of D-glucaric acid by high-performance liquid chromatography. J. Chromatogr. 1983, 278, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Billard, A.; Laval, V.; Fillinger, S.; Leroux, P.; Lachaise, H.; Beffa, R.; Debieu, D. The allele-specific probe and primer amplification assay, a new real-time PCR method for fine quantification of single-nucleotide polymorphisms in pooled DNA. Appl. Environ. Microbiol. 2012, 78, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Marín, F.; García, N.; Muñoz, X.; Capellà, G.; González, C.A.; Agudo, A.; Sala, N. Simultaneous genotyping of GSTT1 and GSTM1 null polymorphisms by melting curve analysis in presence of SYBR Green I. J. Mol. Diagn. 2010, 12, 300–304. [Google Scholar] [CrossRef] [PubMed]

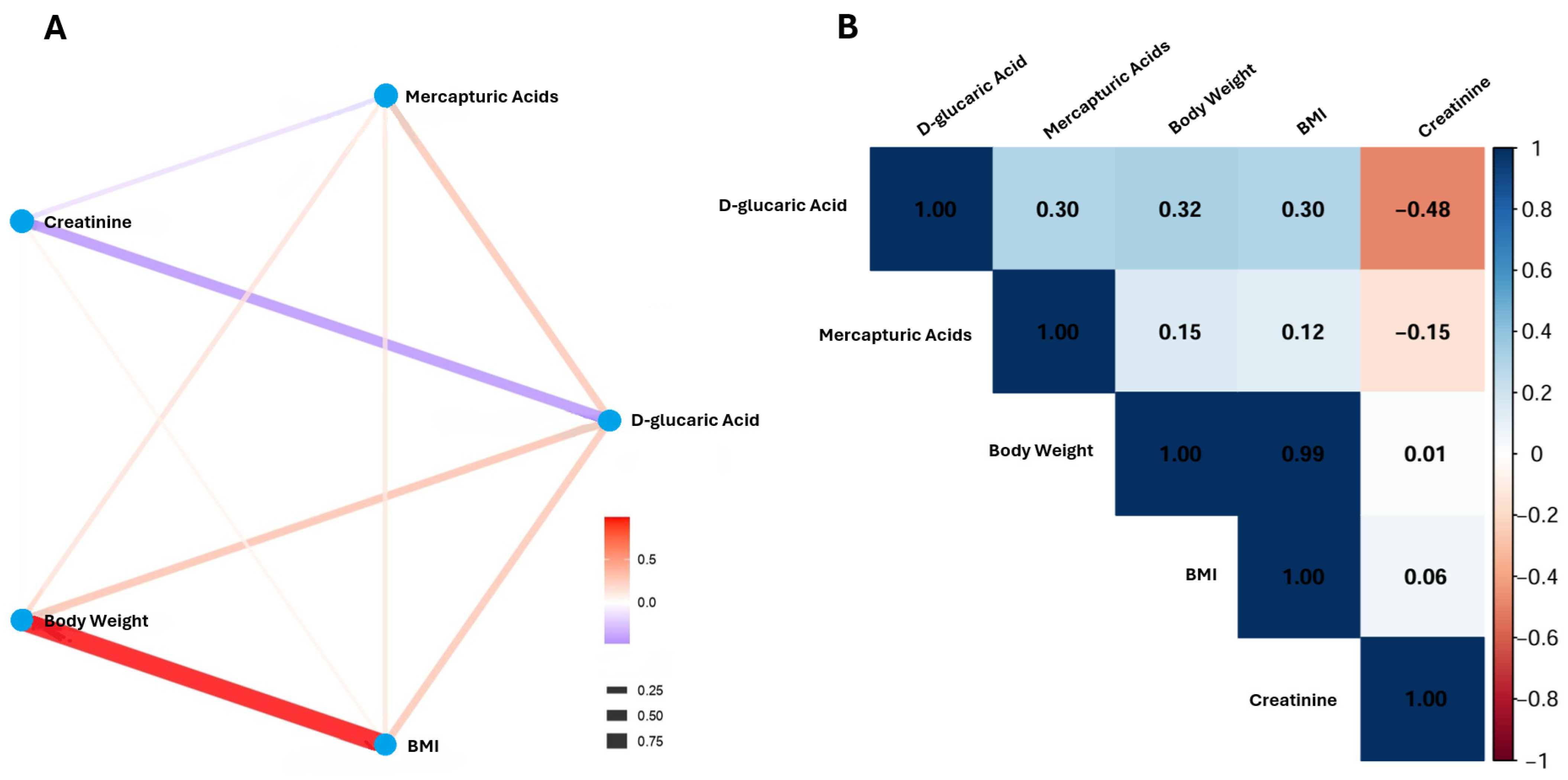
| Total (n = 30) | LDC (n = 18) | NDC (n = 12) | Statistical Difference | ||
|---|---|---|---|---|---|
| Age (years) | 35.77 ± 10.88 | 33.94 ± 9.65 | 38.50 ± 12.44 | p = 0.4 | |
| Sex | Female | 20 (67%) | 13 (72%) | 7 (58%) | p = 0.5 |
| Male | 10 (33%) | 5 (28%) | 5 (42%) | ||
| Body Weight (kg) | 70.91 ± 12.21 | 70.17 ± 12.65 | 72.02 ± 11.96 | p = 0.6 | |
| Height (cm) | 1.71 ± 0.08 | 1.71 ± 0.08 | 1.71 ± 0.07 | p = 0.9 | |
| BMI (kg/m2) | 24.15 ± 3.19 | 23.84 ± 3.05 | 24.61 ± 3.47 | p = 0.7 | |
| D-Glucaric Acid (nmol/mg) | 80.03 ± 50.16 | 78.22 ± 46.69 | 82.75 ± 57.02 | p = 0.9 | |
| D-Glucaric Acid Levels | Low | 4 (13%) | 2 (11%) | 2 (17%) | p = 0.7 |
| Marginally low | 13 (43%) | 9 (50%) | 4 (33%) | ||
| Normal | 13 (43%) | 7 (39%) | 6 (50%) | ||
| Mercapturic Acids (µmol/mmol) | 51.53 ± 15.83 | 50.28 ± 19.49 | 53.42 ± 8.22 | p = 0.4 | |
| Mercapturic Acid Levels | Low | 6 (20%) | 6 (33%) | 0 (0%) | p = 0.8 |
| Marginally low | 5 (17%) | 2 (11%) | 3 (25%) | ||
| Normal | 17 (57%) | 9 (50%) | 8 (67%) | ||
| Marginally high | 1 (3.3%) | 0 (0%) | 1 (8.3%) | ||
| High | 1 (3.3%) | 1 (5.6%) | 0 (0%) | ||
| Creatinine (mg/dL) | 123.73 ± 78.02 | 134.28 ± 88.57 | 107.92 ± 58.92 | p = 0.7 | |
| Estimate (β) | 95% CI | SE | DF | Statistic | p-Value | |
|---|---|---|---|---|---|---|
| Intercept | 66.73 | [61.12, 72.34] | 2.75 | 30.3 | t = 24.27 | <0.001 |
| Time (post vs. pre) | −0.87 | [−1.60, −0.15] | 0.35 | 30 | t = −2.46 | 0.020 |
| Group (NDC vs. LDC) | 0.13 | [−8.02, 8.28] | 3.99 | 30.3 | t = 0.03 | 0.974 |
| Sex (male vs. female) | 12.37 | [3.92, 20.82] | 4.14 | 30 | t = 2.99 | 0.006 |
| Time × Group | 0.30 | [−0.85, 1.44] | 0.56 | 30 | t = 0.53 | 0.600 |
| Estimate (β) | 95% CI | SE | DF | Statistic | p-Value | |
| Intercept | 23.62 | [21.99, 25.26] | 0.80 | 30.4 | t = 29.48 | <0.001 |
| Time (post vs. pre) | −0.31 | [−0.56, −0.06] | 0.12 | 30 | t = −2.49 | 0.019 |
| Group (NDC vs. LDC) | 0.66 | [−1.71, 3.04] | 1.16 | 30.4 | t = 0.57 | 0.572 |
| Sex (male vs. female) | 0.77 | [−1.69, 3.23] | 1.20 | 30 | t = 0.64 | 0.528 |
| Time × Group | 0.10 | [−0.30, 0.50] | 0.20 | 30 | t = 0.52 | 0.610 |
| Estimate (β) | 95% CI | SE | DF | Statistic | p-Value | |
|---|---|---|---|---|---|---|
| Intercept | 90.22 | [65.86, 114.57] | 12.14 | 51.6 | t = 7.43 | <0.001 |
| Time (post vs. pre) | 33.83 | [6.73, 60.93] | 13.27 | 30 | t = 2.55 | 0.016 |
| Group (NDC vs. LDC) | 10.53 | [−25.75, 46.80] | 18.10 | 54.2 | t = 0.58 | 0.563 |
| Sex (male vs. female) | −43.19 | [−74.48, −11.89] | 15.32 | 30 | t = −2.82 | 0.008 |
| Time × Group | −9.25 | [−52.10, 33.60] | 20.98 | 30 | t = −0.44 | 0.662 |
| Estimate (β) | 95% CI | SE | DF | Statistic | p-Value | |
|---|---|---|---|---|---|---|
| Intercept | 48.88 | [42.32, 55.45] | 3.28 | 58.41 | t = 14.91 | <0.001 |
| Time (post vs. pre) | 14.67 | [6.02, 23.32] | 4.24 | 30 | t = 3.46 | 0.002 |
| Group (NDC vs. LDC) | 2.44 | [−7.44, 12.32] | 4.94 | 59.61 | t = 0.49 | 0.623 |
| Sex (male vs. female) | 5.01 | [−2.69, 12.72] | 3.77 | 30 | t = 1.33 | 0.194 |
| Time × Group | −6.42 | [−20.09, 7.26] | 6.70 | 30 | t = −0.96 | 0.346 |
| Estimate (β) | 95% CI | SE | DF | Statistic | p-Value | |
|---|---|---|---|---|---|---|
| Intercept | 126.62 | [42.32, 55.45] | 14.42 | 60 | t = 8.78 | <0.001 |
| Time (post vs. pre) | −42.89 | [6.02, 23.32] | 19.39 | 60 | t = −2.21 | 0.031 |
| Group (NDC vs. LDC) | −30.19 | [−7.44, 12.32] | 21.79 | 60 | t = −1.39 | 0.171 |
| Sex (male vs. female) | 27.56 | [−2.69, 12.72] | 16.10 | 60 | t = 1.71 | 0.092 |
| Time × Group | 76.22 | [−20.09, 7.26] | 30.65 | 60 | t = 2.49 | 0.016 |
| LDC (n = 18) | NDC (n = 12) | Between-Group Difference a | Adjusted Odds Ratio b | ||
|---|---|---|---|---|---|
| D-Glucaric Acid Level Changes | Improved | 10 (56%) | 5 (42%) | p = 0.924 | 2.16 (p = 0.310) |
| Remained marginally low | 1 (6%) | 1 (8%) | |||
| Remained normal | 6 (33%) | 5 (42%) | |||
| Worsened slightly | 1 (6%) | 1 (8%) | |||
| Mercapturic Acid Level Changes | Improved | 13 (72%) | 5 (42%) | p = 0.165 | 3.33 (p = 0.124) |
| Returned to normal | 1 (6%) | 1 (8%) | |||
| Remained normal | 4 (22%) | 6 (50%) | |||
| Overall Response to Intervention | Dual improvement | 7 (39%) | 1 (8%) | p = 0.263 | 2.88 (p = 0.150) |
| Single improvement | 7 (39%) | 8 (67%) | |||
| Stable | 2 (11%) | 2 (17%) | |||
| Mixed responses | 2 (11%) | 1 (8%) | |||
| Supplement | Daily Dosage | Mechanistic Role and Synergistic Potential | References |
|---|---|---|---|
| Liposomal Glutathione | 1 capsule | Acts as a direct antioxidant and key cofactor for Phase II conjugation reactions, facilitating detoxification of reactive species. Enhances overall detoxification capacity and regenerates α-lipoic acid; synergistic with NAC for maintaining intracellular glutathione levels. | [52,53] |
| N-Acetylcysteine (NAC) | 1 capsule | Serves as a cysteine donor and precursor for glutathione synthesis, supporting conjugation of xenobiotics and antioxidant defense. Works synergistically with glutathione and α-lipoic acid to sustain redox balance and detoxification efficiency. | [54,55] |
| R-Alpha Lipoic Acid | 2 capsules | A potent antioxidant that regenerates glutathione and vitamins C and E while supporting mitochondrial energy metabolism. Enhances the intracellular antioxidant network and potentiates the effects of NAC and glutathione. | [56,57] |
| DIM (Diindolylmethane, broccoli extract) | 2 capsules | Modulates Phase I and Phase II detoxification enzymes, promotes balanced estrogen metabolism, and provides antioxidant protection. May enhance glucuronidation and sulfation pathways and act synergistically with NAC and glutathione in detox support. | [58,59] |
| PMA-Zeolith Powder (Zeolite) | 2 × 3 g | Binds heavy metals and toxins in the gastrointestinal tract, reducing enterohepatic recirculation and systemic toxin load. Complements glutathione- and NAC-dependent detoxification pathways for toxin elimination. | [60,61] |
| Liv Ultra Leber Komplex | 3 capsules | Multi-ingredient liver support formulation containing hepatoprotective herbs, choline, vitamins, and antioxidants that enhance Phase II detoxification and hepatic resilience. Acts synergistically with other supplements to strengthen overall liver and metabolic detox pathways. | [62,63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schauer, M.; Mair, S.; Keiner, M.; Werner, C.; Kainz, F.; Motevalli, M. Does Genetic Variation in Detoxification Capacity Influence Hepatic Biomarker Responses to a Liver Support Supplementation Regimen? Int. J. Mol. Sci. 2025, 26, 10209. https://doi.org/10.3390/ijms262010209
Schauer M, Mair S, Keiner M, Werner C, Kainz F, Motevalli M. Does Genetic Variation in Detoxification Capacity Influence Hepatic Biomarker Responses to a Liver Support Supplementation Regimen? International Journal of Molecular Sciences. 2025; 26(20):10209. https://doi.org/10.3390/ijms262010209
Chicago/Turabian StyleSchauer, Markus, Susanne Mair, Michael Keiner, Christian Werner, Florian Kainz, and Mohamad Motevalli. 2025. "Does Genetic Variation in Detoxification Capacity Influence Hepatic Biomarker Responses to a Liver Support Supplementation Regimen?" International Journal of Molecular Sciences 26, no. 20: 10209. https://doi.org/10.3390/ijms262010209
APA StyleSchauer, M., Mair, S., Keiner, M., Werner, C., Kainz, F., & Motevalli, M. (2025). Does Genetic Variation in Detoxification Capacity Influence Hepatic Biomarker Responses to a Liver Support Supplementation Regimen? International Journal of Molecular Sciences, 26(20), 10209. https://doi.org/10.3390/ijms262010209









